Abstract
Hydrophobins are small (ca. 100 amino acids) secreted fungal proteins that are characterized by the presence of eight conserved cysteine residues and by a typical hydropathy pattern. Class I hydrophobins self-assemble at hydrophilic-hydrophobic interfaces into highly insoluble amphipathic membranes, thereby changing the nature of surfaces. Hydrophobic surfaces become hydrophilic, while hydrophilic surfaces become hydrophobic. To see whether surface properties of assembled hydrophobins can be changed, 25 N-terminal residues of the mature SC3 hydrophobin were deleted (TrSC3). In addition, the cell-binding domain of fibronectin (RGD) was fused to the N terminus of mature SC3 (RGD-SC3) and TrSC3 (RGD-TrSC3). Self-assembly and surface activity were not affected by these modifications. However, physiochemical properties at the hydrophilic side of the assembled hydrophobin did change. This was demonstrated by a change in wettability and by enhanced growth of fibroblasts on Teflon-coated with RGD-SC3, TrSC3, or RGD-TrSC3 compared to bare Teflon or Teflon coated with SC3. Thus, engineered hydrophobins can be used to functionalize surfaces.
Hydrophobins are moderately hydrophobic proteins secreted by fungi. They are characterized by the presence of eight cysteine residues in conserved spacing and by similar hydropathy patterns (30, 32, 34). Hydrophobins fulfill a broad spectrum of functions. They mediate escape of hyphae from a hydrophilic environment (41) and are involved in the formation of hydrophobic aerial structures such as aerial hyphae (4, 23, 28, 36, 38), spores (2, 3, 10, 21, 22), and fruiting bodies (5, 13). Moreover, they provide gas channels in fruiting bodies and lichens with a hydrophobic layer, which probably provides for more efficient gas exchange under wet conditions (14, 16, 27, 29, 31). Hydrophobins are also involved in the attachment of hyphae to hydrophobic surfaces (27, 37) and in the sensing thereof (22), which are important initial steps of pathogenic interactions before penetration and infection of the host can occur.
The basidiomycete Schizophyllum commune contains at least four class I hydrophobin genes: SC1, SC3, SC4, and SC6 (6, 11, 17, 33). SC3 is the best-characterized class I hydrophobin. It is involved in the formation of aerial hyphae (28, 36, 38, 41) and in the attachment of hyphae to hydrophobic surfaces (37). Upon contact with hydrophilic-hydrophobic interfaces SC3 monomers self-assemble into a highly insoluble amphipathic membrane (36, 37, 40). This is accompanied with an increase in β-sheet structure at the water-air interface and by increase in α-helical structure at the water-hydrophobic solid interface (6). The latter form is an intermediate to the form with increased β-sheet structure (35). Upon self-assembly of SC3 at the water-air interface the water surface tension is reduced to as low as 24 mJ m−2, which makes assembled SC3 one of the most powerful biosurfactants known to date (26, 41). When a hydrophobic solid is incubated in an aqueous hydrophobin solution, an amphipathic membrane of the protein is formed at the surface of the hydrophobic solid. The hydrophobic side of the membrane strongly interacts with the solid, while the hydrophilic side is exposed, thus making the surface wettable (37, 40). Conversely, by self-assembly of the class I hydrophobin, the surface of a hydrophilic solid can be made hydrophobic (12, 13, 36). Class II hydrophobins differ from class I hydrophobins in the stability of the assembled form. In contrast to assemblages of class I hydrophobins, those of class II hydrophobins dissociate in 60% ethanol and 1% sodium dodecyl sulfate (SDS) (35).
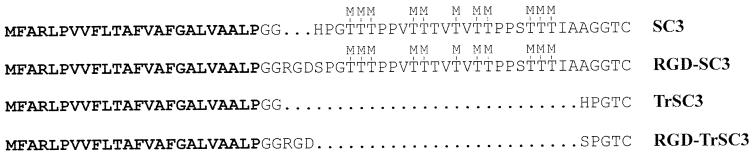
The class I hydrophobins SC4 of S. commune and ABH1 and ABH3 of Agaricus bisporus have similar properties as SC3 (12, 13, 14). The mature forms of these hydrophobins have 8 to 12 amino acids preceding the first cysteine residue and are not glycosylated. In contrast, mature SC3 has 31 amino acids preceding the first cysteine residue and contains 16 to 22 mannose residues most probably linked to one or more of the 12 threonine residues contained in this long N-terminal stretch (6) (Fig. 1). We report here that deletion of 25 of the 31 N-terminal amino acids of mature SC3 (Gly29-Gly53) and/or introduction of a cell-binding domain of human fibronectin (RGD) at the N terminus of SC3 does not affect self-assembly but changes the physiochemical properties of the hydrophilic side of the assembled hydrophobin.
FIG. 1.
N-terminal sequences preceding the first cysteine residues of SC3 and its derivatives. In RGD-SC3 the RGD sequence is introduced behind Gly26 and His27 is replaced by serine, creating the GRGDSP sequence of fibronectin. In TrSC3 the amino acids Gly29-Gly53 of SC3 are removed. RGD-TrSC3 is a derivative of TrSC3 in which the RGD sequence is introduced behind Gly26 and His27 is replaced by serine. −M indicates putative O glycosylation sites. The signal sequences that are cleaved off upon entry in the endoplasmic reticulum are indicated in boldface.
MATERIALS AND METHODS
Strains.
Cloning was done in Escherichia coli JM83. S. commune strains 4-39 (CBS 341.18) and 72-3 were used. S. commune strain 72-3 is identical to the wild-type strain 4-39 apart from a disrupted SC3 gene (ΔSC3 strain) (28, 37).
Growth conditions and media.
S. commune was grown in minimal medium (MM) (7) either solidified or not solidified with 1.5% agar. For transformation, the ΔSC3 strain was grown from a mycelial homogenate for 2 days at 24°C and 225 rpm in 100 ml of MM in 250-ml flasks. For analysis of proteins secreted into the media or for purification of hydrophobins S. commune was grown in 1-liter shaken cultures (225 rpm) for 5 to 7 days. In the latter case, the fungus was grown in production medium, i.e., MM in which asparagine is replaced by 1.3 g of ammonium sulfate liter−1 and containing 36 mM phosphate. For RNA isolation, colonies were grown for 2 to 3 days on the surface of a perforated polycarbonate (PC) membrane (diameter 76 mm, 0.1-μm pores; Poretics) that was positioned on solidified MM.
Construction of derivatives of the SC3 gene.
Three derivatives of the SC3 gene were constructed (Fig. 1). (i) The RGD-SC3 gene contains an insertion of the coding sequence of the RGD peptide between Gly26 and His27 and a replacement of His27 by serine, creating the GRGDSP sequence of human fibronectin. (ii) In the TrSC3 gene, the coding sequence of Gly29-Gly53 of SC3 is deleted. (iii) In the RGD-TrSC3 gene, the coding sequence of the RGDS peptide is introduced in TrSC3 as in RGD-SC3. In all cases the coding sequence of the signal peptide (Met1-Pro24) remained intact to allow secretion of the modified hydrophobins into the medium. To construct RGD-SC3, a 618-bp fragment of the genomic sequence of SC3 was amplified by PCR. Oligonucleotide primers were used corresponding to sense nucleotides 82 to 101 (6) (accession number M32329), containing additional codons for GRGDS (taking into account the preferred codon usage of S. commune) and antisense nucleotides 660 to 679, containing an additional BamHI site. The 618-bp fragment was used as a template in a second PCR with the antisense primer described above and a primer corresponding to the sense codons of GRGDS containing nucleotides 46 to 75 of the genomic SC3 sequence as a linker (containing a NarI site). The resulting 645-bp fragment encompasses nucleotides 49 to 679 of genomic SC3 with codons for RGD incorporated behind the codon of Gly26 and a codon replacement of His27 by serine. This fragment was cloned in pSC3.1 cut with NarI/BamHI, resulting in construct pSC3rgd.1. pSC3.1 is a derivative of pGEM-T (Promega) encompassing the genomic coding sequence of SC3 with a generated NcoI site in the start codon and a BamHI site directly downstream of the stop codon. To construct TrSC3, a 422-bp fragment of the genomic sequence of SC3 was amplified by PCR with an oligonucleotide primer corresponding to sense nucleotides 270 to 287, containing an XmaI site, and the antisense primer described above. This fragment was cloned in pSC3.1 cut with XmaI/BamHI, resulting in pSC3s.1. pSC3s.1 thus contains the truncated SC3 gene in which the codons for Gly29-Gly53 have been deleted. To construct RGD-TrSC3, a 411-bp fragment of the genomic sequence of TrSC3 was amplified by PCR with oligonucleotide primers corresponding to sense nucleotides 83 to 102 of TrSC3, containing additional codons for GRGDS, and antisense nucleotides 475 to 494, containing an additional BamHI site. This fragment was used as a template in a second PCR with a primer corresponding to the sense codons of GRGDS, while containing nucleotides 49 to 75 of the genomic SC3 sequence as a linker (containing a NarI site) and the antisense primer used to amplify the template. The resulting 460-bp fragment encompasses the nucleotides encoding TrSC3 with the codons of RGD incorporated behind the codon for Gly26 and containing a substitution of the codon for His27 for serine. This fragment was cloned in pSC3.1 cut with NarI/BamHI, resulting in construct pSC3srgd.1.
The three derivatives of the coding sequence of the SC3 gene were cloned as NcoI/BamHI fragments behind a 1,022-bp HindIII/NcoI SC3 promoter fragment in pUC20 (3HNCI [20]), resulting in the constructs pSC3rgd.2, pSC3s.2, and pSC3srgd.2. The HindIII/BamHI fragments encompassing the SC3 promoter and the coding sequence of RGD-SC3, TrSC3, or RGD-TrSC3 were then cloned in front of a 1,699-bp SC3 terminator sequence contained in the BamHI/EcoRI sites of pT3P. This derivative of pUC20 also contains a 1,300-bp phleomycin resistance cassette (18) cloned in the EcoRI site. This resulted in plasmids pSC3rgd.3, pSC3s.3, and pSC3srgd.3, which were used to transform the S. commune ΔSC3 strain. The sequences of the PCR fragments were confirmed by sequence analysis.
Transformation of S. commune.
S. commune strain 72-3 (ΔSC3) was transformed as described previously (18), except that it was protoplasted in 1 M MgSO4 containing 1 mg of lysing enzymes from Trichoderma harzianum (Sigma) ml−1. Then, 5 to 10 μg of DNA was added to 3 × 107 protoplasts in 100 μl of 1 M sorbitol. Transformants were selected on MM plates containing 40 μg of phleomycin (Cayla, Toulouse, France) ml−1 and 500 μg of caffeine (Sigma) ml−1, allowing selection of transformants despite the phleomycin resistance cassette already contained in the ΔSC3 strain (28, 37).
Northern blot analysis.
RNA was isolated from mycelium that had been ground in liquid nitrogen by using Trizol reagent (Gibco-BRL). It was separated on a 1% formaldehyde gel and blotted to Hybond-N+ membrane (Amersham). The RNA was hybridized with a 32P-labeled DNA fragment encompassing the coding sequence of SC3.
Isolation of secreted proteins and purification of hydrophobins.
Mycelium was separated from the culture medium of shaken cultures by using Nylon gauze (200-μm mesh). SC3 and RGD-SC3 were purified from the medium as described previously (32, 36). The purification procedures for TrSC3 and RGD-TrSC3 were essentially the same. Briefly, SC3 and its derivatives were assembled on small hydrogen bubbles released from a Pt cathode that was positioned at the bottom of a container (the anode was placed a few millimeters under the surface of the medium) by using a current of 150 mA for 3 h (12). The coated hydrogen bubbles formed a foam, which was freeze-dried after two washes with water. The material was treated with trifluoroacetic acid (TFA) and, after removal of the solvent with a stream of nitrogen gas, was taken up in water (±500 mg of material in 10 ml of water) and adjusted to pH 6 with concentrated ammonia. After the insoluble material was removed by centrifugation at 5,000 × g for 30 min, ethanol was added to a final concentration of 60% (vol/vol). Precipitates were removed by centrifugation, followed by the addition of 2 volumes of 0.3 M NH4 acetate in pure ethanol. This resulted in the precipitation of SC3 and RGD-SC3 but not of TrSC3 and RGD-TrSC3. SC3 and RGD-SC3 were collected by centrifugation and, after being washed with 100% ethanol, were taken up in water. These hydrophobins were then reassembled by electrobubbling. For TrSC3 and RGD-TrSC3, the ethanol was removed by film evaporation under reduced pressure, after which the material was freeze-dried. The freeze-dried material was washed with 100% ethanol, treated with TFA, solubilized in water, and reassembled by electrobubbling.
Protein analysis.
Samples were taken up in SDS sample buffer (containing β-mercaptoethanol) after treatment with TFA and adjusted to pH 6.8 with concentrated ammonia, if necessary. SDS-polyacrylamide gel electrophoresis (PAGE) was performed on 12.5 or 15% (wt/vol) polyacrylamide gels (9). The gels were fixed in 10% trichloroacetic acid for 1 h and stained with colloidal Coomassie brilliant blue (G-250) for 16 h (15) or blotted onto polyvinylidene difluoride (PVDF) for immunodetection with SC3 antibodies (38) or for staining with periodic acid-Schiff reagent (Sigma) to detect glycoproteins.
Chemical deglycosylation of SC3.
Deglycosylation of SC3 was achieved with trifluoromethanesulfonic acid (TFMSA) (8). A mixture of 0.5 ml of TFMSA and 0.25 ml of anisole was cooled on ice and added to 5 mg of lyophilized protein. After incubation for 3 h under N2 on ice, the reaction was terminated by the cautious addition of 1.5 ml 50% (vol/vol) aqueous pyridine. The mixture was then extracted three times with diethyl ether. The aqueous phase was dialyzed exhaustively against water and lyophilized.
Hydrophobicity and surface tension measurements.
The degree of hydrophobicity was expressed as the contact angle of 1-μl water droplets (25). The hydrophobicity of the hydrophobic side of the hydrophobin membrane was measured by ascending a solution of hydrophobin in a Whatman paper strip (13). In this way hydrophobin assembled at the paper-air interface (i.e., “in vitro”). Hydrophobicity was also assessed by measuring the water contact angle of colonies of strains expressing SC3 or one of its derivatives after they had formed aerial hyphae (“in vivo”). The hydrophilicity of the hydrophilic side of the hydrophobin membrane was measured after assembly of the hydrophobin on hot sulfuric acid-KNO3 cleaned Teflon (37). Lowering of the surface tension upon assembly of hydrophobins at the water-air interface was measured as described previously (41).
CD measurements.
Circular dichroism (CD) spectra were recorded over the wavelength region from 190 to 250 nm on an Aviv 62A DS CD spectrometer by using a 5-mm quartz cuvette. The temperature was kept at 25°C, and the sample compartment was continuously flushed with N2. Spectra were the averages of 10 scans, with a bandwidth of 1 nm and a 5-s averaging per point, and corrected for the signal of the solvent. Typically, a protein concentration of 3.5 μM in 20 mM phosphate buffer at pH 7.0 was used. For the spectra of monomeric protein, the protein was simply dissolved in buffer. To obtain spectra of hydrophobin assembled at the water-air interface, the solution was vigorously shaken on a vortex or heated at 95°C. To obtain spectra of hydrophobin assembled at the water-Teflon interface, colloidal Teflon was added to the solution. The protein concentration was determined by total amino acid analysis (19) and used to calculate the mean residue molar ellipticity. The spectra were not smoothed.
MALDI-TOF measurements.
MALDI-TOF (matrix-assisted laser desorption ionization-time of flight) mass spectroscopy was done by drying 1 μl of a solution of hydrophobin (100 μg ml−1, TFA treated) in 0.1% aqueous TFA on a target, together with 1 μl of matrix (20 mg of sinapinic acid in aqueous acetonitrile [40%, vol/vol] ml−1 in the presence of 0.1% TFA). The spectra were recorded in a TofSpec E&SE Micromass mass spectrometer.
Electron microscopic techniques.
Surface shadowing was done with Pt/C at an angle of 45°. Aqueous solutions of hydrophobin (10 μg ml−1) were dried down directly on Formvar-coated nickel grids and shadowed. Colony surfaces were shadowed after being dried on glass. Replicas of Pt/C were cleaned in K2Cr2O7-saturated H2SO4 for 2 days and transferred to a Formvar-coated nickel grid (12). Samples were examined in a Philips CM10 electron microscope.
Biocompatibility assay.
SC3 and its derivatives were assembled on 70% ethanol-cleaned, steam-sterilized (20 min, 120°C) Teflon (37) by placing these sheets overnight in aqueous solutions of all types of hydrophobins (50 μg ml−1). Fibroblasts (cell line L292, from mouse alveolar adipose tissue) were seeded at 7,500 cells/cm2 in wells of 24-well plates in which coated or bare Teflon sheets had been placed and containing RPMI 1690 medium supplemented with 10% fetal calf serum, 104 U of penicillin ml−1, 104 U of streptomycin ml−1, and glutamine solution (1:100; Gibco-BRL). As a positive control, fibroblasts were grown in the absence of a Teflon sheet. Growth processes were evaluated at 24, 48, 72, and 96 h. Biocompatibility was assessed by microscopically estimating the confluence of the cultures at the various surfaces.
RESULTS
Analysis of ΔSC3 strains transformed with derivatives of the SC3 hydrophobin gene.
Three derivatives of the SC3 gene were constructed by PCR (see Materials and Methods). The RGD-SC3 gene contains an insertion encoding the RGD tripeptide following Gly26 of SC3, while His27 is replaced by serine. This gene thus encodes a mature hydrophobin (i.e., after cleavage of the signal sequence) starting at Gly25 or Gly26, followed by the RGDS sequence and Pro28 (see Fig. 1). Coincidently, the RGDS sequence in human fibronectin (accession number K00055) is also preceded by G and is followed by P. The TrSC3 gene encodes a hydrophobin in which the coding sequence of Gly29-Gly53 of SC3 is deleted. Thus, mature TrSC3 is expected to have 6 instead of 31 amino acids preceding the first cysteine residue. The RGDS sequence was introduced in a similar way in TrSC3 as in SC3, resulting in RGD-TrSC3.
The coding sequences of the SC3 derivatives were placed between the regulatory sequences of SC3. Plasmids pSC3rgd.3, pSC3s.3, and pSC3srgd.3, containing the RGD-SC3 gene, the TrSC3 gene, and the RGD-TrSC3 gene, respectively, were used to transform the ΔSC3 strain. Transformants containing either construct were grown on PC membranes placed on MM agar. After 3 days, the PC membranes were transferred to fresh agar plates, on top of which PVDF membranes were placed. After 2 h these membranes were used for immunodetection of SC3. A total of 4 out of 30 strains transformed with the RGD-SC3 gene gave a positive immunosignal on the PVDF membrane that was indicative for secretion of the SC3 derivative. No signals were obtained with strains transformed with the TrSC3 or RGD-TrSC3 genes. This can be explained by the fact that the antiserum reacts with the 16 to 22 mannose residues of SC3 (6) most probably linked to one or more of the 12 threonine residues preceding the first cysteine residue of SC3. These residues are absent in TrSC3 and RGD-TrSC3. Therefore, in order to screen for transformants producing TrSC3 and RGD-TrSC3, RNA was isolated from these transformants and hybridized to the coding sequence of SC3. A total of 7 of 14 transformants containing construct pSC3s.3 (TrSC3) and 5 of 11 transformants containing construct pSC3srgd.3 (RGD-TrSC3) showed mRNA levels of the derivatives similar to that of SC3 in the wild-type strain (results not shown). Colonies that showed a high accumulation of the TrSC3 or RGD-TrSC3 mRNA formed hydrophobic aerial hyphae. In contrast, aerial hyphae of colonies that expressed TrSC3 or RGD-TrSC3 at a low level were hydrophilic. A similar correlation was found for colonies either or not secreting RGD-SC3. Thus, TrSC3, RGD-SC3 and RGD-TrSC3 can complement SC3 in making aerial hyphae hydrophobic.
Purification and characterization of SC3 derivatives.
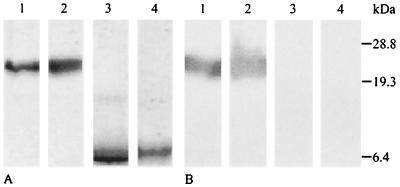
One transformant each, showing high expression of RGD-SC3, TrSC3, or RGD-TrSC3, was selected for further analysis. Strains were grown for 5 days in liquid shaken cultures, and the SC3 derivatives contained in the culture medium were purified in milligram quantities. The purification method was essentially the same as that described for wild-type SC3 (32, 36) (see Materials and Methods). In contrast to SC3 and RGD-SC3, TrSC3 and RGD-TrSC3 did not precipitate in 83% ethanol but required 90 to 100% ethanol at the concentrations used (50 to 100 μg ml−1). SDS-PAGE revealed a protein band at 24 kDa in the RGD-SC3 strain that was similar to the position of SC3 in the wild-type strain (Fig. 2A). As expected, both proteins reacted with the SC3 antiserum (Fig. 2B). Proteins with apparent molecular masses of 6.4 kDa (Fig. 2A) were secreted by the TrSC3 and RGD-TrSC3 strains and did not react with the SC3 antiserum (Fig. 2B). The higher mobility in SDS-PAGE and the fact that the antiserum did not react with these proteins can be explained by the absence of glycosylation in these derivatives of SC3. N-terminal sequencing confirmed the identity of the proteins. The predicted N-terminal sequences for RGD-SC3, TrSC3, and RGD-TrSC3 were obtained: (G)GRGDSP, (G)GHPGTXTTGS, and GGRGDSPGTX, respectively, in which “X” most probably represents a cysteine residue (see Fig. 1). In all cases an additional form of SC3 was identified containing the two last amino acids of the leader sequence (Leu23Pro24). MALDI-TOF mass spectroscopy of SC3 and its derivatives were in good agreement with the expected masses based on N-terminal sequencing and assuming the absence of mannose residues in TrSC3 and RGD-TrSC3 and the presence of 16 to 22 mannose residues in SC3 and RGD-SC3. Indeed, in contrast to the latter hydrophobins, the former did not react with periodic acid-Schiff staining (not shown).
FIG. 2.
SDS-PAGE of SC3 and its derivatives purified from the culture medium. (A and B) Coomassie brilliant blue staining (A) and Western blot analysis (B) with antibodies raised against SC3. Lanes: 1, wild-type SC3; 2, RGD-SC3; 3, TrSC3; 4, RGD-TrSC3.
The surface activities of RGD-SC3, TrSC3, and RGD-TrSC3 were similar to that of SC3 (Table 1). In all cases the water surface tension was lowered from 72 to 30 to 32 mJ m−2 at 100 μg ml−1. Modification of the N terminus also did not affect the wettability of the hydrophobic side of assembled hydrophobins (Table 1). However, the wettability of the hydrophilic side of the assembled hydrophobin did change. Assembled TrSC3 and RGD-TrSC3 had water contact angles of 73 ± 3 and 68 ± 3 at their hydrophilic sides, respectively, which is considerably higher than that of SC3 (44 ± 1) and RGD-SC3 (44 ± 2). The reduced wettability of the truncated SC3 forms is probably due to the absence of the mannose residues because chemically deglycosylated SC3 had a water contact angle (66 ± 6) similar to that of TrSC3 and RGD-TrSC3.
TABLE 1.
Surface activity (γlv) of SC3 and its derivatives at the water-air interface (100 μg ml−1) and wettability at both sides of the hydrophobin membranesa
| Hydrophobin | Mean degrees ± SEM
|
γlv (mJ m−2) | Rodlet diam (nm) | |
|---|---|---|---|---|
| θ hydrophobic side (in vitro, in vivo) | θ hydrophilic side | |||
| SC3 | 115 ± 12, 121 ± 6 | 44 ± 1 | 32 | 10 |
| RGD-SC3 | 122 ± 4, 120 ± 4 | 44 ± 2 | 32 | 10 |
| TrSC3 | NDd, 119 ± 3 | 73 ± 3 | 32 | 8-10 |
| RGD-TrSC3 | 120 ± 3, 123 ± 1 | 68 ± 3 | 30 | 8-10 |
| SC3−Glyb | ND, ND | 66 ± 6 | 32c | -e |
Wettability is expressed as the water contact angle (θ). Note that the surface tension of water is 72 mJ m−2.
That is, chemically deglycosylated SC3.
The surface activity was measured at 22 μg ml−1.
ND, not determined.
-, No rodlets were observed.
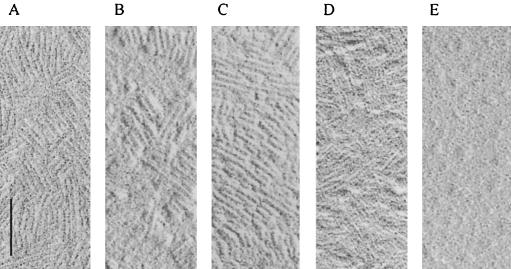
Surface shadowing revealed that RGD-SC3, TrSC3, and RGD-TrSC3 dried down on Formvar-coated grids are characterized by rodlets similar to those formed by SC3 (Fig. 3). The diameter of the TrSC3 and RGD-TrSC3 rodlets, however, ranged from 8 to 10 nm instead of the 10-nm diameter observed for SC3 and RGD-SC3. These rodlets were also shown at the surface of aerial hyphae (not shown). No rodlets were observed after drying down a solution of chemical deglycosylated SC3 (Fig. 3).
FIG. 3.
Rodlets of SC3 (A), RGD-SC3 (B), TrSC3 (C), and RGD-TrSC3 (D) as visualized by surface shadowing. (E) No rodlets were shown in case of chemically deglycosylated SC3. Bar, 100 nm.
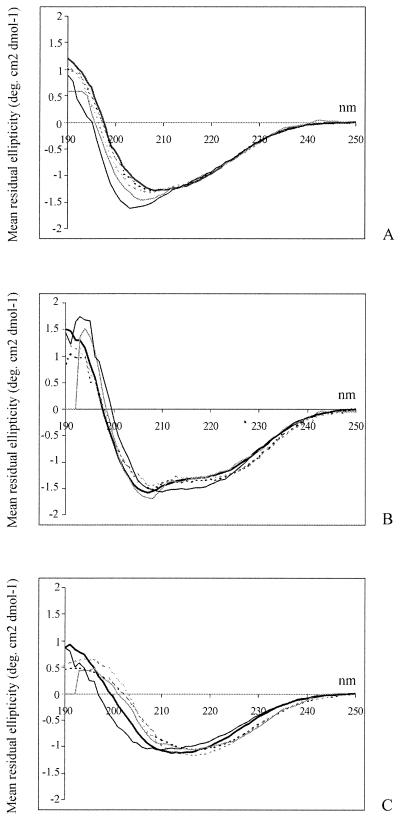
The conformational changes that RGD-SC3, TrSC3, and RGD-TrSC3 undergo during self-assembly were analyzed by using CD and compared to those observed for SC3 and deglycosylated SC3 (6). The spectra indicate that all monomeric forms are rich in β-sheet (Fig. 4A). Maximal absorbance of the proteins in water was seen at slightly different wavelengths; 208 nm for SC3, 209 nm for RGD-SC3, 205 nm for TrSC3, 207 nm for RGD-TrSC3, and 203 nm for deglycosylated SC3. Assembly of RGD-SC3, TrSC3, and RGD-TrSC3 and deglycosylated SC3 on colloidal Teflon was accompanied by an increase in the α-helix structure (Fig. 4B), while at a water-air interface the β-sheet structure was increased (Fig. 4C). These forms were previously observed for SC3 (6).
FIG. 4.
CD spectra of SC3 and its derivatives. (A) Spectra of SC3 and its derivatives in the monomeric form. (B) Same as panel A but assembled at the water-Teflon interface. (C) Same as panel A but assembled at the water-air interface. SC3 is indicated by a thick black line; RGD-SC3 is indicated by a thin, broken black line; TrSC3 is indicated by a thin gray line; RGD-SC3 is indicated by a thin, broken gray line; and deglycosylated SC3 is indicated by a thin black line.
Biocompatibility can be modified by changing the N-terminal part of the mature SC3 hydrophobin.
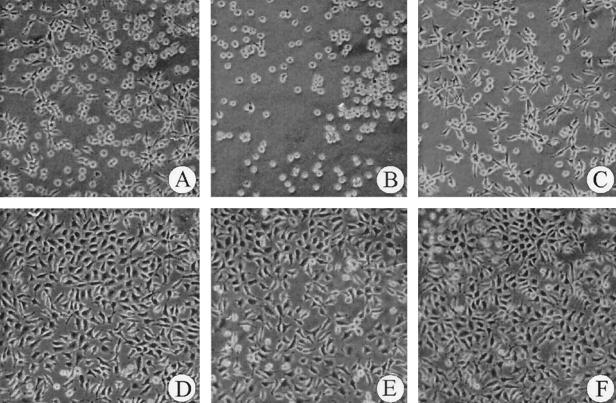
Fibroblasts were grown on surfaces of bare Teflon or Teflon coated with SC3 or one of its derivatives. The growth of fibroblasts was monitored microscopically and compared to the growth of cells at the bottom of a 24-well tissue culture plate that served as a positive control. After 48 h of growth, the confluence of fibroblasts on Teflon coated with the SC3-derivatives was higher (30 to 40%) than that on bare Teflon or SC3-coated Teflon (20%). After 96 h the confluence of fibroblasts on SC3-coated Teflon was lower (70%) compared to bare and RGD-SC3-coated Teflon (80%). However, confluences of 95% were observed on TrSC3- and RGD-TrSC3-coated Teflon, which was similar to the confluence in wells without Teflon that served as a positive control (Fig. 5). Significantly, the morphology of the cells grown on TrSC3 and RGD-TrSC3 coated surfaces was also similar to that of the control well. In contrast, an abberrant morphology was observed in the case of bare Teflon or Teflon coated with SC3 or RGD-SC3. These data show that coating with hydrophobins modified in the N-terminal part can modulate the biocompatibility of surfaces.
FIG. 5.
Growth of fibroblasts at bare Teflon (A) and Teflon coated with SC3 (B), RGD-SC3 (C), TrSC3 (D), and RGD-TrSC3 (E). (F) The growth of fibroblasts at the bottom of a 24-well tissue culture plate served as a control. Photographs were taken after 96 h of growth.
DISCUSSION
The property of hydrophobins to self-assemble at hydrophobic-hydrophilic interfaces makes them interesting candidates for use in medical and technical applications (32). For instance, hydrophobins could be used to increase the biocompatibility of medical implants or to prevent microbial cell adhesion to, e.g., catheter surfaces. Hydrophobins could also be used as an intermediate to attach cells, proteins (e.g., antibodies), or other molecules to hydrophobic surfaces, as in biosensors. In addition, hydrophobic solids or hydrophobic liquids such as oils can be stably dispersed in water by coating them with hydrophobin (37). The property of hydrophobins to coat a surface with a 10-nm thin film also makes these proteins interesting candidates for use in nanotechnology as defined by Thomas (24). In all applications, surfaces are primed with a strongly binding membrane making hydrophobic surfaces hydrophilic and hydrophilic surfaces hydrophobic (see, for example, references 9 and 31). We investigated here whether physiochemical properties (such as the degree of wettability) can be changed by genetic engineering.
The number of amino acids preceding the first cysteine residue in hydrophobins is highly variable. For instance, in mature SC6 of S. commune this N terminus is expected to contain 85 amino acids (11), whereas in SC4 of S. commune (17) and POH1 of P. ostreatus (1) it contains only 9 and 5 amino acids, respectively. X-ray photoelectron spectroscopy indicated that the N-terminal part of the SC3 hydrophobin is exposed at the hydrophilic side after self-assembly (39). These data therefore suggest that the peptide chain at the hydrophilic side of assembled hydrophobins can be highly variable. In the present study, derivatives of the SC3 gene were made in which 25 of 31 amino acids preceding the first cysteine residue of mature SC3 were deleted (TrSC3) and/or in which the cell-binding domain of human fibronectin (i.e., RGD) was introduced at the N-terminal end of the mature hydrophobin (RGD-SC3 and RGD-TrSC3, respectively). These genes were introduced in a ΔSC3 strain of S. commune that does not secrete hydrophobin in its culture medium (the SC1, SC4, and SC6 hydrophobin genes are only active in the dikaryon and not in the monokaryon used here). All three derivatives of the SC3 hydrophobin were recovered from the culture medium in milligram liter−1 quantities. The nature of the proteins was confirmed by N-terminal sequencing and mass spectroscopy. Biophysical characterization of the SC3 derivatives showed that the gross properties of the hydrophobins were unaffected. They self-assembled at the water-air and Teflon-water interface and showed conformational changes similar to SC3, as indicated by their CD spectra. The wettability of the hydrophobic sides of the assembled hydrophobins and the surface activity were also unaffected. They also formed the characteristic rodlet layer, although the rodlets formed by the truncated forms of SC3 were smaller than those of SC3 and RGD-SC3. Interestingly, the biophysical properties of the hydrophilic side of the assembled hydrophobin did change. TrSC3 and RGD-TrSC3 showed a decreased wettability at their hydrophilic side compared to that of SC3 and RGD-SC3. A similar decrease in wettability was observed for chemically deglycosylated SC3, indicating that the mannose residues are responsible for the high wettability at the hydrophilic side of assembled SC3. The inability to form rodlets after deglycosylation of SC3 also indicates that the mannose residues are essential for proper folding of this hydrophobin that is distinct from nonglycosylated hydrophobins in containing a long N-terminal stretch preceding the first cysteine residue. Changes of the biophysical properties of the exposed side of the assembled SC3 had an effect on the growth of fibroblasts. Biocompatibility (defined as coverage by growing fibroblasts) of Teflon coated with RGD-SC3 was improved compared to bare Teflon or Teflon coated with unmodified SC3. This increase in biocompatibility is likely due to the presence of the cell-binding domain (RGD) of fibronectin, which is specifically recognized by integrins in the fibroblast plasma membrane. However, the highest compatibility was found for Teflon coated with TrSC3 and RGD-TrSC3. In a quantitative analysis these differences in cell growth have been confirmed (M. I. Janssen and B. M. van Leeuwen, unpublished results). In the case of TrSC3 the absence of mannose residues and/or the decreased hydrophilicity at the coated surface probably stimulates cell interaction. Currently, qualitative studies are being performed to analyze the biocompatibility of hydrophobin-coated surfaces.
The reported data show that, by changing the N-terminal part of a hydrophobin, the physiochemical nature of the hydrophilic side of the assembled form of a hydrophobin can be altered and new functionalities can be introduced. The fact that hydrophobins self-assemble at any hydrophilic-hydrophobic surface, irrespective of the chemical nature of the surface, thus provides a generic approach to modifying surfaces. By generating a library of engineered hydrophobins, any hydrophobic surface can be provided with a coating with defined physiochemical properties (e.g., a certain wettability) or exposing a certain peptide. We are currently investigating whether mixed membranes can be deposited on solid surfaces, which would further extend the possibilities of surface modification.
Acknowledgments
We thank Henriek Beenen for making some of the plasmid constructs.
K. Scholtmeijer was financially supported by The Netherlands Technology Foundation (STW), coordinated by the Life Sciences Foundation (SLW).
REFERENCES
- 1.Ásgeirsdóttir, S. A., O. M. H. de Vries, and J. G. H. Wessels. 1998. Identification of three differentially expressed hydrophobins in Pleurotus ostreatus (oyster mushroom). Microbiology 144:2961-2969. [DOI] [PubMed] [Google Scholar]
- 2.Beever, R. E., and G. P. Dempsey. 1978. Function of rodlets on the surface of fungal spores. Nature 272:608-610. [DOI] [PubMed] [Google Scholar]
- 3.Bell-Pedersen, D., J. C. Dunlap, and J. J. Loros. 1992. The Neurospora circadian clock-controlled gene, ccg-2, is allelic to eas and encodes a fungal hydrophobin required for formation of the conidial rodlet layer. Genes Dev. 6:2382-2394. [DOI] [PubMed] [Google Scholar]
- 4.Bowden, C. G., E. Smally, R. P. Guries, M. Hubbes, B. Temple, and P. A. Horgen. 1996. Lack of association between cerato-ulmin production and virulence in Ophiostoma ulmi. Mol. Plant-Microbe Interact. 9:556-564. [DOI] [PubMed] [Google Scholar]
- 5.de Groot, P. W. J., P. J. Schaap, A. S. M. Sonneberg, J. Visser, and L. J. L. D. van Griensven. 1996. The Agaricus bisporus hypA gene encodes a hydrophobin and specifically accumulates in peel tissue of mushroom caps during fruit body development. J. Mol. Biol. 257:1008-1019. [DOI] [PubMed] [Google Scholar]
- 6.de Vocht, M. L., K. Scholtmeijer, E. W. van der Vegte, O. M. H. de Vries, N. Sonveaux, H. A. B. Wösten, J.-M. Ruysschaert, G. Hadziioannou, J. G. H. Wessels, and G. T. Robillard. 1998. Structural characterization of the hydrophobin SC3., as a monomer and after self-assembly at hydrophobic/hydrophilic interfaces. Biophys. J. 74:1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dons, J. J. M., O. M. H. de Vries, and J. G. H. Wessels. 1979. Characterisation of the genome of the basidiomycete Schizophyllum commune. Biochim. Biophys. Acta 563:100-112. [DOI] [PubMed] [Google Scholar]
- 8.Edge, A. S. B., C. R. Faltynek, L. Hof, L. E. Reichert, Jr., and P. Weber. 1981. Deglycosylation of glycoproteins by trifluoromethanesulfonic acid. Anal. Biochem. 118:131-137. [DOI] [PubMed] [Google Scholar]
- 9.Laemmli, U. K. 1970. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 10.Lauter, F.-R., V. E. A. Russo, and C. Yanofsky. 1992. Developmental and light regulation of eas, the structural gene for the rodlet protein of Neurospora. Genes Dev. 6:2373-2381. [DOI] [PubMed] [Google Scholar]
- 11.Lugones, L. G. 1998. Function and expression of hydrophobins in the basidiomycetes Schizophyllum commune and Agaricus bisporus. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 12.Lugones, L. G., H. A. B. Wösten, and J. G. H. Wessels. 1998. A hydrophobin (ABH3) specifically secreted by growing hyphae of Agaricus bisporus (common white button mushroom). Microbiology 144:2345-2353. [DOI] [PubMed] [Google Scholar]
- 13.Lugones, L. G., J. S. Bosscher, K. Scholtmeijer, O. M. H. de Vries, and J. G. H. Wessels. 1996. An abundant hydrophobin (ABH1) forms hydrophobic rodlet layers in Agaricus bisporus fruiting bodies. Microbiology 142:1321-1329. [DOI] [PubMed] [Google Scholar]
- 14.Lugones, L. G., H. A. B. Wösten, K. U. Birkenkamp, K. A. Sjollema, J. Zagers, and J. G. H. Wessels. 1999. Hydrophobins line air channels in fruiting bodies of Schizophyllum commune and Agaricus bisporus. Mycol. Res. 103:635-640. [Google Scholar]
- 15.Neuhof, V., N. Arold, D. Taube, and W. Erhardt. 1988. Improved staining of proteins in polyacrylamide gels including isoelectric focussing gels with clear background at nanogram sensitivity using Coomassie brilliant blue G-250 and R-250. Electrophoresis 9:255-262. [DOI] [PubMed] [Google Scholar]
- 16.Scherrer, S., O. M. H. de Vries, R. Dudler, J. G. H. Wessels, and R. Honegger. 2000. Interfacial self-assembly of fungal hydrophobins of the lichen forming ascomycetes Xantoria parietina and X. etaneoides. Fungal Genet. Biol. 30:81-93. [DOI] [PubMed] [Google Scholar]
- 17.Schuren, F. H. J., and J. G. H. Wessels. 1990. Two genes specifically expressed in fruiting dikaryons of Schizophyllum commune: homologies with a gene not regulated by mating-type genes. Gene 90:199-205. [DOI] [PubMed] [Google Scholar]
- 18.Schuren, F. H. J., and J. G. H. Wessels. 1994. Highly efficient transformation of the homobasidiomycete Schizophyllum commune to phleomycin resistance. Curr. Genet. 26:179-183. [DOI] [PubMed] [Google Scholar]
- 19.Schuster, R. 1988. Determination of amino acids in biological, pharmaceutical, plant and food samples by automated precolumn derivatization and high-performance liquid chromatography. J. Chromatogr. 431:271-284. [DOI] [PubMed] [Google Scholar]
- 20.Schuurs, T. A. 1998. Regulation of hydrophobin genes in Schizophyllum commune. Ph.D. thesis. University of Groningen, Groningen, The Netherlands.
- 21.Stringer, M. A., R. A. Dean, T. C. Sewall, and W. E. Timberlake. 1991. Rodletness, a new Aspergillus developmental mutant induced by directed gene inactivation. Genes Dev. 5:1161-1171. [DOI] [PubMed] [Google Scholar]
- 22.Talbot, N. J., M. Kershaw, G. E. Wakley, O. M. H. de Vries, J. G. H. Wessels, and J. E. Hamer. 1996. MPG1 encodes a fungal hydrophobin involved in surface interactions during infection related development of the rice blast fungus Magnaporthe grisea. Plant Cell 8:985-999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Temple, B., P. A. Horgen, L. Bernier, and W. E. Hintz. 1997. Cerato-ulmin, a hydrophobin secreted by the causal agents of Dutch elm disease, is a parasitic fitness factor. Fungal Genet. Biol. 22:39-53. [DOI] [PubMed] [Google Scholar]
- 24.Thomas, D. 1995. Nanotechnology's many disciplines. Bio/Technology 13:439-443. [Google Scholar]
- 25.van der Mei, H. C., M. Rosenberg, and H. J. Busscher. 1991. Assessment of microbial cell surface hydrophobicity, p. 261-287. In N. Mozes, P. S. Handly, H. J. Busscher, and P. G. Rouxhet (ed.), Microbial cell surface analysis. VCH Publishers, New York, N.Y.
- 26.van der Vegt, W., H. C. van der Mei, H. A. B. Wösten, J. G. H. Wessels, and H. J. Busscher. 1996. A comparison of the surface activity of the fungal hydrophobin SC3p with those of other proteins. Biophys. Chem. 57:253-260. [DOI] [PubMed] [Google Scholar]
- 27.van Wetter, M.-A., H. A. B. Wösten, and J. G. H. Wessels. 2000. SC3 and SC4 hydrophobins have distinct roles in formation of aerial structures in dikaryons of Schizophyllum commune. Mol. Microbiol. 36:201-210. [DOI] [PubMed] [Google Scholar]
- 28.van Wetter, M.-A., F. H. J. Schuren, T. A. Schuurs, and J. G. H. Wessels. 1996. Targeted mutation of the SC3 hydrophobin gene of Schizophyllum commune affects formation of aerial hyphae. FEMS Microbiol. Lett. 140:265-2691. [Google Scholar]
- 29.Wessels, J. G. H. 1993. Cell wall growth, protein excretion and morphogenesis in fungi. Transley review no. 45. New Phytol. 123:397-413. [DOI] [PubMed] [Google Scholar]
- 30.Wessels, J. G. H. 1994. Developmental regulation of fungal cell wall formation. Annu. Rev. Phytopathol. 32:413-437. [Google Scholar]
- 31.Wessels, J. G. H. 1996. Fungal hydrophobins: proteins that function at an interface. Trends Plant Sci. 1:9-15. [Google Scholar]
- 32.Wessels, J. G. H. 1997. Hydrophobins: proteins that change the nature of a fungal surface. Adv. Microb. Physiol. 38:1-45. [DOI] [PubMed] [Google Scholar]
- 33.Wessels, J. G. H., S. A. Ásgeirsdóttir, K. U. Birkenkamp, O. M. H. de Vries, L. G. Lugones, J. M. J. Scheer, F. H. J. Schuren, T. A. Schuurs, M.-A. van Wetter, and H. A. B. Wösten. 1995. Genetic regulation of emergent growth in Schizophyllum commune. Can. J. Bot. 73:S273-S281.
- 34.Wösten, H. A. B., and J. G. H. Wessels. 1997. Hydrophobins, from molecular structure to multiple functions in fungal development. Mycoscience 38:363-374. [Google Scholar]
- 35.Wösten, H. A. B., and M. L. de Vocht. 2000. Hydrophobins: the fungal coat unravelled. Biochim. Biophys. Acta 1469:79-86. [DOI] [PubMed] [Google Scholar]
- 36.Wösten, H. A. B., O. M. H. de Vries, and J. G. H. Wessels. 1993. Interfacial self-assembly of a fungal hydrophobin into a hydrophobic rodlet layer. Plant Cell 5:1567-1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wösten, H. A. B., F. H. J. Schuren, and J. G. H. Wessels. 1994. Interfacial self-assembly of a hydrophobin into an amphipathic membrane mediates fungal attachment to hydrophobic surfaces. EMBO J. 13:5848-5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wösten, H. A. B., S. A. Ásgeirsdóttir, J. H. Krook, J. H. H. Drenth, and J. G. H. Wessels. 1994. The SC3p hydrophobin self-assembles at the surface of aerial hyphae as a protein membrane constituting the hydrophobic rodlet layer. Eur. J. Cell Biol. 63:122-129. [PubMed] [Google Scholar]
- 39.Wösten, H. A. B., O. M. H. de Vries, H. C. van der Mei, H. J. Busscher, and J. G. H. Wessels. 1994. Atomic composition of the hydrophobic and hydrophilic sides of self-assembled SC3P hydrophobin. J. Bacteriol. 176:7085-7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wösten, H. A. B., T. G. Ruardy, H. C. van der Mei, H. J. Busscher, and J. G. H. Wessels. 1995. Interfacial self-assembly of a Schizophyllum commune hydrophobin into an insoluble amphipathic protein membrane depends on surface hydrophobicity. Colloids Surf. B Biointerf. 5:189-195. [Google Scholar]
- 41.Wösten, H. A. B., M.-A. van Wetter, L. G. Lugones, H. C. van der Mei, H. J. Busscher, and J. G. H. Wessels. 1999. How a fungus escapes the water to grow into the air. Curr. Biol. 9:85-88. [DOI] [PubMed] [Google Scholar]