Abstract
Acinetobacter sp. strain M-1 accumulated a large amount of wax esters from an n-alkane under nitrogen-limiting conditions. Under the optimized conditions with n-hexadecane as the substrate, the amount of hexadecyl hexadecanoate in the cells reached 0.17 g/g of cells (dry weight). Electron microscopic analysis revealed that multilayered disk-shaped intracellular inclusions were formed concomitant with wax ester formation. The contribution of acyl-CoA reductase to wax ester synthesis was evaluated by gene disruption analysis.
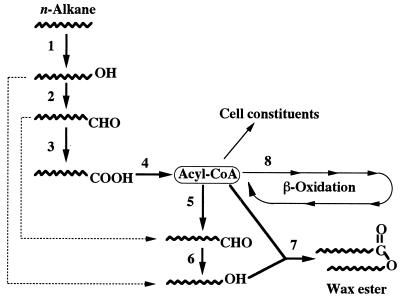
Various types of wax ester are widely used in the manufacture of fine chemicals, i.e., cosmetics, candles, printing inks, lubricants, coating stuffs, etc. Acinetobacter spp. are known to accumulate wax ester as a storage material (10). The pathway for wax ester synthesis of Acinetobacter spp. is assumed to start from acyl coenzyme A (acyl-CoA), which is reduced to the corresponding alcohol via acyl-CoA reductase (6) and aldehyde reductase (12) (Fig. 1). The microbial production of wax ester has advantages over other biological sources since the wax ester composition can be controlled by the choice of starting material or growth conditions (1).
FIG. 1.
The proposed carbon flow from n-alkanes to wax esters in Acinetobacter spp. Dotted lines indicate alternative ways to wax ester synthesis. Enzymes: 1, alkane hydroxylase complex; 2, NAD(P)-dependent alcohol dehydrogenase; 3, NAD(P)-dependent aldehyde dehydrogenase; 4, acyl-CoA synthetase; 5, acyl-CoA reductase; 6, aldehyde reductase; 7, acyl-CoA: alcohol transacylase; 8, acyl-CoA dehydrogenase.
Acinetobacter sp. strain M-1 has characteristic properties as to n-alkane metabolism, such as the ability to grow on a broad range of n-alkanes (7, 8), enzymatic diversity of related enzymes (12), and a notable level of wax ester accumulation. Based on the obtained results, we report here the novel ultrastructure of intracellular inclusion bodies and a metabolic pathway for wax ester synthesis from n-alkanes.
MATERIALS AND METHODS
Chemicals and enzymes.
Restriction enzymes, alkaline phosphatase (calf intestine), T4 DNA ligase, and Ex Taq DNA polymerase were products of Takara Shuzo Co. (Kyoto, Japan). A Thermo Sequence fluorescence-labeled primer cycle sequencing kit with 7-deaza-dGTP was purchased from Amersham Pharmacia Biotech (Little Chalfont, Buckinghamshire, England), and [α-32P]dATP was from Amersham Corp. (Arlington Heights, Ill.).
Organism, culture conditions, and vectors.
Acinetobacter sp. strain M-1 (7) was used throughout this work. To induce wax ester accumulation, n-hexadecane or the oxidized compounds were added at a concentration of 0.5% (vol/vol) to the nitrogen-limited medium (2). Growth condition of Escherichia coli JM109 and its transformants and vectors for gene cloning and expression were principally the same as described previously (4).
Assay.
Determination of protein concentration and sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) were performed as described previously (4). Acyl-CoA reductase activity was assayed by a previously described method (6). Wax ester was determined by gas chromatography (1).
Electron microscopy.
For the preparation of thin sections, the bacterial cells were fixed with glutaraldehyde as described previously (9). The ultrathin sections were observed under an electron microscope (JEM-1220; JEOL Ltd., Tokyo, Japan). For the quick-freezing replica method, the washed cells were quickly frozen using the technique of Heuser (3). The replicas, which were obtained by rotary shadowing with Pt and C, were examined under a Hitachi H-7000 electron microscope (Hitachi, Japan) operating at 100 kV.
Cloning of acrM from the genome of Acinetobacter sp. strain M-1.
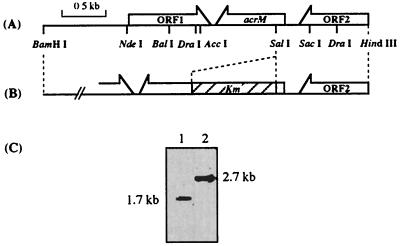
To amplify the fragment DNA encoding acyl-CoA reductase (AcrM) from the chromosomal DNA of Acinetobacter sp. strain M-1, the following primers were synthesized based on the acr1 gene from Acinetobacter calcoaceticus BD413: BDN, 5′-GTGAACAAAAAACTTGAAGCTCTC-3′, and BDC, 5′-TTACCAGTGTTCGCCTGGGAA-3′. The PCR product was approximately 2.8 kb long and contained an open reading frame (ORF), which showed high homology with the original gene, acr1. A new primer, BDN2, 5′-AGAGAAGTTAGATGAGGTC-3′, was also synthesized to amplify only a portion of the objective ORF. PCR with the BDC and BDN2 primers produced a gene of 0.7 kb, and the resulting fragment was used as a probe. Through Southern hybridization (11) and colony hybridization, one positive clone which comprised a 4.0-kb BamHI-HindIII fragment in pBluescript II SK(+) was isolated. The whole nucleotide sequence of this plasmid (pBM4) was determined. A restriction map of pBM4 is shown in Fig. 2A.
FIG. 2.
Genetic organization of the cloned region including the acrM gene and that of the disruptant. (A) ORFs and restriction map of the cloned BamHI-HindIII 4.0-kb cloned fragment. (B) Organization of the disruption vector, pDAR1, derived from pBM4. The hatched box represents the kanamycin resistance gene. (C) Genomic Southern analysis of DraI-digested total DNA (5.0 μg per each lane) from the wild-type strain (lane 1) and the acrM-KO strain (lane 2), using the 32P-labeled acrM gene fragment as a probe.
Expression of the acrM gene in E. coli JM109.
The following primers were designed for overexpression of the acrM gene in a heterologous host, E. coli JM109: acrM-N, 5′-GAATTCTAAGGAGGTTTTTATATGAATGCAAAACTCAAAAAACTT-3′, and acrM-C, 5′-AAGCTTTTACCAGTGTTCGCCTGGG-3′. acrM-N contained an EcoRI site and the ribosomal binding sequence, and acrM-C contained a HindIII site. The PCR product was ligated with the pT7Blue vector and then with pUC118. A pUC118-derived plasmid, named pMR1, was introduced into E. coli JM109. Expression was confirmed by activity measurement and the protein band on SDS-PAGE.
Construction of the acrM disruptant.
The kanamycin resistance gene (Km) was amplified with the primers described previously (4), except for the flanking site of SalI. The amplified Km gene was inserted at the SalI site in the acrM gene of pBM4, yielding pDAR1 (Fig. 2B). The acrM gene disruption plasmid, pDAR1, was linearized, dephosphorylated, and then introduced into Acinetobacter sp. strain M-1 by electroporation. The kanamycin resistance clone was selected, and the gene disruption was confirmed by Southern analysis (Fig. 2C).
Nucleotide sequence accession number.
The nucleotide sequence data reported in this paper have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under accession number AB070446.
RESULTS AND DISCUSSION
Wax ester production.
The production of wax ester was optimized with respect to cell number, substrate concentration, and incubation time. As a result, using 1.85 × 1010 cells/ml with 3% n-hexadecane as the substrate, hexadecyl hexadecanoate was accumulated up to the amount of 0.17 g/g of cells (dry weight) in a 10-h reaction. The produced wax ester comprised hexadecyl hexadecanoate (98%) and hexadecyl myristate (2%). The by-product was two carbon atoms shorter than the main product. This suggested that a part of palmitoyl-CoA was converted to myristoyl-CoA through the β-oxidation cycle, which was then utilized by acyl-CoA:alcohol transacylase. The level of by-product formation depends on the substrate used for the reaction. When n-hexadecanol was used instead of n-hexadecane, 9.2% of hexadecyl myristate was formed as a by-product. From n-octadecane, octadecyl octadecanoate and octadecyl hexadecanoate were accumulated in the ratio of 67:33, and a negligible amount of wax ester with 32 carbon atoms was also detected. One of the major determinants for by-product formation may be the substrate specificities of the enzymes that participate in wax ester synthesis.
Ultrastructure of intracellular wax ester.
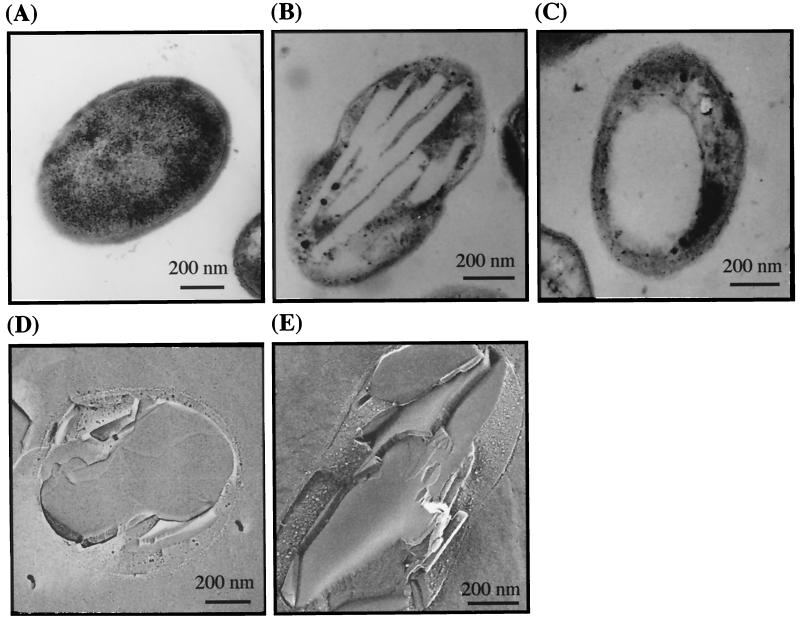
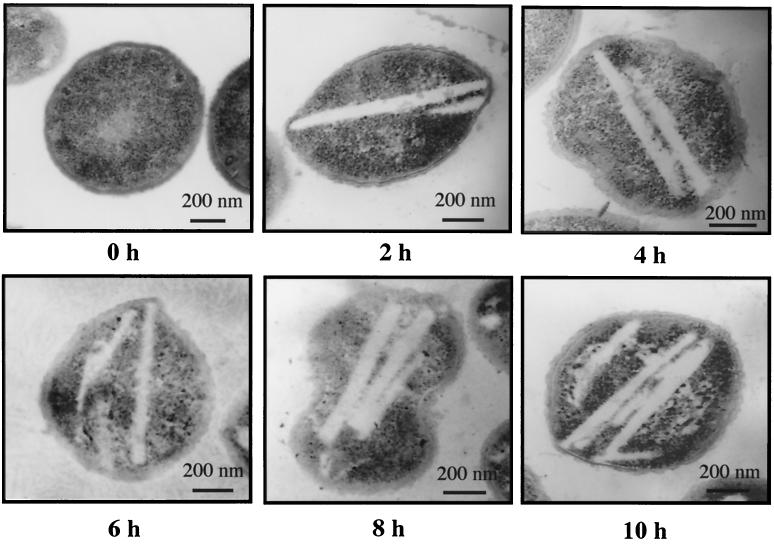
Thin sections of Acinetobacter sp. strain M-1 cells, which had been incubated in the nitrogen-limited medium with 0.5% n-hexadecane, contained multiple, long and narrow, electron-transparent, intracellular inclusion bodies (Fig. 3B). Figure 3C shows cells grown under the same conditions as in Fig. 3B, but the cells were sliced in another direction. On the other hand, cells grown on 2× yeast extract-tryptone (YT) medium (4) showed no characteristic intracellular structures (Fig. 3A). Quick-freezing replica microscopy showed the clearer structure of the inclusion bodies, which were disk-shaped, had a smooth surface, and grew to almost the same diameter as the cells (Fig. 3D). No intracytoplasmic membrane structures or limiting membranes surrounding these inclusions were observed. These ultrastructures were considerably distinct from those in the case of Acinetobacter sp. strain HO1-N, which accumulated rectangular inclusion bodies surrounded by a phospholipid-rich membrane. The wax ester accumulation proceeded with the sequential formation of disks, i.e., the completion of one disk led to another one (Fig. 4). We suggested that the inclusion bodies represent the accumulated form of wax ester based on the results that the intracellular amount of wax ester was exactly proportional to the volume of the inclusion bodies.
FIG. 3.
Electron microscopy of accumulated wax ester in Acinetobacter sp. strain M-1. Thin sections of cells grown on nutritious medium (A) and of cells after a shift to nitrogen-limited medium containing 0.5% n-hexadecane and incubation for 72 h at 30°C (B and C). (D and E) Images obtained by the quick-freezing replica method for cells prepared under the same conditions as in panel B.
FIG. 4.
Time course of wax ester accumulation in Acinetobacter sp. strain M-1. Cells were precultured in 2× YT medium (4), shifted to nitrogen-limited medium containing 0.5% n-hexadecane for the indicated incubation periods, and then subjected to electron microscopy.
Cloning of the acyl-CoA reductase gene from Acinetobacter sp. strain M-1.
acrM comprised 888 bp, corresponding to 295 amino acid residues with a predicted molecular mass of 32,375 Da. Comparison of AcrM with Acr1 of A. calcoaceticus BD413 (6) showed homology, i.e., identity of 69.8% at the gene level and 78.3% at the amino acid level, although their initiation codons are different, AcrM (ATG) and Acr1 (GTG).
Expression of the acrM gene in E. coli JM109.
Since the amount of acyl-CoA reductase in Acinetobacter sp. strain M-1 was too low, the acrM gene was overexpressed in E. coli JM109 to study its enzymatic properties. The expressed protein was recognized as a protein with an apparent molecular mass of ca. 33 kDa on SDS-PAGE. The protein corresponding to recombinant AcrM was exclusively localized in the membrane fraction that was obtained on ultracentrifugation (105,000 × g for 1 h) of the cell extract, and the specific activity of this fraction toward palmitoyl-CoA was 0.95 U/mg of protein. This was in good accordance with the hydrophobic prediction of cloned AcrM and indicated that the enzyme existed on the membranes in the parental strain. Recombinant AcrM of the membrane fraction showed the highest activity toward myristoyl-CoA (1.32 U/mg of protein), and the following activity toward the other substrates: decyl-CoA (2.5% of the activity toward myristoyl-CoA), lauroyl-CoA (33%), palmitoyl-CoA (90%), stearoyl-CoA (44%), and arachidoyl-CoA (29%). The substrates with carbon chain lengths shorter than 8 did not show any activity, and ones with chains longer than 22 were not tested.
Gene disruption experiments.
Accumulation of wax ester was compared between the wild type and the acrM disruptant (acrM-KO) strain. As shown in Table 1, there was no significant difference in the amount of accumulated intracellular wax ester between the two strains when n-hexadecane or n-hexadecanol was used as the substrate. In contrast, when hexadecanoic acid was used as the substrate, no wax ester formation was detected for the acrM-KO strain. This indicated the critical function of acyl-CoA reductase in wax ester synthesis from carboxylic acid. An acyl-CoA reductase-deficient mutant of A. calcoaceticus BD413 produced wax ester from neither n-hexadecane nor hexadecanoic acid (6). Judging from recent results, both A. calcoaceticus BD413 and Acinetobacter sp. strain M-1 have principally the same pathway for n-alkane oxidation (Fig. 1), whose first step is catalyzed by membrane-bound alkane monooxygenase (5, 12). The high level of production of wax ester from n-alkanes by Acinetobacter sp. strain M-1 can be explained by the fact that the alcohol supply is fairly higher in strain M-1 than in other Acinetobacter strains.
TABLE 1.
Accumulation of wax ester from various carbon sources
| Strain | Hexadecyl hexadecanoate (μmol/mg of protein) accumulated froma:
|
|||
|---|---|---|---|---|
| n-Hexadecane | n-Hexadecanol | cis-11-Hexadecenal | Hexadecanoic acid | |
| Wild type | 9.70 | 4.75 | 2.40 | 7.26 |
| acrM-KO | 8.52 | 4.91 | 7.90 | 0 |
The cells were grown under the nitrogen-limiting conditions with a 0.5% concentration of each substrate.
Acinetobacter sp. strain M-1 is a good candidate for the commercial production of wax ester based on the high productivity from diverse substrates, especially n-alkanes. Furthermore, the alcohol and carboxylic acid compositions of wax ester can possibly be controlled more precisely by alteration of the substrates in the acrM-KO strain, because the alcohol moiety can be completely fixed to the substrate carbon source in the disruptant strain.
Acknowledgments
We thank Yuriko Ikeda and Takako Seki for excellent technical assistance with the quick-freezing-replica electron microscopy.
REFERENCES
- 1.Dewitt, S., J. L. Ervin, D. Howes-Orchison, D. Dalietos, and S. L. Neidleman. 1982. Saturated and unsaturated wax esters produced by Acinetobacter sp. HO1-N grown on C16-C20 n-alkanes. J. Am. Oil Chemists Soc. 59:69-74. [Google Scholar]
- 2.Fixter, L. M., M. N. Nagi, J. G. McCormack, and C. A. Fewson. 1986. Structure, distribution and function of wax esters in Acinetobacter calcoaceticus. J. Gen. Microbiol. 132:3147-3157. [Google Scholar]
- 3.Heuser, J. 1981. Preparing biological samples for stereo-microscopy by the quick-freeze, deep-etch, rotary-replication technique. Methods Cell Biol. 22:97-122. [DOI] [PubMed] [Google Scholar]
- 4.Ishige, T., A. Tani, Y. Sakai, and N. Kato. 2000. Long-chain aldehyde dehydrogenase that participates in n-alkane utilization and wax ester synthesis in Acinetobacter sp. strain M-1. Appl. Environ. Microbiol. 66:3481-3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ratajczak, A., W. Geissdorfer, and W. Hillen. 1998. Alkane hydroxylase from Acinetobacter sp. strain ADP1 is encoded by alkM and belongs to a new family of bacterial integral-membrane hydrocarbon hydroxylases. Appl. Environ. Microbiol. 64:1175-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiser, S., and C. Somerville. 1997. Isolation of mutants of Acinetobacter calcoaceticus deficient in wax ester synthesis and complementation of one mutation with a gene encoding a fatty acyl coenzyme A reductase. J. Bacteriol. 179:2969-2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sakai, Y., J. H. Maeng, S. Kubota, A. Tani, and Y. Tani. 1996. A non-conventional dissimilation pathway for long chain n-alkanes in Acinetobacter sp. M-1 that starts with a dioxygenase reaction. J. Ferment. Bioeng. 81:286-291. [Google Scholar]
- 8.Sakai, Y., J. H. Maeng, Y. Tani, and N. Kato. 1994. Use of long-chain n-alkanes (C13-C44) by an isolate, Acinetobacter sp. M-1. Biosci. Biotechnol. Biochem. 58:2128-2130. [Google Scholar]
- 9.Sakai, Y., A. Saiganji, H. Yurimoto, K. Takabe, H. Saiki, and N. Kato. 1996. The absence of Pmp47, a putative yeast peroxisomal transporter, causes a defect in transfer and folding of a specific matrix enzyme. J. Cell Biol. 134:37-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Singer, M. E., S. M. Tyler, and W. R. Finnerty. 1985. Growth of Acinetobacter sp. strain HO1-N on n-hexadecanol: physiological and ultrastructural characteristics. J. Bacteriol. 162:162-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Southern, E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503-517. [DOI] [PubMed] [Google Scholar]
- 12.Tani, A., T. Ishige, Y. Sakai, and N. Kato. 2001. Gene structures and regulation of the alkane hydroxylase complex in Acinetobacter sp. strain M-1. J. Bacteriol. 183:1819-1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tani, A., Y. Sakai, T. Ishige, and N. Kato. 2000. Thermostable NADP+-dependent medium-chain alcohol dehydrogenase from Acinetobacter sp. strain M-1: purification and characterization, and gene expression in Escherichia coli. Appl. Environ. Microbiol. 66:5231-5235. [DOI] [PMC free article] [PubMed] [Google Scholar]