The nitrogen (N) cycle is composed of multiple transformations of nitrogenous compounds, catalyzed primarily by microbes. The N cycle controls the availability of nitrogenous nutrients and biological productivity in marine systems (84) and thus is linked to the fixation of atmospheric carbon dioxide and export of carbon from the ocean's surface (30). Human activities are influencing the N cycle even in the oceans (100), and some of the nitrogenous gaseous products of microbial metabolism are greenhouse gases that are potentially involved in controlling Earth's climate.
The last decade has brought the discovery of several new links and changes in our understanding of components of the marine N cycle. Much of our basic information about the N cycle is derived from measurements of transformation rates or from experiments with cultivated isolates. Generalization from the behavior and physiology of cultivated isolates can be misleading, since it appears that many marine microorganisms in situ have yet to be obtained in culture (33). The rapid increase in knowledge of genes and molecular biology has had an enormous impact on our understanding of the N cycle by making it possible to study the ecological underpinnings and diversity of microorganisms involved in specific N cycle components. For example, nitrate assimilation by photosynthetic picoplankton was assumed to be universal but has now been shown to be absent from some of the most abundant photosynthetic organisms on the planet. Conversely, nitrate assimilation by heterotrophic bacteria was largely ignored, but it has recently been shown by both gene probes and physiological experiments that this capability is widespread in bacteria. Genetic and biochemical investigations have also changed our understanding of processes such as nitrification and denitrification, which were thought to be restricted to very specific habitats and microbes but in fact are more widely distributed.
The N cycle is composed of oxidation-reduction reactions, many of which are used in the energy metabolism of microbes. Specific enzymes catalyze many of these reactions, and the enzymes and genes are useful targets for studying microbial processes such as assimilatory nitrate reduction, dissimilatory nitrate reduction, and N2 fixation (Table 1). Knowledge of the genes encoding enzymes involved in biogeochemical transformations provides useful tools not only for assaying gene expression but also for determining the diversity of microorganisms involved in specific N cycle transformations. We still know very little about the link between the ecology of the N cycle and the redundancy of microbes and genes in the marine environment. Understanding these links is important for determining the role of microbial diversity in ecosystem processes and the sensitivity of the environment to perturbations. Regulatory proteins are also useful for investigating natural populations for “N status,” the degree of cellular N deficiency or sufficiency.
TABLE 1.
Nitrogen cycle gene probes
| Transformation | Gene(s)a | Protein | Reference(s) |
|---|---|---|---|
| N2 fixation | nifHDK | Nitrogenase | 117 |
| Nitrite assimilation | nir | Nitrite reductase | 102 |
| Nitrate assimilation | narB, nasA | Assimilatory nitrate reductase | 1 |
| Ammonium assimilation | glnA | Glutamine synthetase | 54 |
| Nitrate respiration and denitrification | nirS | Nitrate reductase | 8, 18, 87 |
| nirK | Nitrite reductase | ||
| norB | Nitric oxide reductase | ||
| nosZ | Nitrous oxide reductase | ||
| Organic N metabolism | ure | Urease | 25 |
| Ammonium oxidation/nitrification | amo | Ammonia monooxygenase | 2, 83 |
| Nitrogen regulation (cyanobacteria) | ntcA | Nitrogen regulatory protein | 58 |
Nitrogen cycle genes for which probes or PCR primers have been designed.
In the following sections, we review some important aspects of the N cycle that have undergone major reevaluation in recent years. Rather than present a detailed review of the entire cycle, we focus on important recent changes in our understanding of these aspects and specific components of the cycle. Understanding these features of the nitrogen cycle is critical for understanding the global biogeochemical cycles. Here we focus on the microbiological underpinnings of these processes; the global implications for the biogeochemical cycling of nitrogen are beyond the scope of this review.
INORGANIC AND ORGANIC N UPTAKE
It is usually assumed that most microorganisms can use inorganic N in the form of nitrate, nitrite, and ammonium. Particularly in the oligotrophic open ocean gyres, low concentrations of these compounds (<0.03 to 0.1 μM [12]) can limit the rate of productivity in the surface layer (0- to 200-m depth), but N can regulate productivity even in coastal upwelling regions (55). In some regions, including coastal regions and the high-nutrient, low-chlorophyll regions, upwelling or runoff can supply N in concentrations that exceed phytoplankton demand. Thus, there are large geographical variations in the sources and fluxes of nitrate and ammonium. The geographic aspects of the marine N cycle are beyond the scope of this review but are covered elsewhere (12, 16, 103).
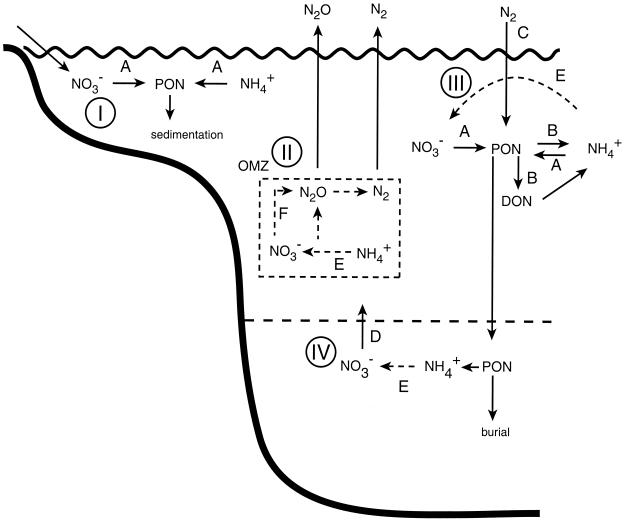
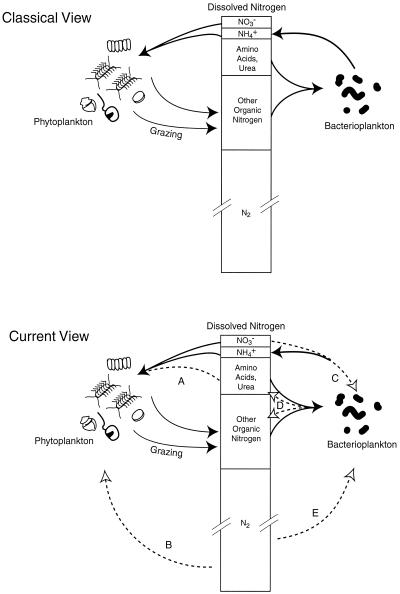
The first comprehensive view of the N cycle in the surface ocean proposed that inorganic N was taken up by phytoplankton and that the N was subsequently recycled from phytoplankton cells by heterotrophs, both large grazers (e.g., planktonic invertebrates) and microbial decomposers (Fig. 1, pathway A; Fig. 2). The death, lysis, and decay of the phytoplankton, either after ingestion by “herbivores” or because of physiological stressors (such as nutrient limitation, temperature, or other factors), liberated N in the form of dissolved organic N or ammonium, which was termed “regenerated” N (Fig. 1, pathway B; Fig. 2) (28). Biological N2 fixation, the reduction of atmospheric dinitrogen gas (N2) to ammonia (Fig. 1, pathway C), was thought to be insignificant in the open ocean, and essentially all pelagic N fixation was ascribed to two genera of N2-fixing microbes. Nitrate was believed to be supplied to the upper ocean primarily by mixing, advection and diffusion from deep ocean water, or terrestrial runoff (Fig. 1, pathway D). Because nitrifying bacteria are inhibited by light, it was assumed that nitrification (Fig. 1, pathway E) proceeded only in deep water (Fig. 1, pathway E); therefore, the only source of nitrate in surface waters was water mixing from the deep ocean reservoir (Fig. 1, pathway D).
FIG. 1.
Conceptual diagram of major features of the nitrogen cycle in coastal shelf and upwelling (I), OMZs (II), surface waters of the open ocean (III), and deep water (IV). PON, particulate organic nitrogen. Dashed lines indicate transformations involving multiple steps. Pathways: A, DIN assimilation; B, ammonium regeneration; C, nitrogen fixation; D, nitrate diffusion/advection from deep water; E, nitrification; F, nitrification; G, denitrification.
FIG. 2.
Diagrams of classical and present views of the N cycle in the surface waters of oligotrophic oceans. The composition of the dissolved N pool is shown with approximate relative concentrations of inorganic and organic constituents indicated by the size of the box. Dashed lines indicate transformations and processes included in the newer view of nitrogen cycling. (A) Some phytoplankton use simple organic compounds as a source of nitrogen. (B) There are multiple species of phytoplankton (cyanobacteria) in the open ocean that fix N2. (C) Bacteria can compete for nitrate and ammonium. (D) Bacteria can excrete urea and can also be a source of high-molecular-weight DON. (E). Some oceanic bacterioplankton appear to fix N2.
Nitrate, nitrite, and ammonium, called dissolved inorganic nitrogen (DIN), can be taken up (via membrane transporters) and assimilated by many microorganisms. Nitrate is assimilated after sequential reduction to nitrite (assimilatory nitrate reductase) and ammonium (assimilatory nitrite reductase). Phytoplankton (both eukaryotes and cyanobacteria), in general, prefer ammonium to nitrate, presumably due to the additional energy and reductant necessary to reduce nitrate to ammonium. However, this preference is not universal, as the reductant requirement for nitrate does not necessarily result in decreased growth rate (95). Although it is still likely that most microorganisms prefer ammonium to nitrate, it has recently become clear that (i) not all phytoplankton use nitrate and (ii) some bacteria can use nitrate and some cannot. While it was previously assumed that eukaryotic phytoplankton dominated both photosynthesis and DIN assimilation, it is now known that two major groups of small unicellular cyanobacteria (the Synechococcus and Prochlorococcus groups) are extremely abundant in surface waters and contribute a large fraction of photosynthesis and DIN demand (19). Some of these microbes do not contain assimilatory nitrate reductase genes (G. Rocap, F. W. Larimer, J. Lamerdin, E., S. Stilwagon, and S. W. Chisholm, Abstr. Aquatic Sci. Meet., p. 120, 2001). The high- and low-light-adapted Prochlorococcus spp. (67), divinyl chlorophyll a-containing cyanobacteria common in oceanic waters (22), differ in their abilities to use N sources (G. Rocap, F. W. Larimer, J. Lamerdin, E., S. Stilwagon, and S. W. Chisholm, Abstr. Aquatic Sci. Meet., 2001). The high-light strain (strain MED4) lacks both nitrate reductase genes and nitrite reductase genes and can grow only on ammonium, whereas the low-light strain (strain MIT9313) lacks only nitrate reductase genes and can grow on nitrite or ammonium (G. Rocap, F. W. Larimer, J. Lamerdin, E., S. Stilwagon, and S. W. Chisholm, Abstr. Aquatic Sci. Meet., 2001). Previously, it was assumed that microorganisms generally could use either nitrate or ammonium and that organisms differed in kinetics of nitrate or ammonium uptake and utilization (29), but now it is clear that some microorganisms may not be able to use nitrate.
It is now recognized that bacteria also can play a role in nitrate uptake. It is now known that assimilatory nitrate reductase genes are dispersed among marine bacterial strains as well as among phytoplankton (1). Thus, in complex microbial communities in situ, the roles of different microorganisms in uptake of nitrate and ammonium differ due to genetic as well as biochemical and physiological constraints.
The major source of nitrate in the ocean surface is diffusion and upwelling of nitrate-rich deep ocean water. In recent years, it has been shown that even this process, which would appear to be driven largely by physical forcing, has an important microbiological component. Many large oceanic diatoms that sometimes form large mats migrate to great depth to obtain nutrients from the nutrient-rich deep water (66), only to return to the surface carrying with them nitrate (99). Migrating Rhizosolenia mats may transport an average of 20%, ranging up to 78%, of the upward diffusive flux of nitrate (99). Thus, biological controls are involved even in the upward movement of nitrate from deep water (Fig. 1, pathway D).
N LIMITATION
It is often assumed that N limits the productivity of phytoplankton in the oceans. This conclusion is based on relatively few studies, most of which assess N limitation indirectly (39). It is quite difficult to demonstrate N limitation experimentally. Ideally, one would determine this from a simple bottle experiment in which different nutrients are added to each bottle. In ocean studies this can be difficult, since microbial communities change during bottle incubations (26). Techniques that directly assay biochemical or physiological targets can provide incubation-independent information on the nutritional status of assemblages and even individual cells. A number of cellular and molecular markers for nutritional deficiencies have been developed (36, 56, 88). Molecular and immunological techniques have provided ways of investigating natural communities, for example, by using probes for the N regulator in cyanobacteria, ntcA (57), or cell surface proteins that are expressed under N limitation (76). Detection of transcripts for ntcA indicated that marine Synechococcus spp. in the Gulf of Aqaba were not nitrogen stressed and were using primarily regenerated nitrogen. This approach has not yet been applied in open-ocean oligotrophic environments but could be used to determine the seasonal and depth variations in nitrogen limitation of cyanobacteria in the open ocean.
HETEROTROPHIC BACTERIA
The primary role of bacteria in the N cycle was presumed to be the release of inorganic N (NH4+) during the decomposition of organic matter, thereby recycling N (and other nutrients) to phytoplankton (Fig. 1, pathway B; Fig. 2). There is now an appreciation for the role of bacteria in the “microbial loop” (3, 4), which emphasizes the role of bacteria in trophic transfers through a micrograzer (protozoan) food chain. This food chain results in transfer of carbon, N, and other nutrients from dissolved organic matter into the food web. However, this picture is complicated by recent findings regarding the metabolism of heterotrophic marine bacteria (for a review, see reference 49) and the composition and sources of organic matter (62, 63). Bacteria can use DIN as well as organic matter (Fig. 1, pathway A; Fig. 2) and thus might even compete with phytoplankton for inorganic N (50). Whether or not bacteria take up inorganic N probably depends on the C:N ratio of the substrates being used for growth (34). Bacterial regeneration of N during the mineralization of organic matter also depends on the C:N ratio of cell material relative to substrate availability (49), and thus, whether bacteria provide N or compete with primary producers for N varies substantially in space and time (49). Interestingly, growth rates of bacteria are consistent with the uptake of N from dissolved free amino acids and ammonium, indicating that the larger pool of dissolved organic N is not a major source of N for growth (49). Bacteria can take up DIN while simultaneously liberating NH4+ in decomposition (96). Thus, bacteria in the ocean can be competing for NH4+, regenerating NH4+, or both (Fig. 2). It is unclear how both processes are occurring simultaneously, but one explanation is that different members of the microbial assemblage are responsible for different processes (49).
DON
Dissolved organic N (DON) compounds include a wide range of chemical compounds varying in size, complexity, and resilience to degradation. The characterization of dissolved organic matter has long been a challenge to the chemist but perhaps even more so to the microbial ecologist, since it is impossible to trace the metabolism of all of the different compounds even if all of the chemical structures and concentrations are known. DON can be a large pool in the oceans (3 to 7 μM [12]) and an even larger one in coastal waters (90). Important, but usually minor, constituents of DON are amino acids and urea [CO(NH2)2], which are readily used by bacteria and some phytoplankton. Amino acids, either dissolved and free or combined in oligopeptides, are important sources of organic C and N for bacteria (49). Although rates of urea production and catabolism have been measured in marine environments, relatively little is known about the microbiology of urea metabolism in marine systems. Bacteria can be a source or a sink for urea (Fig. 2). Urease is a nickel-containing multisubunit metalloprotein encoded by the ure genes (38), which have been characterized in eukaryotes, cyanobacteria, and heterotrophic bacteria and recently reported in autotrophic nitrifiers (T.E. Koper, J. M. Norton, and M. G. Klotz, Abstr. 9th Int. Symp. Microbial Ecol., P.08.059, 2001). Studies of the diversity of urease genes in the environment are likely to provide interesting information on the distribution of urea utilization capabilities in natural assemblages (K. Baker and J. Collier, unpublished data).
The definition of “dissolved” organic N has always been a tenuous distinction from that of “particulate” organic nitrogen. This has now been shown to be an artificial distinction, with interactions between organic molecules creating a complete continuum from small dissolved molecules to polymers, colloids, and particles. Chemical interactions among these molecules can result in rearrangements and interactions that form macromolecular gel structures (21). These networked structures, which can form and dissociate biologically or abiologically, can make complex microenvironments that have been suggested to be important for ecological interactions at some microbial scales (3).
DON was previously assumed to be primarily high-molecular-weight condensation products, including heterocyclic compounds, with a smaller fraction composed of more degradable compounds, including protein, and dissolved free and combined amino acids (Fig. 2). Recent studies indicate that a significant fraction of DON is composed of amide N (62) and that a fraction originates from bacterial cell walls (63). This finding reverses the perception that phytoplankton are the direct primary source for organic matter through excretion, decomposition, and lysis. Thus, bacteria metabolize organic matter that originates from photosynthetic microorganisms but are now both a source of and a sink for the DON pool (Fig. 2). Chemical and photochemical reactions have been found to be important in influencing the quantity and quality of organic matter. Rates of photolysis can equal or exceed bacterial decomposition rates (10).
Despite this new perspective on the source and nature of the bulk DOM in the sea, it is abundantly clear that some small fraction of the total DON pool is rapidly cycled by phytoplankton and bacteria in the photic zone (Fig. 2). A large fraction of the DIN assimilated by phytoplankton can be released as DON within a few hours, probably due to grazing disruption of cells (Fig. 2) (9, 104). To complicate matters further, there is now evidence that phytoplankton and cyanobacteria can assimilate some small labile components of DON. Thus, the primary producers may not be restricted to exploitation of the DIN pool but may be an important sink for DON as well (Fig. 2) (77, 78).
Release and uptake of DON on a scale of hours or less are probably important for the more labile DON fractions such as protein, peptides, and free amino acids, which are the DON components on which metabolic studies have focused in the past. Bacterioplankton can excrete proteolytic enzymes that digest peptides and proteins so that the monomers or oligonucleotides can be taken up by the cell and metabolized (41, 42, 91). There are substantial differences in activities and temperature responses of these enzymes in different ocean regions, with implications for how and where bacteria are regenerating inorganic N (24). These studies emphasize that the composition of organic matter (e.g., polysaccharides and sugars versus proteins and amino acids) and the variability among different ocean regions may be important in understanding the role of bacteria and dissolved organic matter in ocean carbon cycling (24) but also in N regeneration. DON and its metabolism are probably among the most poorly understood parts of the marine N cycle. New methods that combine the use of radiotracers and autoradiography with fluorescence in situ hybridization have begun to provide information on which types of bacteria are metabolizing different organic compounds (75). Given the magnitude of the DON pool and the dependence of heterotrophic bacteria on dissolved organic matter, this topic deserves much more attention. Since DON is composed of many different organic substrates, the uptake and metabolism of DON must involve multiple transport proteins and extracellular and intracellular enzymes. The study of DON metabolism is likely to profit from advances in genomic research from the information gained about the diversity of catabolic pathways in cultivated and uncultivated microorganisms.
N2 FIXATION
Biological N2 fixation, the reduction of atmospheric N2 to ammonia, is catalyzed by a diverse set of microorganisms. In the marine environment, N2 fixation rates are highest in a few specific habitats such as benthic cyanobacterial mats (40). In the oceanic biome, there are relatively few known N2 fixers despite the fact that DON concentrations are extremely low in many parts of the ocean. Blooms of filamentous N2-fixing cyanobacteria often exploit N-limiting conditions in lakes and sometimes estuaries, and so it is a curious paradox that there are so few obvious nitrogen-fixing microorganisms in the ocean (43). An exception is the Baltic Sea, where extensive blooms of filamentous heterocyst-forming cyanobacteria (Aphanizomenon and Nodularia) frequently occur and have occurred for thousands of years (5). Within the past decade, research on N2 fixation in the sea has received increasing attention (47), since the role of the ocean in C flux is linked to nutrient cycling.
In the open ocean, the filamentous nonheterocystous cyanobacterium Trichodesmium is common in tropical and subtropical waters (13, 74). This organism is particularly interesting because it forms macroscopic aggregates of filaments, is buoyant due to gas vacuoles, and fixes N2 only in the light. Most cyanobacteria segregate O2-sensitive N2 fixation from O2 evolved through photosynthesis by fixing N2 during the night or in specialized cells, called heterocysts, where photosystem II activity is reduced or absent. Trichodesmium is one of a few known species that evolve O2 simultaneously with N2 fixation without an obvious mechanism to avoid O2 inactivation. Trichodesmium fixes N2 only during the day, and this cycle is regulated by the synthesis of nitrogenase under the control of a circadian clock (20). There are various theories and hypotheses regarding the mechanisms involved in simultaneous N2 fixation and photosynthesis, including the possible division of labor among cells that are morphologically similar (31, 45), but the mechanisms whereby Trichodesmium fixes N2 aerobically are still not completely understood (13). The Trichodesmium nitrogenase protein is similar phylogenetically to that of other cyanobacterial diazotrophs and is not likely to be more O2 resistant than other nitrogenases (116). The molecular biology of N2 fixation in Trichodesmium has been reviewed elsewhere (116).
Perhaps the next most abundant diazotrophs in oceanic waters are the heterocyst-forming cyanobacterial symbionts of diatoms (98). These symbionts have not been successfully maintained in culture for extended periods of time, and so relatively little is known about the biology of the symbiotic interactions between the diatom and cyanobacteria. However, the symbiont-containing diatoms can form large aggregates that can be abundant in oligotrophic waters. These diatoms can form extensive blooms (17) that can be significant sources of N in the mixed layer of the ocean.
For years, it was believed that Trichodesmium and the symbionts of diatoms were the major N2 fixers in the open ocean. However, a number of recent studies have highlighted imbalances in N budgets that indicate that higher rates of N2 fixation are occurring in the open ocean than was previously estimated (37, 59, 64). This conclusion is based on biogeochemical calculations rather than direct measurements of N2 fixation rates or observed distributions of microorganisms but has led to a reevaluation of N2 fixation in the sea. Evidence of diverse bacterial and cyanobacterial N2-fixing microorganisms in the Atlantic and Pacific Oceans based on amplification of nitrogenase genes from bulk water samples has recently been reported (115, 118). Nitrogenase genes obtained from the Hawai'i Ocean Time (HOT) series long-term monitoring site at Station ALOHA in the North Pacific Subtropical Gyre were most closely related to nitrogenase genes of unicellular cyanobacterial genera that are not typically reported in oceanic waters. The cyanobacterial nifH phylotypes were detected at several times of year and were ultimately shown to be expressing nitrogenase genes (119). The finding of the genes led to the microscopic observation of cells that looked similar to the expected morphology (3 to 8 μm in diameter, spherical) and subsequent cultivation of N2-fixing isolates (119). Unicellular cyanobacteria with this morphology have been reported from the South Pacific Ocean (71), the Baltic Sea (108), and, previously, from Station ALOHA (11). Estimates of the concentrations and nitrogen fixation activity of these organisms at Station ALOHA indicate that they could equal or exceed the contribution of Trichodesmium (119), and they have been observed elsewhere in the world's oceans as well.
It is not well understood what controls the distribution and activity of diazotrophs in the sea. The distributions of Trichodesmium and some other cyanobacteria appear to be correlated with water temperature (15). It could be that diazotrophs are limited by the availability of Fe, a metal that is a component of many proteins in addition to nitrogenase (both the MoFe and Fe components). Fe distributions in the world's oceans are controlled to a large extent by aeolian transport of dust. Temporal and spatial variation in Fe supply may result in oscillation between N and P limitation of the oceans through its effect on N2 fixation (111). However, there does not appear to be a correlation between Fe concentration and Trichodesmium abundance and activity, at least in the Atlantic Ocean near Bermuda (85).
NITRIFICATION/DENITRIFICATION
Isolations of nitrifying and denitrifying bacteria using conventional enrichment techniques have provided extensive culture collections on which our understanding of the biochemistry and ecology of these processes is based. Chemolithoautotrophic nitrification is rather restricted in its occurrence and is represented in culture by 25 species of ammonia oxidizers in the beta and gamma subdivisions of the Proteobacteria and by eight species of nitrite oxidizers in the alpha, beta, and gamma subdivisions of the Proteobacteria (51). Molecular phylogeny has supported this generalization, showing that both ammonia-oxidizing bacteria (AOB) and nitrite-oxidizing bacteria belong to a small number of coherent groups (94).
The first AOB to be cultivated from the marine environment was Nitrosocystis oceanus (109), now called Nitrosococcus oceani. Strains of N. oceani have been obtained from several locations (14), and it has been detected in seawater by immunofluorescence (105) and by PCR amplification (B. B. Ward and G. O’Mullan unpublished data) and in Antarctic lakes by immunofluorescence hybridization and fluorescence in situ hybridization (101). Members of the Nitrosomonas/Nitrosospira group appear to dominate most terrestrial and aquatic environments, including marine sediments (52, 73, 107, 110). Purkhold et al. (79) surveyed all published 16S rRNA and amoA sequences, from both cultures and environmental clones, and concluded that although much diversity among AOB remains to be cultured, it is unlikely that entirely novel species will be discovered, at least in the case of the beta-subdivision AOB. However, this does not preclude the existence of entirely novel AOB that are not detected by probes based on known groups.
Nitrite oxidizers have received somewhat less attention, and culture collections are dominated by Nitrobacter strains. Recent work in various wastewaters, however, has shown that Nitrospira is the dominant group in these environments (32). For both AOB and nitrite-oxidizing bacteria, phylogeny and functionality appear to be well correlated, making these groups attractive for molecular phylogeny studies despite their slow and fastidious growth habits in culture. Research on nitrifiers, particularly AOB, based on 16S rRNA genes and functional genes has proliferated in recent years and was recently reviewed (53).
Denitrifying bacteria are the opposite of nitrifiers in many ways; denitrification ability is found in heterotrophic opportunists and in chemoautotrophs, is widespread among Bacteria and Archaea, and has even been reported in Eukarya(120). 16S rRNA and functional gene phylogenies are not congruent for the denitrifiers, implying substantial horizontal gene transfer of the functional genes over time (D. P. Martino and B. B. Ward, Aquatic Sciences Meeting: Limnology and Oceanography: navigating into the next century, abstract book, p. 117, 1999; B. Song and B. B. Ward, submitted for publication). Environmental research on the diversity of denitrifiers has focused, therefore, on the functional genes involved in the denitrification pathway, mainly nitrite reductase (7, 8) and nitrous oxide reductase (86, 87).
Dissimilatory reduction of nitrate to ammonium is often ignored in the marine realm but could be important in sediments in which fermentative bacteria, with whose metabolism it is often associated, are likely to be found. Christensen et al. (23) found it to be a significant nitrate sink only in sediments with very high organic carbon loading, and Bonin et al. (6) suggested that it could be important in coastal sediments, and it is unlikely to occur in the water column. Thus, we refer here mainly to respiratory denitrification, in which nitrate is reduced sequentially to nitrite, nitric oxide, nitrous oxide, and nitrogen gas.
Chemical distributions in marine sediments and the water column indicate that nitrification is an obligately aerobic process and that denitrification is an obligately anaerobic process. Although the oxygen requirements and tolerances vary among isolates, these requirements are reflected in the physiology of cultivated nitrifiers and denitrifiers, which are predominantly obligate aerobes and facultative anaerobes, respectively. Nitrification and denitrification are often coupled across oxic/anoxic interfaces in both sediments and suboxic waters, leading to the loss of fixed nitrogen via mineralization, oxidation, and denitrification (72). Because the deep ocean contains high nitrate concentrations, it was long assumed that nitrification occurred in that environment. However, direct rate measurements using 15N tracer techniques have consistently shown that most water column nitrification occurs in the lower portion of the euphotic zone and that the nitrate produced there can supply a large fraction of phytoplankton nitrate demand (27, 106).
The recent description of aerobic denitrification by novel strains and “conventional” denitrifying bacteria introduces a new link in the N cycle. Aerobic denitrification was first described in Paracoccus pantotrophus (formerly Thiosphaera pantotropha [81]), and like others possessing this ability, P. pantotrophus is also a heterotrophic nitrifier. The nitrite generated by the oxidation of ammonia can be released into the medium or denitrified to N2 gas. Denitrification of nitrite or nitrate to N2 can occur with atmospheric levels of O2 (82). P. pantotrophus was originally isolated from wastewater; its ability to denitrify aerobically in batch culture, as well as that of several other conventional heterotrophic denitrifiers, has been confirmed (82). Su et al. (93) reported aerobic denitrification by a strain of Pseudomonas stutzeri at rates and with oxygen tolerance greatly exceeding those reported for P. pantotrophus. The process may be common in isolates, but its significance in the environment remains uncertain, and even in cultures questions still remain. For example, the enzymology of the aerobic pathway is unknown. While P. pantotrophus expresses the nitrite reductase gene (nirS) under anaerobic conditions, it is not expressed under aerobic conditions (65), and thus, the mechanism for aerobic reduction of nitrite remains unknown.
AOB also perform a subset of the conventional set of denitrifying reactions, reducing nitrite to NO and N2O. The process occurs aerobically but apparently is enhanced at low oxygen concentrations (35, 60). These gases are also intermediates in denitrification but could derive from nitrification under a low or nearly zero oxygen concentration. Both N2O and NO are important in atmospheric processes; they contribute to greenhouse warming and to catalytic destruction of stratospheric ozone. Thus, understanding which processes are responsible for their production could prove to be important in understanding or potentially regulating their fluxes.
It appears that chemoautotrophic AOB produce N2O and NO by using a pathway that is essentially identical to the classical denitrification pathway. First, ammonia is oxidized to nitrite and some of the nitrite is reduced to N2O. The reductions are catalyzed by enzymes that are encoded by nitrite reductase and NO reductase genes that are homologous to the nirK and norB genes of conventional denitrifying bacteria (reference 18 and our unpublished data). Even methylotrophs, to which some marine nitrifiers are related, possess genes with homology to nirK and norB (112). The significance of this denitrifying metabolism to the physiological ecology of nitrifying bacteria is unknown. However, its discovery casts uncertainty on the roles of “nitrifiers” and “denitrifiers” in trace gas metabolism in the oceans and may complicate the use of molecular approaches for studying and detecting their presence and activity in the environment.
Suboxic and anoxic waters and sediments tend to have large fluxes in, and sometimes large accumulations of, the gaseous intermediates of nitrification and denitrification. Trace levels of N2O in oxic ocean waters are thought to arise from nitrification (113) and show a stoichiometric relationship to oxygen utilization. Oxygen minimum zones (OMZs) show depth zones of N2O accumulation and depletion. Nitrous oxide is depleted in the core of the OMZ, where denitrification rates are thought to be greatest. N2O maxima typically occur both above and below the minimum. Stable-isotope measurement of nitrous oxide from O2-depleted waters in the Arabian Sea indicates that both nitrification and denitrification may contribute to the signal (70) (Fig. 1). The surface waters of the ocean are generally slightly supersaturated with N2O, and the ocean constitutes a significant source of atmospheric N2O, especially in regions containing OMZs (69, 80).
Autotrophic nitrifying bacteria exhibit some abilities in anaerobic metabolism as well. Enrichment cultures under chemolithotrophic conditions and with very low O2 concentrations catalyzed the net removal of ammonium as N2 gas (68). Schmidt and Bock have shown that Nitrosomonas eutropha produces gaseous products, mainly NO and N2, during growth on nitrogen dioxide gas (NO2) and ammonia (89). The process proceeds at a lower rate than ammonia oxidation in the presence of a normal air atmosphere and supports cell growth. Additions of NO2 and NO enhanced the complete removal of N in the form of ammonia and organic N without the addition of organic carbon substrates (114). The relevance of these observations to marine N cycling is unknown, but the phylogenetic homogeneity of the beta-subdivision ammonium oxidizers suggests that analogous metabolic capabilities may exist in marine strains as well.
A completely novel process, in which ammonia and nitrite are converted anaerobically to dinitrogen gas, has recently been reported from anaerobic wastewater systems (92, 97), and the organisms responsible for this novel metabolism have been identified as relatives of Planctomyces (46). Referred to as “anammox,” the process probably involves a consortium of the planctomycete organism and an autotrophic ammonia oxidizer such as Nitrosomonas europaea or N. eutropha. The planctomycete oxidizes ammonium to N2 by using NO2−, which is produced by the conventional ammonia oxidizer, as an oxidant. Both oxygen and nitrite concentrations are maintained at nearly undetectable levels by the metabolism of the members of the consortium, and while both organisms grow quite slowly (generation times for the planctomycete of two weeks or more are reported), the net removal of ammonium occurs at a rate 25 times faster than that reported (91) for N removal by N. eutropha growing anaerobically in pure culture (97). Still, it can require months to establish an anammox enrichment, and the consortium is stable only in bioreactors with long retention times; thus, it seems unlikely that this anammox consortium is active in natural environments (97).
Anammox would constitute a shortcut in the conventional N cycle, in which nitrification and denitrification are linked at the level of nitrite, without going through nitrate. Therefore, natural environments where ammonium and nitrite both occur in the presence of low oxygen concentrations might be suitable habitats for anammox-like reactions. Such environments include oxic/anoxic interfaces, such as those found at sediment/water interfaces in hemipelagic and shallow sediments and in stratified lakes and water columns of stratified basins such as the Black Sea and the Cariaco Basin. Upon initial consideration, anaerobic ammonium oxidation seems unlikely to dominate processes in these environments, based on observation of chemical distributions. Microaerophilic autotrophic nitrification, linked to anaerobic denitrification across the oxic/anoxic interface, has been used to interpret the chemical distributions, which typically show depletion of oxygen and nitrate above the interface and accumulation of ammonium below it. Denitrification produces a net loss of fixed N, which would presumably be much larger if anammox were also involved. Mass balance measurements might provide insight into the potential for anammox involvement in natural systems. Dalsgaard and Thamdrup (Abstr. Am. Soc. Limnol. Oceanogr., 2001) recently reported evidence from 15N tracer experiments that an anammox-like process may be at work in marine sediments and at rates that should be evident in mass balance experiments.
In addition to the unconventional activities of “conventional” nitrifiers and denitrifiers and the discovery of novel N metabolic pathways in new organisms, it has also been proposed recently that a short circuit of the nitrification/denitrification couple can also be accomplished abiotically. In marine sediments, which typically contain relatively high manganese levels, N2 gas can be produced by the oxidation of ammonia and organic N by manganese dioxide in air (61) or linked in series to anoxic organic matter oxidation through several biogeochemical reductants, including iron and hydrogen sulfide (44).
Anoxic ammonia oxidation, whether it results directly in N2 formation (as in anammox) or in nitrate production (when linked to manganese reduction), would introduce new links into the aquatic and sediment N cycle. Failure to account for anoxic ammonia oxidation might lead to an underestimate of ammonium removal, because the products do not accumulate; they are either lost to the atmosphere immediately or rapidly reduced by the next step in the anaerobic cycling of organic matter.
SHIFTING PARADIGMS AND MOVING TARGETS
Although we have learned much about the N cycle in marine systems, it must be remembered that our knowledge is superimposed on a dynamic and spatially variable biome. Knowledge gained by long-term studies demonstrates that our previous models of N cycling in the ocean are insufficient and that there are ecosystem- and global-scale changes that have occurred and are occurring that have implications for the N cycle in the oceans. For example, the conceptual model of Dugdale and Goering (28) was developed during an era when nitrification and N2 fixation were assumed to be minor N fluxes in surface waters. At station ALOHA, the long-term biogeochemical monitoring site of the HOT program, both of these processes have been shown to be more important than was previously believed at that site, but analyses of time-series data at the monitoring station in the North Pacific Subtropical Gyre suggest ecosystem shifts at annual, decade-long, and possibly longer time scales (48). These ecosystem shifts are linked to climatic physical forcing events, including the El Niño-Southern Oscillation, because physical processes determine the depth of the mixed layer and upwelling or mixing with nutrient-rich deep water. The consequences of the ecosystem shifts are reflected in the linked ecosystem properties of community structure and nutrient fluxes and ultimately determine the constraints on ocean-atmosphere carbon flux. At station ALOHA, an increase in primary productivity, changes in community structure, and changes in N dynamics have been observed over the past decade (48). One of the features of this change seems to be an increased importance of N2 fixation as a source of nitrogen. These dynamic ecosystem changes highlight the importance of understanding the N cycle in the marine environment and challenge us to reexamine our conceptual understanding of marine N fluxes and transformations.
REFERENCES
- 1.Allen, A. E., M. G. Booth, M. E. Frischer, P. G. Verity, J. P. Zehr, and S. Zani. 2001. Diversity and detection of nitrate assimilation genes in marine bacteria. Appl. Environ. Microbiol. 67:5343-5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzerreca, J. J., J. M. Norton, and M. G. Klotz. 1999. The amo operon in marine, ammonia-oxidizing gamma-Proteobacteria. FEMS Microbiol. Lett. 180:21-29. [DOI] [PubMed] [Google Scholar]
- 3.Azam, F. 1998. Microbial control of oceanic carbon flux: the plot thickens. Science 280:694-696. [Google Scholar]
- 4.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, and L. A. T. F. Meyer-Reil. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 5.Bianchi, T. S., E. Engelhaupt, P. Westman, T. Andren, C. Rolff, and R. Elmgren. 2000. Cyanobacterial blooms in the Baltic Sea: natural or human-induced? Limnol. Oceanogr. 45:716-726. [Google Scholar]
- 6.Bonin, P., P. Omnes, and A. Chalamet. 1998. Simultaneous occurrence of denitrification and nitrate ammonification in sediments of the French Mediterranean Coast. Hydrobiologia 389:169-182. [Google Scholar]
- 7.Braker, G., H. L. Ayala-del-Rio, A. H. Devol, A. Fesefeldt, and J. M. Tiedje. 2001. Community structure of denitrifiers, Bacteria, and Archaea along redox gradients in Pacific Northwest marine sediments by terminal restriction fragment length polymorphism analysis of amplified nitrite reductase (nirS) and 16S rRNA genes. Appl. Environ. Microbiol. 67:1893-1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braker, G., J. Z. Zhou, L. Y. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific Northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bronk, D. A., and B. B. Ward. 1999. Gross and net nitrogen uptake and DON release in the euphotic zone of Monterey Bay, California. Limnol. Oceanogr. 44:573-585. [Google Scholar]
- 10.Bushaw-Newton, K. L., and M. A. Moran. 1999. Photochemical formation of biologically available nitrogen from dissolved humic substances in coastal marine systems. Aquat. Microb. Ecol. 18:285-292. [Google Scholar]
- 11.Campbell, L., H. Liu, H. A. Nolla, and D. Vaulot. 1997. Annual variability of phytoplankton and bacteria in the subtropical North Pacific Ocean at station ALOHA during the 1991-1994 ENSO event. Deep-Sea Res. I 44:167-192. [Google Scholar]
- 12.Capone, D. G. 2000. The marine microbial nitrogen cycle, p. 455-494. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 13.Capone, D. G., J. P. Zehr, H. W. Paerl, B. Bergman, and E. J. Carpenter. 1997. Trichodesmium: a globally significant marine cyanobacterium. Science 276:1221-1229. [Google Scholar]
- 14.Carlucci, A. F., and J. D. H. Strickland. 1968. The isolation, purification and some kinetic studies of marine nitrifying bacteria. J. Exp. Mar. Biol. Ecol. 2:156-166. [Google Scholar]
- 15.Carpenter, E. J. 1983. Nitrogen fixation by marine Oscillatoria (Trichodesmium) in the world's oceans, p. 65-103. In E. J. Carpenter and D. G. Capone (ed.), Nitrogen in the marine environment. Academic Press, New York, N.Y.
- 16.Carpenter, E. J., and D. G. Capone (ed.). 1983. Nitrogen in the marine environment. Academic Press, New York, N.Y.
- 17.Carpenter, E. J., J. P. Montoya, J. Burns, M. Mulholland, A. Subramaniam, and D. G. Capone. 1999. Extensive bloom of a N2 fixing symbiotic association in the tropical Atlantic Ocean. Mar. Ecol. Prog. Ser. 185:273-283. [Google Scholar]
- 18.Casciotti, K. L., and B. B. Ward. 2001. Nitrite reductase genes in ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 67:2213-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavendar-Bares, K. K., D. M. Karl, and S. W. Chisholm. 2001. Nutrient gradients in the western North Atlantic Ocean: relationship to microbial community structure and comparison to patterns in the Pacific Ocean. Deep-Sea Res. I 48:2373-2395. [Google Scholar]
- 20.Chen, Y.-B., B. Dominic, M. T. Mellon, and J. P. Zehr. 1998. Circadian rhythm of nitrogenase gene expression in the diazotrophic filamentous nonheterocystous cyanobacterium Trichodesmium sp. strain IMS 101. J. Bacteriol. 180:3598-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin, W., M. V. Orellana, and P. Verdugo. 1998. Spontaneous assembly of marine dissolved organic matter into polymer gels. Nature 391:568-572. [Google Scholar]
- 22.Chisholm, S. W., S. L. Frankel, R. Goericke, R. J. Olson, B. Palenik, J. B. Waterbury, L. West-Johnsrad, and E. R. Zettler. 1992. Prochorococcus marinus nov. gen. nov. sp.: an oxyphototrophic marine prokaryote containing divinyl chlorophyll a and b. Arch. Microbiol. 157:297-300. [Google Scholar]
- 23.Christensen, P. B., S. Rysgaard, N. P. Sloth, T. Dalsgaard, and S. Schwaerter. 2000. Sediment mineralization, nutrient fluxes, denitrification and dissimilatory nitrate reduction to ammonium in an estuarine fjord with sea cage trout farms. Aquat. Microb. Ecol. 21:73-84. [Google Scholar]
- 24.Christian, J., and D. Karl. 1995. Bacterial ectoenzymes in marine waters: activity ratios and temperature responses in three oceanographic provinces. Limnol. Oceanogr. 40:1042-1049. [Google Scholar]
- 25.Collier, J. L., B. Brahamsha, and B. Palenik. 1999. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology (United Kingdom) 145:447-459. [DOI] [PubMed] [Google Scholar]
- 26.Dore, J. E., and D. M. Karl. 2001. Microbial ecology at sea: sampling, subsampling and incubation considerations, p. 13-42. In J. H. Paul (ed.), Methods in microbiology: marine microbiology, vol. 30. Academic Press, London, England. [Google Scholar]
- 27.Dore, J. E., and D. M. Karl. 1996. Nitrification in the euphotic zone as a source for nitrite, nitrate, and nitrous oxide at Station ALOHA. Limnol. Oceanogr. 41:1619-1628. [Google Scholar]
- 28.Dugdale, R. C., and J. J. Goering. 1967. Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol. Oceanogr. 12:196-206. [Google Scholar]
- 29.Eppley, R. W., J. L. Coatsworth, and L. Solorzano. 1969. Studies of nitrate reductase in marine phytoplankton. Limnol. Oceanogr. 14:194-205. [Google Scholar]
- 30.Falkowski, P. G., R. T. Barber, and V. Smetacek. 1998. Biogeochemical controls and feedbacks on ocean primary production. Science 281:200-206. [DOI] [PubMed] [Google Scholar]
- 31.Fredriksson, C., and B. Bergman. 1997. Ultrastructural characterisation of cells specialised for nitrogen fixation in a non-heterocystous cyanobacterium Trichodesmium spp. Protoplasma 197:76-85. [Google Scholar]
- 32.Gieseke, A., L. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Giovannoni, S., and M. Rappe. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 34.Goldman, J. C., and M. R. Dennett. 2000. Growth of marine bacteria in batch and continuous culture under carbon and nitrogen limitation. Limnol. Oceanogr. 45:789-800. [Google Scholar]
- 35.Goreau, T. J., W. A. Kaplan, S. C. Wofsy, M. B. McElroy, F. W. Valois, and S. W. Watson. 1980. Production of NO2−and N2O by nitrifying bacteria at reduced concentrations of oxygen. Appl. Environ. Microbiol. 40:526-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graziano, L. M., R. J. Geider, W. K. W. Li, and M. Olaizola. 1996. Nitrogen limitation of North Atlantic phytoplankton: analysis of physiological condition in nutrient enrichment experiments. Aquat. Microb. Ecol. 11:53-64. [Google Scholar]
- 37.Gruber, N., and J. L. Sarmiento. 1997. Global patterns of marine nitrogen fixation and denitrification. Global Biogeochem. Cycles 11:235-266. [Google Scholar]
- 38.Hausinger, R. P., G. J. Colpas, and A. Soriano. 2001. Urease: a paradigm for protein-assisted metallocenter assembly. ASM News 67:78-84. [Google Scholar]
- 39.Hecky, R. E., and P. Kilham. 1988. Nutrient limitation of phytoplankton in freshwater and marine environments: a review of recent evidence on the effects of enrichment. Limnol. Oceanogr. 33:796-822. [Google Scholar]
- 40.Herbert, R. A. 1999. Nitrogen cycling in coastal marine ecosystems. FEMS Microbiol. Rev. 23:563-590. [DOI] [PubMed] [Google Scholar]
- 41.Hollibaugh, J. T., and F. Azam. 1983. Microbial degradation of dissolved proteins in seawater. Limnol. Oceanogr. 28:1104-1116. [Google Scholar]
- 42.Hoppe, H.-G. 1983. Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylumbelliferyl substrates. Mar. Ecol. Prog. Ser. 11:299-308. [Google Scholar]
- 43.Howarth, R. W., and R. Marino. 1988. Nitrogen fixation in freshwater, estuarine, and marine ecosystems. 2. Biogeochemical controls. Limnol. Oceanogr. 33:688-701. [Google Scholar]
- 44.Hulth, S., R. C. Aller, and F. Gilbert. 1999. Coupled anoxic nitrification/ manganese reduction in marine sediments. Geochim. Cosmochim. Acta 63:49-66. [Google Scholar]
- 45.Janson, S., E. J. Carpenter, and B. Bergman. 1994. Compartmentalisation of nitrogenase in a non-heterocystous cyanobacterium: Trichodesmium contortum. FEMS Microbiol Lett. 118:9-14. [Google Scholar]
- 46.Jetten, M. S. M., M. Wagner, J. Fuerst, M. C. M. van Loosdrecht, G. Kuenen, and M. Strous. 2001. Microbiology and application of the anaerobic ammonium oxidation ("anammox") process. Curr. Opin. Biotechnol. 12:283-288. [DOI] [PubMed] [Google Scholar]
- 47.Karl, D., A. Michaels, B. Bergman, D. Capone, E. Carpenter, R. Letelier, F. Lipschultz, H. Paerl, D. Sigman, and L. Stal. Dinitrogen fixation in the world's oceans. Biogeochemistry, in press.
- 48.Karl, D. M. 1999. A sea of change: biogeochemical variability in the North Pacific Subtropical Gyre. Ecosystems 2:181-214. [Google Scholar]
- 49.Kirchman, D. L. 2000. Uptake and regeneration of inorganic nutrients by marine heterotrophic bacteria, p. 261-288. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 50.Kirchman, D. L., and P. A. Wheeler. 1998. Uptake of ammonium and nitrate by heterotrophic bacteria and phytoplankton in the sub-Arctic Pacific. Deep-Sea Res. I 45:347-365. [Google Scholar]
- 51.Koops, H.-P., and A. Pommerening-Roser. 2001. Distribution and ecophysiology of the nitrifying bacteria emphasizing cultured species. FEMS Microbiol. Ecol. 37:1-9. [Google Scholar]
- 52.Kowalchuk, G. A., P. L. E. Bodelier, G. H. J. Heilig, J. R. Stephen, and H. J. Laanbroek. 1998. Community analysis of ammonia-oxidising bacteria, in relation to oxygen availability in soils and root-oxygenated sediments, using PCR, DGGE and oligonucleotide probe hybridisation. FEMS Microbiol. Ecol. 27:339-350. [Google Scholar]
- 53.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 54.Kramer, J. G., M. Wyman, J. P. Zehr, and D. G. Capone. 1996. Diel variability in transcription of the structural gene for glutamine synthetase (glnA) in natural populations of the marine diazotrophic cyanobacterium Trichodesmium thiebautii. FEMS Microbiol. Ecol. 21:187-196. [Google Scholar]
- 55.Kudela, R. M., and R. C. Dugdale. 2000. Nutrient regulation of phytoplankton productivity in Monterey Bay, California. Deep-Sea Res. II 47:1023-1053. [Google Scholar]
- 56.LaRoche, J., R. J. Geider, L. M. Graziano, H. Murray, and K. Lewis. 1993. Induction of specific proteins in eukaryotic algae grown under iron-, phosphorus-, or nitrogen-deficient conditions. J. Phycol. 29:767-777. [Google Scholar]
- 57.Lindell, D., E. Padan, and A. F. Post. 1998. Regulation of ntcA expression and nitrite uptake in the marine Synechococcus sp. strain WH 7803. J. Bacteriol. 180:1878-1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lindell, D., and A. F. Post. 2001. Ecological aspects of ntcA gene expression and its use as an indicator of the nitrogen status of marine Synechococcus spp. Appl. Environ. Microbiol. 67:3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lipschultz, F., and N. J. P. Owens. 1996. An assessment of nitrogen fixation as a source of nitrogen to the North Atlantic Ocean. Biogeochemistry 35:261-274. [Google Scholar]
- 60.Lipschultz, F., O. C. Zafiriou, S. C. Wofsy, M. B. Melroy, F. W. Valois, and S. W. Watson. 1981. Production of NO and N2O by soil nitrifying bacteria. Nature 294:641-643. [Google Scholar]
- 61.Luther, G. W., B. Sundgy, G. L. Lewis, P. G. Brendel, and N. Silverberg. 1997. Interactions of manganese with the nitrogen cycle: alternative pathways to dinitrogen. Biochim. Biophys. Acta 61:4043-4053. [Google Scholar]
- 62.McCarthy, M., T. Pratum, J. Hedges, and R. Benner. 1997. Chemical composition of dissolved organic nitrogen in the ocean. Nature 390:150-154. [Google Scholar]
- 63.McCarthy, M. D., J. I. Hedges, and R. Benner. 1998. Major bacterial contribution to marine dissolved organic nitrogen. Science 281:231-234. [DOI] [PubMed] [Google Scholar]
- 64.Michaels, A. F., D. Olson, J. L. Sarmiento, J. W. Ammerman, K. Fanning, R. Jahnke, A. H. Knap, F. Lipschultz, and J. M. Prospero. 1996. Inputs, losses and transformations of nitrogen and phosphorus in the pelagic North Atlantic Ocean. Biogeochemistry 35:181-226. [Google Scholar]
- 65.Moir, J. W. B., D. J. Richardson, and S. J. Ferguson. 1995. The expression of redox proteins of denitrification in Thiosphaera pantotropha grown with oxygen, nitrate, and nitrous-oxide as electron-acceptors. Arch. Microbiol. 164:43-49. [Google Scholar]
- 66.Moore, J. K., and T. A. Villareal. 1996. Size-ascent rate relationships in positively buoyant marine diatoms. Limnol. Oceanogr. 41:1514-1520. [Google Scholar]
- 67.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 68.Muller, E. B., A. H. Stouthamer, and H. W. van Verseveld. 1995. Simultaneous NH3 oxidation and N2 production at reduced O2 tensions by sewage sludge subcultured with chemolithotrophic medium. Biodegradation 6:339-349. [DOI] [PubMed] [Google Scholar]
- 69.Naqvi, S. W. A., and R. J. Noronha. 1991. Nitrous-oxide in the Arabian Sea. Deep-Sea Res. 38:871-890. [Google Scholar]
- 70.Naqvi, S. W. A., T. Yoshinari, D. A. Jayakumar, M. A. Altabet, P. V. Narvekar, A. H. Devol, J. A. Brandes, and L. A. Codispoti. 1998. Budgetary and biogeochemical implications of N2O isotope signatures in the Arabian Sea. Nature 391:462-464. [Google Scholar]
- 71.Neveux, J., F. Lantoine, D. Vaulot, D. Marie, and J. Blanchot. 1999. Phycoerythrins in the southern tropical and equatorial Pacific Ocean: evidence for new cyanobacterial types. J. Geophys. Res. 104:3311-3321. [Google Scholar]
- 72.Nielsen, L. P., and R. N. Glud. 1996. Denitrification in a coastal sediment measured in situ by the nitrogen isotope pairing technique applied to a benthic flux chamber. Mar. Ecol. Prog. Ser. 137:181-186. [Google Scholar]
- 73.Nold, S. C., J. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the β-subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Orcutt, K. M., F. Lipschultz, K. Gundersen, R. Arimoto, A. F. Michaels, A. H. Knap, and J. R. Gallon. 2001. A seasonal study of the significance of N2 fixation by Trichodesmium spp. at the Bermuda Atlantic Time-series Study (BATS) site. Deep-Sea Res. II 48:1583-1608. [Google Scholar]
- 75.Ouverney, C. C., and J. A. Fuhrman. 1999. Combined microautoradiography-16S rRNA probe technique for determination of radioisotope uptake by specific microbial cell types in situ. Appl. Environ. Microbiol. 65:1746-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Palenik, B., and J. A. Koke. 1995. Characterization of a nitrogen-regulated protein identified by cell surface biotinylation of a marine phytoplankton. Appl. Environ. Microbiol. 61:3311-3315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Palenik, B., and F. M. M. Morel. 1990. Amino acid utilization by marine phytoplankton—a novel mechanism. Limnol. Oceanogr. 35:260-269. [Google Scholar]
- 78.Palenik, B., and F. M. M. Morel. 1990. Comparison of cell-surface l-amino acid oxidases from several marine phytoplankton. Mar. Ecol. Prog. Ser. 59:195-201. [Google Scholar]
- 79.Purkhold, L., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rahn, T., and M. Wahlen. 2000. A reassessment of the global isotopic budget of atmospheric nitrous oxide. Global Biogeochem. Cycles 14:537-543. [Google Scholar]
- 81.Rainey, F. A., D. P. Kelly, E. Stackebrandt, J. Burghardt, A. Hiraishi, Y. Katayama, and P. M. Wood. 1999. A re-evaluation of the taxonomy of Paracoccus denitrificans and a proposal for the creation of Paracoccus pantotrophus comb. nov. Int. J. Syst. Evol. Bacteriol. 49:645-651. [DOI] [PubMed] [Google Scholar]
- 82.Robertson, L. A., T. Dalsgaard, N.-P. Revsbeck, and J. G. Kuenen. 1995. Confirmation of aerobic denitrification in batch cultures, using gas chromatography and 15N mass spectrometry. FEMS Microbiol. Ecol. 18:113-120. [Google Scholar]
- 83.Rotthauwe, J. H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ryther, J. H., and W. M. Dunstan. 1971. Nitrogen, phosphorus, and eutrophication in the coastal marine environment. Science 171:1008-1013. [DOI] [PubMed] [Google Scholar]
- 85.Sanudo-Wilhelmy, S. A., A. B. Kustka, C. J. Gobler, D. A. Hutchins, M. Yang, K. Lwiza, J. Burns, D. G. Capone, J. A. Raven, and E. J. Carpenter. 2001. Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean. Nature 411:66-69. [DOI] [PubMed] [Google Scholar]
- 86.Scala, D. J., and L. J. Kerkhof. 2001. Horizonal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scala, D. J., and L. J. Kerkhof. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol Lett. 162:61-68. [DOI] [PubMed] [Google Scholar]
- 88.Scanlan, D. J., and W. H. Wilson. 1999. Application of molecular techniques to addressing the role of P as key effector in marine ecosystems. Hydrobiologia 401:151-177. [Google Scholar]
- 89.Schmidt, I., and E. Bock. 1997. Anaerobic ammonia oxidation with nitrogen dioxide by Nitrosomonas eutropha. Arch. Microbiol. 167:106-111. [PubMed] [Google Scholar]
- 90.Sharp, J. H. 1983. The distribution of inorganic nitrogen and dissolved and particulate organic nitrogen in the sea, p. 1-36. In E. J. Carpenter and D. G. Capone (ed.), Nitrogen in the marine environment. Academic Press, New York, N.Y.
- 91.Stepanauskas, R., H. Edling, and L. J. Tranvik. 1999. Differential dissolved organic nitrogen availability and bacterial aminopeptidase activity in limnic and marine waters. Microbial Ecol. 38:264-272. [DOI] [PubMed] [Google Scholar]
- 92.Strous, M., G. Kuenen, and M. S. M. Jetten. 1999. Key physiology of anaerobic ammonium oxidation. Appl. Environ. Microbiol. 65:3248-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Su, J.-J., B.-Y. Liu, and D.-Y. Liu. 2001. Comparison of aerobic denitrification under high oxygen atmosphere by Thiosphaera pantotropha ATCC 35512 and Pseudomons stutzeri SU2 newly isolated from the activated sludge of a piggery wastewater treatment system. J. Appl. Microbiol. 90:457-462. [DOI] [PubMed] [Google Scholar]
- 94.Teske, A., E. Alm, J. M. Regan, S. Toze, B. E. Rittmann, and D. A. Stahl. 1994. Evolutionary relationships among ammonia- and nitrite-oxidizing bacteria. J. Bacteriol. 176:6623-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thompson, P. A., M. E. Levasseur, and P. J. Harrison. 1989. Light-limited growth on ammonium vs. nitrate—what is the advantage for marine phytoplankton? Limnol. Oceanogr. 34:1014-1024. [Google Scholar]
- 96.Tupas, L. M., I. Koike, D. M. Karl, and O. Holmhansen. 1994. Nitrogen metabolism by heterotrophic bacterial assemblages in Antarctic coastal waters. Polar Biol. 14:195-204. [Google Scholar]
- 97.Van Loosdrecht, M. C. M., and M. S. M. Jetten. 1998. Microbiological conversions in nitrogen removal. Water Sci. Technol. 38:1-7. [Google Scholar]
- 98.Villareal, T. A., and E. J. Carpenter. 1989. Nitrogen fixation, suspension characteristics and chemical composition of Rhizosolenia mats in the central North Pacific Gyre. Biol. Oceanogr. 6:387-405. [Google Scholar]
- 99.Villareal, T. A., C. Pilskaln, M. Brzezinski, F. Lipschultz, M. Dennett, and G. B. Gardner. 1999. Upward transport of oceanic nitrate by migrating diatom mats. Nature 397:423-425. [DOI] [PubMed] [Google Scholar]
- 100.Vitousek, P. M., J. D. Aber, R. W. Howarth, G. E. Likens, P. A. Matson, D. W. Schindler, W. H. Sclesinger, and D. G. Tilman. 1997. Human alterations of the global nitrogen cycle: sources and consequences. Ecol. Appl. 7:737-750. [Google Scholar]
- 101.Voytek, M. A., B. B. Ward, and J. C. Priscu. 1998. The abundance of ammonium-oxidizing bacateria in Lake Bonney, Antarctica determined by immunofluorescence, PCR and in situ hybridization, p. 217-228. In J. C. Priscu (ed.), Ecosystem/dynamics in a polar desert: the McMurdo Dry Valley, Antarctica. American Geophysical Union, Washington, D.C.
- 102.Wang, Q. F., H. Li, and A. F. Post. 2000. Nitrate assimilation genes of the marine diazotrophic, filamentous cyanobacterium Trichodesmium sp. strain WH9601. J. Bacteriol. 182:1764-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ward, B. B. 2000. Nitrification and the marine nitrogen cycle, p. 427-454. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 104.Ward, B. B., and D. A. Bronk. 2001. Net nitrogen uptake and DON release in surface waters: importance of trophic interactions implied from size fractionation experiments. Mar. Ecol. Prog. Ser. 219:11-24. [Google Scholar]
- 105.Ward, B. B., and A. F. Carlucci. 1985. Marine ammonia- and nitrite-oxidizing bacteria: serological diversity determined by immunofluorescence in culture and in the environment. Appl. Environ. Microbiol. 50:194-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ward, B. B., K. A. Kilpatrick, E. Renger, and R. W. Eppley. 1989. Biological nitrogen cycling in the nitracline. Limnol. Oceanogr. 34:493-513. [Google Scholar]
- 107.Ward, B. B., D. P. Martino, C. M. Diaz, and S. B. Joye. 2000. Analysis of ammonia-oxidizing bacteria from hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl. Environ. Microbiol. 66:2873-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wasmund, N., M. Voss, and K. Lochte. 2001. Evidence of nitrogen fixation by non-heterocystous cyanobacteria in the Baltic Sea and re-calculation of a budget of nitrogen fixation. Mar. Ecol. Prog. Ser. 214:1-14. [Google Scholar]
- 109.Watson, S. W. 1965. Characteristics of a marine nitrifying bacterium. Nitrosocystis oceanus sp. n. Limnol. Oceanogr. :R274-R289.
- 110.Whitby, C. B., J. R. Saunders, J. Rodriguez, R. W. Pickup, and A. McCarthy. 1999. Phylogenetic differentiation of two closely related Nitrosomonas spp. that inhabit different sediment environments in an oligotrophic freshwater lake. Appl. Environ. Microbiol. 65:4855-4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu, J., W. Sunda, E. A. Boyle, and D. M. Karl. 2000. Phosphate depletion in the western North Atlantic Ocean. Science 289:759-762. [DOI] [PubMed] [Google Scholar]
- 112.Ye, R. W., and S. M. Thomas. 2001. Microbial nitrogen cycles: physiology, genomics and applications. Curr. Opin. Microbiol. 4:307-312. [DOI] [PubMed] [Google Scholar]
- 113.Yoshinari, T. 1976. Nitrous oxide in the sea. Mar. Chem. 4:189-202. [Google Scholar]
- 114.Zart, D., and E. Bock. 1998. High rate of aerobic nitrification and denitrification by Nitrosomonas eutropha grown in a fermentor with complete biomass retention in the presence of gaseous NO2 or NO. Arch. Microbiol. 169:282-286. [DOI] [PubMed] [Google Scholar]
- 115.Zehr, J. P., E. J. Carpenter, and T. A. Villareal. 2000. New perspectives on nitrogen-fixing microorganisms in tropical and subtropical oceans. Trends Microbiol. 8:68-73. [DOI] [PubMed] [Google Scholar]
- 116.Zehr, J. P., B. Dominic, Y. B. Chen, M. Mellon, and J. C. Meeks. 1999. Nitrogen fixation in the marine cyanobacterium Trichodesmium: a challenging model for ecology and molecular biology, p. 485-500. In G. A. Peschek, W. Loffelhardt, and G. Schmetterer (ed.), The phototrophic prokaryotes. Kluwer Academic/Plenum Publishers, New York, N.Y.
- 117.Zehr, J. P., and L. A. McReynolds. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium spp. Appl. Environ. Microbiol. 55:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by the amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zehr, J. P., J. B. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Karl. 2001. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412:635-638. [DOI] [PubMed] [Google Scholar]
- 120.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]