Abstract
A combination of fluorescence in situ hybridization, microprofiles, denaturing gradient gel electrophoresis of PCR-amplified 16S ribosomal DNA fragments, and 16S rRNA gene cloning analysis was applied to investigate successional development of sulfate-reducing bacteria (SRB) community structure and in situ sulfide production activity within a biofilm growing under microaerophilic conditions (dissolved oxygen concentration in the bulk liquid was in the range of 0 to 100 μM) and in the presence of nitrate. Microelectrode measurements showed that oxygen penetrated 200 μm from the surface during all stages of biofilm development. The first sulfide production of 0.32 μmol of H2S m−2 s−1 was detected below ca. 500 μm in the 3rd week and then gradually increased to 0.70 μmol H2S m−2 s−1 in the 8th week. The most active sulfide production zone moved upward to the oxic-anoxic interface and intensified with time. This result coincided with an increase in SRB populations in the surface layer of the biofilm. The numbers of the probe SRB385- and 660-hybridized SRB populations significantly increased to 7.9 × 109 cells cm−3 and 3.6 × 109 cells cm−3, respectively, in the surface 400 μm during an 8-week cultivation, while those populations were relatively unchanged in the deeper part of the biofilm, probably due to substrate transport limitation. Based on 16S rRNA gene cloning analysis data, clone sequences that related to Desulfomicrobium hypogeium (99% sequence similarity) and Desulfobulbus elongatus (95% sequence similarity) were most frequently found. Different molecular analyses confirmed that Desulfobulbus, Desulfovibrio, and Desulfomicrobium were found to be the numerically important members of SRB in this wastewater biofilm.
While in marine sediments the dominating role of sulfate reduction in the anaerobic mineralization of organic matter is well established (17, 18, 19), its role in freshwater habitats is considered to be less important due to low sulfate concentrations (16, 45). The importance of sulfate reduction in wastewater biofilms has lately attracted considerable attention (23, 35, 36, 38, 44). Successive vertical zonations of predominant respiratory processes occur simultaneously in close proximity in wastewater biofilms with a typical thickness of only a few millimeters, and sulfate reduction can be found in the deeper anaerobic biofilm strata depending on the oxygen penetration depth (23, 36, 38, 44). Typical domestic wastewater contains high sulfate concentrations (ca. 100 to 1,000 μM) relative to organic carbon concentrations, but almost no nitrate or nitrate. Thus, sulfate reduction could be the dominant terminal electron accepting process and may account for up to 50% of the mineralization of organic matter in some wastewater treatment biofilms (23, 35). Accordingly, internal sulfide reoxidation in the biofilms accounted for a substantial part of oxygen consumption (23, 36, 38, 44). A major drawback of sulfate reduction in wastewater treatment systems is the production of toxic H2S, which is also a potential precursor of odorants and is a major cause of extensive microbially mediated corrosion of concrete sewage pipes (31) and wastewater treatment facilities (15, 37).
Although sulfate-reducing bacteria (SRB) play such crucial roles in organic matter mineralization and biocorrosion in wastewater treatment systems, the microbial diversity and population dynamics of SRB at the genus level in wastewater biofilms in relation to in situ sulfate reducing activity remains mostly unknown. This is partly due to the limitations in methodology and the presence of the complex internal sulfur cycle in the biofilm. Mass balance of sulfide and/or sulfate flux across a biofilm-liquid interface cannot describe sulfur transformations occurring within the biofilm. Furthermore, the versatile metabolic ability of SRB leads to more-complicated questions. Although sulfate reduction is generally considered to be an anaerobic process, the abundance and metabolic activity of SRB in oxic zones were frequently determined to be higher than those in neighboring anoxic zones in wastewater treatment biofilms (36, 44), microbial mats (5, 21, 29, 30, 41), oligotrophic lake sediment (45), and marine sediments (19). However, none of the SRB isolated so far can either grow aerobically or reduce sulfate under high oxygen concentrations. To accurately elucidate whether sulfate reduction indeed occurs at high rates in the oxic zones, the in situ detection of sulfate-reducing activity and SRB populations with the needed spatial and temporal resolution is essential.
A few studies relating SRB community structure to SRB activity in wastewater biofilms (36, 38, 44) and upflow anaerobic sludge blanket granules (43) were conducted by combining microelectrode measurements and molecular approaches. However, these studies were performed with well-established and mature biofilms or granules. Therefore, in the present study, the questions were addressed as to which SRB subgroups are numerically important and how these SRB are able to develop their populations and sulfate-reducing activity, in time and space, in mixed-population wastewater biofilms growing under microaerophilic conditions. Successional development of SRB populations in a developing biofilm was analyzed by fluorescence in situ hybridization (FISH) and denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S ribosomal DNA (rDNA) fragments and was directly related to development of in situ sulfide production activity as determined by microelectrodes. Furthermore, phylogenetic diversity of numerically important SRB subgroups, Desulfobulbus and Desulfovibrionaceae, was investigated by 16S rRNA gene cloning analysis.
MATERIALS AND METHODS
Biofilm samples.
Microaerophilic mixed-population biofilms were grown in fully submerged rotating disk reactors consisting of 10 poly-methyl-methacrylate disks as previously described (36). Eight removable slides (1 by 6 cm) were installed in each disk for sampling biofilms. The dilution rate in the reactors was kept at 0.2 h−1. Disk rotational speed was fixed at 14 rpm. An average dissolved organic carbon (DOC) loading rate was 11.2 g of DOC m−2 of biofilm h−1. The primary settling tank effluent from Soseigawa municipal wastewater treatment plant in Sapporo, Japan, was fed into the reactor. To facilitate sulfide denitrification, KNO3 solution was supplemented into the influent to give a final concentration of 350 μM NO3−. Since the bulk water was not aerated, average dissolved oxygen concentration in the bulk water was 40 ± 30 μM during the experiment.
Biofilm fixation and cryosectioning.
Biofilm samples were taken at regular time intervals during an 8-week cultivation. Biofilm samples were fixed by incubation in paraformaldehyde (4% [wt/vol] in phosphate-buffered saline) at 4°C for 8 h and subsequently washed twice in phosphate-buffered saline for in situ hybridization (2). After fixation, biofilm samples were embedded in Tissue-Tek OCT compound (Miles, Elkhart, Ind.) and frozen at −20°C. Vertical thin sections (20 μm thick) of the fixed samples were prepared as described by Ramsing et al. (38). Vertical thin slices were fixed on gelatin-coated microscope slides, air dried, and dehydrated in a series of increasing concentrations of ethanol (50, 80, and 96% [vol/vol]).
Oligonucleotide probes.
The following 16S rRNA-targeted oligonucleotide probes were used: (i) EUB338 (3), (ii) NON338 as a negative control (25), (iii) SRB385 (4), and (iv) five group-specific probes (SRB687, 660, 129, 804, and 221) (12). All probe sequences, their specificities, hybridization conditions, and references are given in Table 1. All probes were synthesized and labeled with tetramethylrhodamine-5-isothiocyanate (TRITC) or fluorescein isothiocyanate at the 5′ end by TaKaRa Shuzou Co., Ltd. (Shiga, Japan).
TABLE 1.
16S rRNA-targeted oligonucleotide probes used in this study
| Probe | Specificity | Sequence of probe (5′-3′) | Target sitea | FAb concn (%) | NaClc concn (mM) | Reference |
|---|---|---|---|---|---|---|
| EUB338 | Domain Bacteria | GCTGCCTCCCGTAGGAGT | 338-355 | 20 | 0.166 | 3 |
| NON338 | None (negative control) | ACTCCTACGGGAGGCAGC | 338-355 | 0 | 0.900 | 25 |
| SRB385 | SRB of the δ subdivision of the proteobacteria plus several gram-positive bacteria (e.g., Clostridium spp.) | CGGCGTCGCTGCGTCAGG | 385-402 | 30 | 0.071 | 4 |
| SRB687 | Desulfovibrio spp. plus members of the genera Geobacter, Desulfomonas, Desulfuromonas, Desulfomicrobium, Bilophila, and Pelobacter | TACGGATTTCACTCCT | 687-702 | 10 | 0.386 | 12 |
| 660 | Desulfobulbus spp. | GAATTCCACTTTCCCCTCTG | 660-679 | 30 | 0.071 | 12 |
| 221 | Desulfobacterium spp. | TGCGCGGACTCATCTTCAAA | 221-240 | 10 | 0.386 | 12 |
| 129 | Desulfobacter spp. | CAGGCTTGAAGGCAGATT | 129-146 | 20 | 0.166 | 12 |
| 804 | Desulfobacterium spp., Desulfobacter, Desulfococcus, Desulfosarcina, and Desulfobotulus spp. | CAACGTTTACTGCGTGGA | 804-821 | 10 | 0.386 | 12 |
16S rRNA position according to E. coli numbering.
Formamide concentration in the hybridization buffer.
sodium chloride concentration in the washing buffer.
In situ hybridization.
All in situ hybridizations were performed according to the procedure described by Amann (2) in 8 μl of hybridization buffer (0.9 M NaCl, 20 mM Tris hydrochloride [pH = 7.2], 0.01% sodium dodecyl sulfate [SDS]) and variable formamide concentrations as described in Table 1 with 1 μl of probe solution at 46°C for 2 h in an equilibrated sealed moisture chamber. The final probe concentration was approximately 5 ng μl−1. Subsequently, a stringent washing step was performed at 48°C for 20 min in 50 ml of prewarmed washing solution (variable NaCl concentration as described in Table 1, 20 mM Tris hydrochloride [pH = 7.2], 0.01% SDS). Washing buffer was removed by rinsing the slides with double-distilled H2O. Some biofilm samples were simultaneously stained with 4′6′-diamidino-2-phenylindole (DAPI) (1 μg ml−1) for 10 min in the dark. The slides were then rinsed briefly with double-distilled H2O, allowed to air dry, and mounted in antifading solution (Slow fade light; Molecular Probe Co. Ltd., Eugene, Oreg.).
Vertical distribution of SRB.
Since no clear in situ hybridization was observed when probes of SRB687, 221, 129, and 804 were used, we used SRB385, SRB660, and NON338 for the following FISH cell counting. To quantify changes in the vertical distributions of SRB populations in the biofilm, we directly counted positively probe-hybridized cells along five vertical transects through the biofilm for each slide as described previously (36, 38). Three randomly chosen slides were prepared for each probe. The counts were recalculated to absolute cell density (unit: cells per centimeter3 of biofilm) from the scan frame area and the scan depth, and average cell densities were obtained. The specimens were examined with an LSM 510 confocal laser scanning microscope (Carl Zeiss) equipped with an argon laser (488 nm), a HeNe laser (543 nm), and a UV laser (364 nm). Images were recorded by using simultaneous excitation of 488- and 543-nm lasers to distinguish the probe-stained cells from other debris and minerals (36). All image combining, processing, and analysis were performed with the standard software package provided by Zeiss. Processed images were printed out using the software package Adobe Photoshop (version 3.0J; Adobe Systems Incorporated, Mountain View, Calif.).
Microelectrode measurements.
Clark-type O2 microelectrode (39), H2S microelectrode (40), and liquid ion-exchanging membrane microelectrodes for pH and NO3− (11) were prepared and calibrated as previously described. The tip diameters of the O2 and H2S microelectrodes were about 15 μm. The tip diameters of the liquid ion-exchanging membrane microelectrodes were about 5 to 10 μm. Concentration profiles in the biofilm were measured using a motor-driven micromanipulator (ACV-104-HP; Chuo seiki, Tokyo, Japan) in intervals of 25 to 100 μm from the bulk liquid into the biofilm. The biofilm-liquid interface was determined using a dissection microscope (Stemi 2000; Carl Zeiss). The concentration of total dissolved sulfide (H2S, HS−, and S2−) was determined by a spectrophotometric method (6). Since the pH profiles showed a significant variation (>0.1 pH unit) throughout the biofilm, pH correction of the measured sulfide profiles was performed to obtain total sulfide concentration profiles (23). The dissociation constants for sulfide used were a pK1 of 7.05 and a pK2 of 17.1 (28). All measurements were performed as described previously in a flow cell reactor at 20°C, with an average liquid velocity of 2 to 3 cm s−1, by blowing N2 gas on the liquid surface (36). The concentrations of O2, NO3−, and NH4+ and the pH and DOC of the medium used for microprofile measurements were adjusted to the rotating disk reactor conditions (36). The medium contained the following (micromolar concentrations shown in parentheses): NaNO3 (70 to 320), MgSO4 · 7H2O (300), sodium propionate (600), NH4Cl (600), Na2HPO4 (570), MgCl2 · 6H2O (84), CaCl2 (200), and EDTA-2Na (270).
Based on the oxygen and sulfide profiles measured, diffusive fluxes of oxygen into the biofilm (net oxygen respiration rate) and sulfide from the anaerobic zone (net sulfate reduction rate [SRR]) were calculated by using Fick's first law, J = −D × (dC/dx), where J is flux (in micromoles meter−2 second−1), D is the diffusion coefficient (in meters2 second−1), and dC/dx is the concentration gradient (in micromoles meter−3 meter−1) which was determined at the steepest gradient of oxygen and sulfide concentration profiles obtained by the microelectrode. Values of 2.12 × 10−5 cm2 s−1 and 1.39 × 10−5 cm2 s−1 were used for the diffusion coefficients of oxygen and total sulfide at 20°C (23, 34), respectively.
Chemical analyses.
Influent and effluent water samples were collected at regular time intervals. The concentrations of NO3−, NO2−, SO42−, elemental sulfur (S0), acid-volatile sulfides (AVS) (S2−, HS−, H2S, and FeS), and chromium-reducible sulfide (CRS) (FeS2), were analyzed. The concentrations of NO3−, NO2−, and SO42− were determined by ion chromatography (model DX-100 with an AS4A column; Nippon DIONEX, Osaka, Japan) after prefiltering with 0.45-μm-pore-size membrane filter. S0 was extracted in 96% ethanol and analyzed by high-performance liquid chromatography using a UV detector. After distillation from the remaining samples from ethanol extraction in 2 N HCl, the acid-volatile sulfide was measured colorimetrically by the methylene blue method (6). After the AVS distillation, the remaining FeS2 was reduced in CrCl2 and determined as sulfide. The details of this method have been previously described (36).
Nucleic acid extraction.
Approximately, 1 g of each wet biofilm sample was mixed with 0.5 ml of AE buffer (20 mM sodium acetate [pH 5.5], 1 mM EDTA) in sterile 1.5-ml tubes. After mechanical disruption (1 min at the maximum speed with 1.0-mm-diameter glass beads) by a Mini Bead Beater (Biospec Products, Bartlesville, Okla.), total bacterial DNA was extracted from biofilm samples by a combined freeze-thaw (three cycles of freezing in liquid nitrogen and thawing at 37°C), 1% (wt/vol) SDS treatment, and hot phenol-chloroform-isoamyl alcohol treatment (47).
PCR amplification of 16S rDNA fragments.
The 16S rDNA from the mixed bacterial DNA was amplified by PCR as described by Muyzer et al. (32, 33). For DGGE analysis, a primer set of primer SRB385 (4) with a GC-clamp (33) and the universal primer 907R (33) was used. For comparative 16S rDNA cloning analysis of Desulfovibrionaceae and Desulfobulbus family-related groups, selective primer sets of DSV230f and DSV838r and DBB121f and DBB1237r were used, respectively, as described by Daly et al. (8). To minimize nonspecific annealing of the primers to nontarget DNA, a hot-start PCR program was used for all amplification (32). The PCR products were evaluated on a 1% (wt/vol) agarose gel.
DGGE analysis of PCR fragments.
DGGE was performed with D-Gene system (Bio-Rad) and the following ingredients and conditions: 1× TAE (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA at pH 8.3), 1.0-mm-thick gels, and a denaturant gradient containing 35 to 65% urea-formamide at 60°C, with 100 V constantly for 17 h version. DGGE gels were stained in an ethidium bromide solution (0.5 μg ml−1) and documented on a UV transilluminator (302 nm) with a Polaroid camera. Photos were scanned, and inverse images were prepared with Adobe Photoshop 3.0J. Some DGGE bands were carefully excised for DNA isolation and reamplification as described previously (14). The PCR products were directly sequenced for phylogenetic analysis as described below.
Cloning, sequencing, and phylogenetic analysis.
Three parallel PCR products were combined and purified with the WIZARD PCR Preps DNA purification systems (Promega, Tokyo, Japan). One microliter of the amplified bacterial 16S rDNA fragments was directly ligated into the pGEM-T vector cloning system (Promega) and transformed into competent cells (high-efficiency Escherichia coli JM109; Promega) as described in the manufacturer's instruction. Plasmids were extracted and purified from clones with the Wizard Plus Minipreps DNA purification system (Promega) in accordance with the manufacturer's instructions. Partial sequencing (575 bases for the Desulfovibrionaceae clone library and 1,093 bases for the Desulfobulbus clone library) of the 16S rDNA inserts was performed with an automatic sequencer (ABI Prism 310 Genetic Analyzer; Applied Biosystems). To avoid redundant sequencing, PCR-amplified rDNA fragments of all clones were placed into categories by restriction fragment length polymorphism analysis after digestion with restriction enzymes of RsaI and MspI (8), respectively, as described in the manufacturer's instruction. The PCR fragments digested were loaded on a 2.0% (wt/vol) agarose gel. Identical fragment migration patterns were defined as identical recombinants, and one representative of each group of recombinants was selected for comparative sequence analysis. All sequences were checked for chimeric artifacts by the CHECK_CHIMERA program in the Ribosomal Database Project (24) and compared with similar sequences of the reference organisms by BLAST search (1). Sequence data were aligned with the CLUSTAL W package (48). Phylogenetic trees were constructed by the neighbor-joining method (42). Tree topology was tested by maximum-parsimony method. Bootstrap resampling analysis for 100 replicates was performed.
Nucleotide sequence accession numbers.
The GenBank/EMBL/DDBJ accession numbers for the 16S rDNA sequences of three DGGE bands and seven clones obtained in this study are AB069765 to AB069774.
RESULTS
Reactor performance.
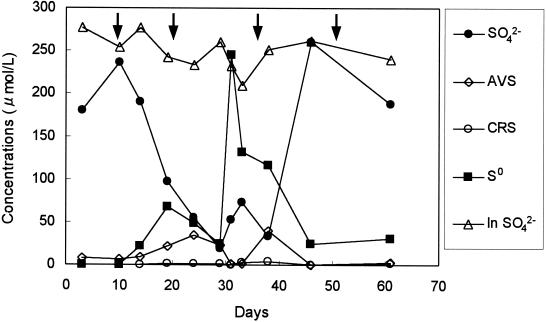
Water quality of the reactor effluent is presented in Fig. 1. The concentration of SO42− started decreasing after 10 days, reached about 50 μM SO42−, and then increased to the influent concentration (ca. 250 μM SO42−) again after 6 weeks. On the other hand, the change in S0 concentration revealed an inverse image of the SO42− concentration change. A maximum concentration of ca. 250 μM S0 was observed in the 5th week (whitish colloidal materials were observed in the bulk liquid during this period), and then the S0 concentration decreased to about 30 μM. This suggested that SO42− was initially reduced to H2S which could be chemically and/or biologically reoxidized to S0, resulting in the accumulation of S0. The accumulated S0 was further oxidized to SO42− after 5 weeks, and the oxidation of S0 was completed after 6 weeks. Thus, sulfur transformation in the reactor could not be seen from the mass balance of SO42− and sulfide in the reactor effluent after 6 weeks. Concentrations of other reduced sulfur compounds (i.e., AVS and CRS) were very low during the experiment. NO3− fed into the reactor was completely utilized within a week (data not shown). The bulk O2 concentration was in the range of 20 to 60 μM during the entire experiment (data not shown).
FIG. 1.
Time courses of water quality (SO42−, S0, AVS, and CRS) of the reactor effluent during start-up. Only SO42− concentration in the reactor influent (In SO42−) was presented because the concentrations of other reduced sulfur compounds were under the detection limit. Arrows indicate biofilm samplings.
Development of sulfate-reducing activity.
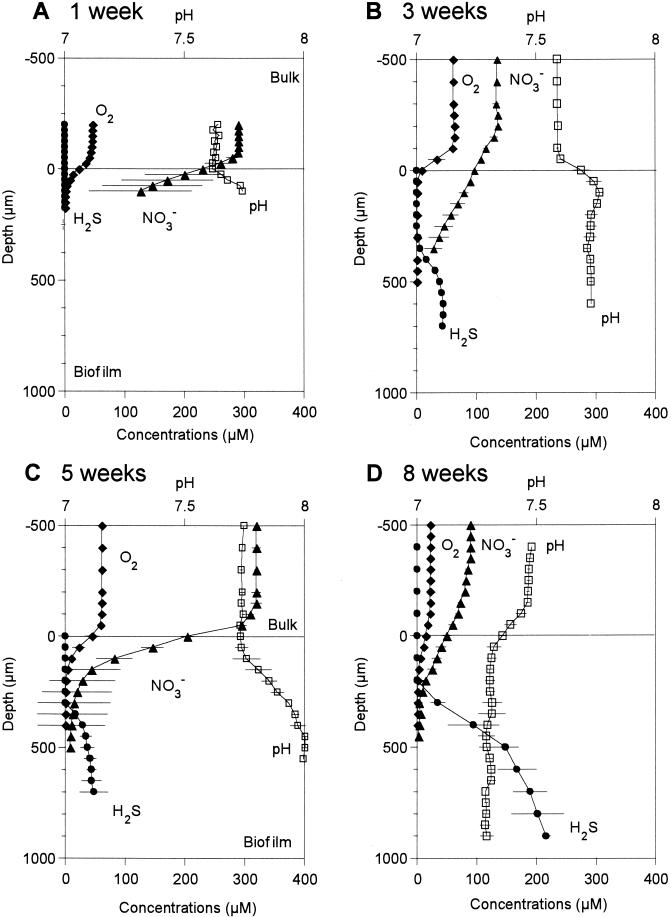
Oxygen, total sulfide, NO3−, and pH profiles were measured at different developmental stages of SRB populations in a wastewater biofilm. Figure 2 shows the mean values of three to five profiles measured at different positions. The biofilm thickness increased from ca. 400 μm in the first week to ca. 1,400 μm in the 5th week and thereafter remained more or less constant. In the first week, oxygen penetrated only 200 μm from the surface, below which anoxia was already developed (Fig. 2A). However, no sulfide was detected yet. This was partly because NO3− fully penetrated through the biofilm. In the 3rd week, O2 was depleted at the surface of the biofilm, but NO3− penetrated ca. 500 μm from the surface. A total sulfide concentration of 45 μM was first detected below ca. 500 μm (Fig. 2B). Thus, O2 and total sulfide profiles were vertically separated more than 200 μm. In the 5th week, the NO3− profile rapidly decreased within 500 μm even though the bulk concentration was 320 μM (Fig. 2C). Total sulfide concentration profile slightly increased to 61 μM and shifted toward the surface, but no net sulfide production was observed below 500 μm as indicated by a constant H2S profile. This is probably due to a carbon limitation caused by competition with denitrifying bacteria. To confirm this, microelectrode measurements were performed without addition of NO3−. Sulfide production significantly increased in the absence of NO3− (data not shown). Total sulfide concentration increased to 216 μM, and the sulfide production zone moved toward the chemocline in the 8th week (Fig. 2D). The pH profile showed a sharp increase just below the surface in the 1st, 3rd, and 5th week. However, pH decreased in the surface zone in the 8th week due to mainly anaerobic oxidation of sulfide.
FIG. 2.
Oxygen, total sulfide, nitrate, and pH microprofiles measured at different developmental stages of wastewater biofilms. The film surface is at a depth of 0 μm. Error bars indicate the standard deviations of measurements.
Development of SRB community structure.
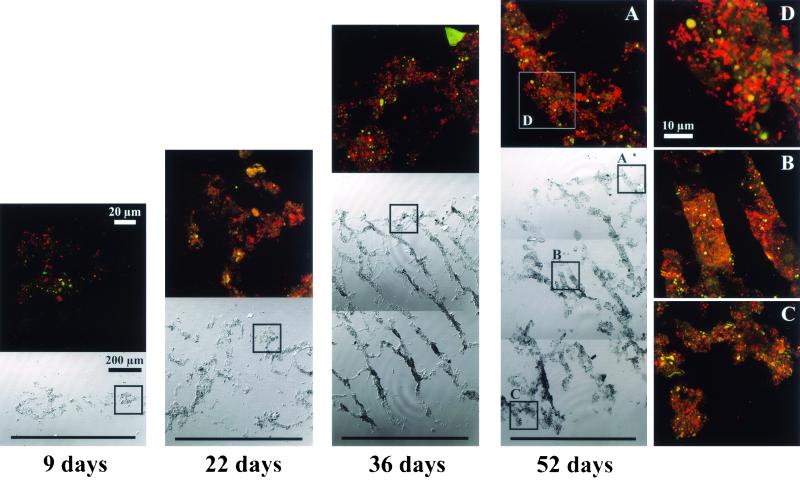
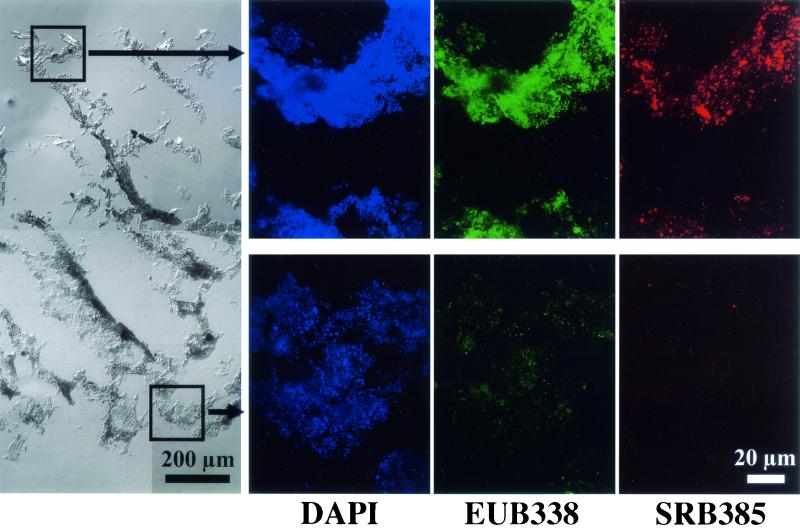
Figure 3 represents a series of composite biofilm cross-sectional differential interference contrast (DIC) images to display successional development of the entire biofilm structure and SRB populations in situ. It is clear that heterogeneous biofilm structure consisting of discrete biomass and interstitial voids was developed with time. The abundance of probe SRB385-hybridized cells was very low at the initial stage and gradually increased with time. The abundance and fluorescent signals of probe SRB385-hybridized cells were higher in the surface than in the deeper part of the biofilm (see the 52-day-old biofilm). The probe SRB385-hybridized cells were present in the forms from single scattered cells to clustered cells. When the biofilm was stained with bacterial probe EUB338 and DAPI, a clear vertical gradient of fluorescent intensity was observed throughout the biofilm (Fig. 4). This indicates that the higher abundance of more-active bacteria was present in the surface part than in the deeper part of the biofilm.
FIG. 3.
Composite DIC images of entire biofilm vertical sections (20 μm thick) and confocal laser scanning microscope projection images of the microscopic fields enclosed by boxes after in situ hybridization with TRITC-labeled SRB385 probe displaying time-dependent development of the entire biofilm structure and the vertical distributions of SRB385-hybridized cells in the biofilm. The biofilm surface is at the top of panel.
FIG. 4.
A composite DIC image of an entire biofilm vertical section and CSLM projection images of the microscopic fields enclosed by boxes after in situ hybridization with TRITC-labeled SRB 385 probe and fluorescein isothiocyanate-labeled EUB338 and staining with DAPI displaying the vertical distributions of total cells, and general bacteria, and SRB385-hybridized cells in the biofilm. The biofilm sample was taken in the 8th week. The biofilm surface is at the top of each panel.
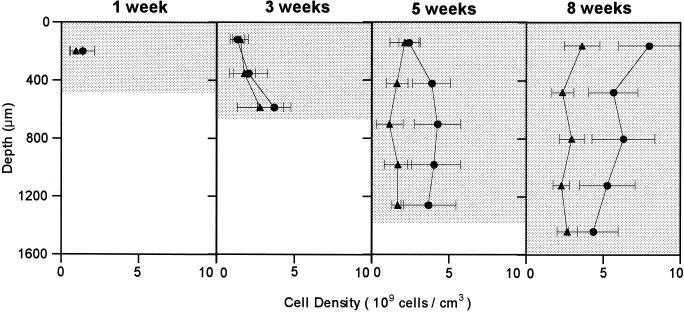
To quantify the development of SRB populations in the biofilm, positively hybridized cells were directly counted at each developmental stage. Figure 5 presents changes in the vertical distributions of probe SRB385-hybridized cells and probe 660-hybridized Desulfobulbus cells in the biofilm during the 8-week cultivation. The number of the probe SRB385-hybridized cells in the surface of the biofilm increased by a factor of 6 (from 1.3 × 109 to 7.9 × 109 cells/cm3 of biofilm) for 8 weeks, whereas the numbers in the deeper part of the biofilm were relatively unchanged. Similarly, the number of the probe 660-hybridized cells increased from 0.9 × 109 to 3.6 × 109 cells cm−3 in the surface zone. The counts of the probe 660-hybridized Desulfobulbus cells accounted for 30 to 65% of the counts of the probe SRB385-hybridized cells.
FIG. 5.
Time-dependent development of the vertical distributions of the probe SRB385-hybridized cells (•) and probe 660-hybridized Desulfobulbus cells (▴) in the biofilm during the 8-week cultivation. The biofilm surface is at a depth of 0 μm. Error bars indicate the standard deviations of measurements.
DGGE analysis of PCR-amplified rDNA fragments.
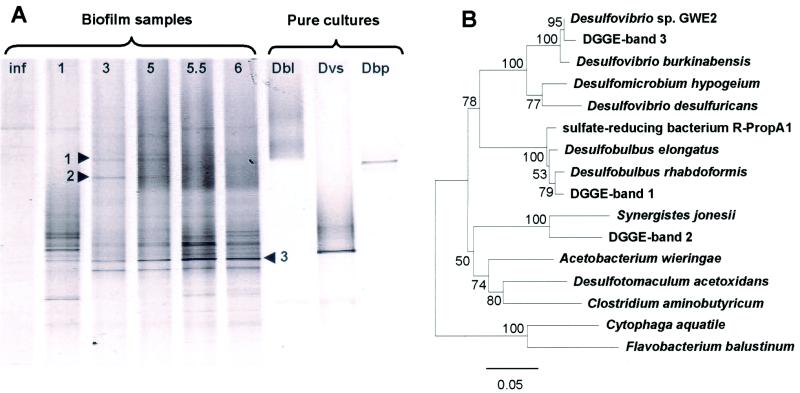
To determine time-dependent development of SRB community structure within the biofilm, DGGE of 16S rRNA gene fragments after PCR amplification with a primer set of SRB385-GC and 907R was performed (Fig. 6A). Most of bands in the DGGE profile remained in all stages of development, and some appeared during the biofilm development. The microbial community changed within the first 3 weeks. Five bands were excised from the DGGE gel, from which three were successfully reamplified and sequenced (numbers are indicated in Fig. 6A). The phylogenetic affiliations of these sequences are depicted in Fig. 6B. The sequence of band 3, whose intensity gradually increased with time, was closely related to Desulfovibrio sp. strain GWE2 with 98% similarity. The sequence of band 1 was related to Desulfobulbus rhabdoformis (95% similarity). The sequence of band 2 resembled an as-yet-undescribed eubacterium clone with 96% similarity.
FIG. 6.
(A) A DGGE profile of PCR-amplified 16S rRNA gene fragments obtained with a primer set of SRB385f-GC and 907R and bacterial genomic DNA extracted from a wastewater biofilm at different developmental stages. Lanes: (1) Inf, influent wastewater; (2) 1-week-, (3) 3-week-, (4) 5-week-, (5) 5.5-week-, (6) 6 week-old biofilm; (7) Dbl: Desulfobacter latus; (8) Dvs: Desulfovibrio salexigens, and (9) Dbp: Desulfobulbus propionicus. The numbers next to the arrows refer to the numbers of excised and successfully sequenced bands. (B) Phylogenetic analysis of the partial 16S rDNA sequences (461 bp) derived from the excised DGGE bands depicted in Fig. 6A. The trees were calculated by neighbor-joining algorithm and a 50% conservation filter. The scale bar denotes 5% sequence divergence, and the values at the nodes represent bootstrap values (resampling analysis performed 100 times).
16S rRNA clone analysis.
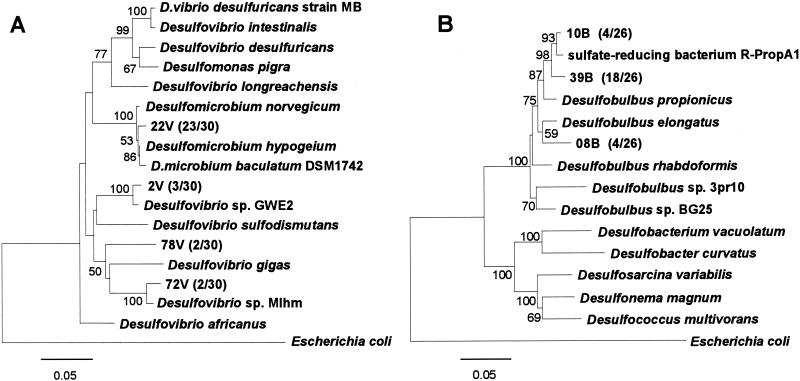
All excised DGGE bands did not yield usable sequence data, and thus the SRB community structure in this biofilm was not completely revealed from the DGGE analysis. Thus, two 16S rDNA clone libraries were constructed from the 52-day-old biofilm sample with the specific primer sets for Desulfovibrio and for Desulfobulbus to further identify the probe SRB385-hybridized Desulfovibrio and the probe 660-hybridized Desulfobulbus, respectively. Thirty and twenty-six clones were randomly selected from the Desulfovibrionaceae and Desulfobulbus clone libraries, respectively. Among the clones analyzed, four phylogenetically distinct sequences were found in the Desulfovibrionaceae clone library (Fig. 7A). Twenty-three clone sequences out of thirty (represented by clone 22V) were closely related to Desulfomicrobium hypogeium (99% similarity). The other three clones of 2V, 78V, and 72V were affiliated with the Desulfovibrio cluster. The clone 72V was most closely related to an as-yet-undescribed SRB clone R-LacA1 with 99% similarity. The clones 2V and 78V were related to Desulfovibrio sp. strain GWE2 with 98% similarity and to Desulfovibrio sp. strain Mlhm with 91% similarity, respectively. The clone sequences of 22V and 72V contained one mismatch with probe SRB385.
FIG. 7.
Phylogenetic distance trees of members of Desulfovibrionaceae family (A) and Desulfobulbus family (B) based on comparative analysis of partial sequences (575 bp for the Desulfovibrionaceae clone library and 1,093 bp for the Desulfobulbus clone library, respectively) of 16S rRNA gene sequences retrieved from a 52-day-old wastewater biofilm. The trees were calculated by neighbor-joining algorithm and a 50% conservation filter. Numbers in parentheses indicate the frequencies of appearance of the completely identical sequences (clones) in the total clones. The scale bar denotes 5% sequence divergence, and the values at the nodes represent bootstrap values (100 times resampling analysis). E. coli is used as the outgroup.
On the other hand, three different representative clone sequences were found in the Desulfobulbus clone libraries (Fig. 7B). The clones 10B and 39B share strong sequence similarities with Desulfobulbus propionicus lineage and are most closely related to an as-yet-undescribed SRB clone R-ProPA1 with 98 and 97% similarities, respectively. A clone 08B was related to Desulfobulbus elongatus and Desulfobulbus rhabdoformis with 95% similarity. These three clones (10B, 39B, and 08B) contained no mismatch with probe SRB385, but the clone 08B had one mismatch with probe 660.
DISCUSSION
Integration of molecular and microsensor approaches.
We have shown the successional development of SRB community structure within a wastewater biofilm growing under microaerophilic conditions, which was monitored by FISH, PCR-amplified DGGE analysis, and comparative 16S rDNA phylogenetic analysis (structural analyses). The development of SRB community structure corresponded well with the development of in situ sulfide production activity determined by microelectrodes (functional analyses). However, these molecular techniques have some inherent biases and limitations, such as DNA extraction efficiency for different SRB species, preferential PCR amplification, specificity of oligonucleotide probes, and so on. These insufficiencies of molecular techniques suggest the importance of combining different molecular approaches and relating them with activity measurements. Therefore, the data sets resulting from different approaches have to be carefully integrated to reveal a valid overall picture of development of SRB community structure and sulfate-reducing activity.
Development of in situ sulfate-reducing activity.
Although an anoxic zone was already developed in the 1st week, sulfide was not detected. A sulfide production of 0.32 μmol of H2S m−2 s−1 was first detected after 3 weeks, and the rate increased gradually to 0.70 μmol of H2S m−2 s−1 in the 8th week. We did not perform microelectrode measurements in the 2nd week. It is more likely that sulfide production occurred in the second week, because sulfate concentration in the bulk liquid began to decrease after about 2 weeks as shown in Fig. 1. This evidence indicates that sulfidogenically inactive SRB could adapt to new biofilm environment within a week. The successional development of in situ sulfide production was coincident with the increase in probe SRB385- and 660-hybridized cells (Fig. 3 and 5).
The SRRs measured in this study were in the range of 0.32 to 0.70 μmol of H2S m−2 s−1 and similar to those reported by Kühl and Jorgensen (23) for a trickling filter biofilm and by Santegoeds et al. (44) for biofilms grown in an activated sludge aeration basin. Sulfide fluxes ranged from 30% in the 3rd week to 79% in the 5th week of the oxygen fluxes (0.76 to 1.08 μmol of O2 m−2 s−1). Since oxidation of 1 mol of sulfide to sulfate requires 2 mol of oxygen, electron acceptors other than oxygen (e.g., nitrate) were used for sulfide reoxidation. In this study, the biofilm was grown under microaerophilic conditions (the bulk oxygen concentration was in the range of 20 to 60 μM), and most of the oxygen was, therefore, consumed for the sulfide reoxidation.
The specific SRR per cell in this biofilm was calculated based on the measured H2S profiles and SRB cell counts by FISH. The calculated specific SRRs were on the order of 10−15 mol H2S cell−1 day−1, which is in the range of the previously reported values of pure cultures on H2, lactate, or pyruvate (17, 49). The in situ SRR determined by microelectrode measurement was in the range of 1.0 to 1.6 μmol of H2S cm−3 h−1, which is in the same range reported in previous microelectrode studies of other wastewater biofilm systems (23, 38, 44) but is lower than that in anaerobic sulfidogenic granules (3.6 to 21.6 μmol of H2S cm−3 h−1) (43). This rate is, however, about 3 orders of magnitude higher than those in marine sediments (ca. 0.1 to 4 nmol of H2S cm−3 h−1) due primarily to the higher SRB populations and influx of organic matter (17, 18, 19).
The SRR calculated from the H2S profiles was prone to associate with some errors. The H2S microelectrodes used in this study could not detect immediate reoxidation of sulfide by O2 or NO3− and immediate precipitation, e.g., as FeS, and are sensitive to oxygen, so that the H2S profile in the zone where O2 and H2S coexist may not be so reliable. In addition, the measured H2S concentration profiles are not profiles that actually occurred under growth conditions in the biofilm reactor, which limits an exact quantitative comparison of the in situ SRR with SRB growth rates in the biofilm as discussed below.
Development of SRB community structure.
Although the number of sequenced clones was too low to draw solid conclusions about SRB community structure, results of in situ hybridization, DGGE, and 16S rDNA clone analysis revealed that Desulfobulbus, Desulfovibrio, and Desulfomicrobium were numerically important SRB species in this wastewater biofilm community. Desulfobulbus and Desulfovibrio species have been found to be the numerically important SRB in wastewater biofilms grown under oxic conditions (36, 44), in activated sludge flocs (26, 46), in the aerobic layers of a stratified fjord (47), in oxic regions of microbial mats (21, 22), and in the permanently oxic region of a hypersaline microbial mat (29, 30). The reason for the higher abundance and activity of these SRB species in the oxic surface and near the chemocline is partly that these species were able to tolerate O2 (7, 27) and to utilize NO3− (9, 50) or even O2 (9, 13, 27) instead of SO42− as a terminal electron acceptor. Furthermore, these species are able to oxidize reduced sulfur compounds (e.g., sulfide and sulfite) with O2 and NO3− as the electron acceptor (9, 13) and, thus, catalyze all reactions of a complete sulfur cycle. Desulfovibrio oxyclinae isolated from a hypersaline microbial mat was capable of aerobic respiration by forming aggregates when subjected to oxygen (21). Thus, the central part of such aggregates could remain anaerobic even under microaerobic and periodically oxic conditions.
Vertical distributions of SRB populations and activity.
The microelectrode data revealed that the active sulfide production zone gradually shifted upward and intensified with time. This result coincided with the increase in the SRB populations near the biofilm surface (Fig. 3 and 5). The number of probe SRB385-hybridized cells in the surface of the biofilm increased by a factor of 6 (from 1.3 × 109 cells cm−3 of biofilm to 7.9 × 109 cells cm−3). Similarly, the number of probe 660-hybridized cells increased from 0.9 × 109 cell cm−3 of biofilm to 3.6 × 109 cells cm−3 in the surface zone. The numbers of both probe-hybridized SRB cells were relatively unchanged in the inner part of the biofilm. These results suggested that since oxygen depletes more rapidly in the surface layer of the biofilm, SRB are forced to proliferate near the oxic and anoxic interface where organic carbon and sulfate are more available. However, the spatial resolution of microelectrode measurements and FISH cell counting method may not be sufficient to exactly reveal that the SRB proliferated in the zones where oxygen was still present in this study. Simultaneous in situ hybridization of vertical sections of the biofilm with probe EUB338 and DAPI clearly revealed that the higher abundance of more-active bacteria including SRB was present in the surface part than in the deeper part of the biofilm due to substrate limitation (Fig. 4). It is most likely that inert matter and dormant bacterial cells were present in the deeper biofilm.
It is noted that the specificity of the SRB385 probe for in situ hybridization is still problematic. However, probe 660 is currently known to be specific for only Desulfobulbus spp., and the vertical profile of the probe 660-hybridized Desulfobulbus is, therefore, more reliable. The FISH counts were also associated with a relatively large standard deviation due to the heterogeneity of the biofilm, but the difference in cell numbers in the surface biofilms between the 1st week and the 8th week was statistically significant.
16S rRNA gene clone analysis revealed at least the presence of some Desulfovibrio species in the biofilm. However, no clear in situ whole-cell hybridization was observed with probe 687, which is reported to hybridize with Desulfovibrio spp. (12). Comparison of the target sequences of probe 687 with the clone sequences indicated that all clones retrieved except the clone 2 V have no mismatch with the probe 687. Thus, we speculated that the probe 687 may not be useful for in situ hybridization or that the number of the probe 687-hybridized Desulfovibrio cells were simply under the detection limit of FISH analysis. The in situ probing technique for detection of some SRB members is not absolute yet, and more development of new specific probes is needed.
Based on the changes in the numbers of SRB385- and 660-hybridized cells at different biofilm developmental stages (9, 22, 36, and 52 days), the net in situ growth rate of SRB cells in the surface zones of the biofilm was roughly estimated by using an equation of k = 0.693 g−1 (where g is the doubling time). The net in situ growth rates of 0.002 to 0.003 h−1 and 0.001 to 0.002 h−1 were determined for the SRB385- and 660-hybridized cells, respectively. These rates were more than 1 or 2 orders of magnitude lower than the maximum specific growth rates of pure cultures of Desulfovibrio and Desulfobulbus species (49, 50). This is probably because of substrate diffusion limitation, presence of nitrate which created a competition for organic matter with denitrifying bacteria, and oxygen inhibition. Furthermore, it is noted that these rates do not take attachment and detachment rates into account. Since the biofilm surface was subjected to the high shear force that resulted from the disk rotation (the disk rotational speed was 14 rpm, which is equivalent to a peripheral speed of ca. 14 cm s−1), the detachment effect on the growth rates might be more significant than anticipated. We also speculated that the numbers of SRB cells were still underestimated due to the presence of SRB species which cannot be detected by the probe SRB385 as demonstrated by 16S rDNA clone analysis (e.g., clones 22V and 72V). In addition, growth rate in the biofilm might decrease after cell attachment to a solid surface due to an increased maintenance coefficient (20) and exopolysaccharide production (10).
Acknowledgments
This research has been supported by the CREST (Core Research for Evolutional Science and Technology) of Japan Science and Technology Corporation (JST) and by Grant-in-Aid 13650593 for Developmental Scientific Research from the Ministry of Education, Science, and Culture of Japan.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R. I. 1995. In situ identification of micro-organisms by whole-cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkerman, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 3.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann, R., J. Stromley, R. Devereux, R. Key, and D. A. Stahl. 1992. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl. Environ. Microbiol. 58:614-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canfield, D. E., and D. J. Des Marais. 1991. Aerobic sulfate reduction in microbial mats. Science 251:1471-1473. [DOI] [PubMed] [Google Scholar]
- 6.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 7.Cypionka, H., F. Widdel, and N. Pfenning. 1985. Survival of sulfate-reducing bacteria after oxygen stress, and growth in sulfate-free oxygen-sulfide gradients. FEMS Microbiol. Ecol. 31:39-45. [DOI] [PubMed] [Google Scholar]
- 8.Daly, K., R. J. Sharp, and A. J. McCarthy. 2000. Development of oligonucleotide probes and PCR primers for detecting phylogenetic subgroups of sulfate-reducing bacteria. Microbiol. 146:1693-1705. [DOI] [PubMed] [Google Scholar]
- 9.Dannenberg, S., M. Kroder, W. Dilling, and H. Cypionka. 1992. Oxidation of H2, organic compounds and inorganic sulfur compounds coupled to reduction of O2 or nitrate by sulfate-reducing bacteria. Arch. Microbiol. 158:93-99. [Google Scholar]
- 10.Davies, D. G., A. M. Chakrabarty, and G. G. Geesey. 1993. Exopolysaccharide production in biofilm: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 59:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Beer, D., A. Schramm, C. M. Santegoeds, and M. Kühl. 1997. A nitrite microsensor for profiling environmental biofilms. Appl. Environ. Microbiol. 63:973-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Devereux, R., M. D. Kane, J. Winfrey, and D. A. Stahl. 1992. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst. Appl. Microbiol. 15:601-609. [Google Scholar]
- 13.Dilling, W., and H. Cypionka. 1990. Aerobic respiration in sulfate-reducing bacteria. FEMS Microbiol. Lett. 71:123-128. [Google Scholar]
- 14.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hamilton, W. A. 1985. Sulfate-reducing bacteria and anaerobic corrosion. Annu. Rev. Microbiol. 39:195-217. [DOI] [PubMed] [Google Scholar]
- 16.Ingvorsen, K., and T. D. Brock. 1982. Electron flow via sulfate reduction and methanogenesis in the anaerobic hypolimnion of Lake Mendota. Limnol. Oceanogr. 27:559-564. [Google Scholar]
- 17.Jorgensen, B. B. 1978. A comparison of methods for the quantification of bacterial sulfate reduction in coastal marine sediments. III. Estimation from chemical and bacteriological field data. Geomicrobiol. J. 1:49-64. [Google Scholar]
- 18.Jorgensen, B. B. 1982. Mineralization of organic matter in the sea bed: the role of sulphate reduction. Nature 296:643-645. [Google Scholar]
- 19.Jorgensen, B. B., and F. Bak. 1991. Pathways and microbiology of thiosulfate transformations and sulfate reduction in a marine sediment (Kattegat, Denmark). Appl. Environ. Microbiol. 57:847-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kieft, T. L., and D. E. Caldwell. 1984. Chemostat and in-situ colonization kinetics of Thermothrix thiopara on calcite and pyrite surfaces. Geomicrobiol. J. 3:217-229. [Google Scholar]
- 21.Krekeler, D., P. Sigalevich, A. Teske, H. Cypionka, and Y. Cohen. 1997. A sulfate-reducing bacterium from the oxic layer of a microbial mat from Solar Lake (Sinai). Desulfovibrio oxyclinae sp. nov. Arch. Microbiol. 167:369-375. [Google Scholar]
- 22.Krekeler, D., A. Teske, and H. Cypionka. 1997. Strategies of sulfate-reducing bacteria to escape oxygen stress in a cyanobacterial mat. FEMS Microbiol. Ecol. 25:89-96. [Google Scholar]
- 23.Kühl, M., and B. B. Jorgensen. 1992. Microsensor measurements of sulfate reduction and sulfide oxidation in compact microbial communities of aerobic biofilms. Appl. Environ. Microbiol. 58:1164-1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maidak, B. L., G. L. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The RDP (Ribosomal Database Project). Nucleic Acids Res. 25:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 26.Manz, W., M. Eisenbrecher, T. R. Neu, and U. Szewzyk. 1998. Abundance and spatial organization of gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol. Ecol. 25:43-61. [Google Scholar]
- 27.Marschall, C., P. Frenzel, and H. Cypionka. 1993. Influence of oxygen on sulfate reduction and growth of sulfate-reducing bacteria. Arch. Microbiol. 159:168-173. [Google Scholar]
- 28.Millero, F. J., and J. P. Hershey. 1989. Thermodynamics and kinetics of hydrogen sulfide in natural waters, p. 282-313. In E. S. Saltzman and W. J. Cooper (ed.), Biogenic sulfur in the environment. American Society for Microbiology, Washington, D. C.
- 29.Minz, D., S. Fishbain, S. J. Green, G. Muyzer, Y. Cohen, B. E. Rittmann, and D. A. Stahl. 1999. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryotic preference for anoxia. Appl. Environ. Microbiol. 65:4659-4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori, T., M. Koga, Y. Hikosaka, T. Nonaka, F. Mishina, Y. Sakai, and J. Koizumi. 1992. Microbial corrosion of concrete sewer pipes, H2S production from sediments and determination of corrosion rate. Water Sci. Technol. 23:1275-1282. [Google Scholar]
- 32.Muyzer, G., T. Brinkhoff, U. Nubel, C. M. Santegoeds, H. Schafer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer, Dordrecht, The Netherlands. [Google Scholar]
- 33.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes encoding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson, D. C., B. B. Jorgensen, and N. P. Revsbech. 1986. Growth pattern and yield of a chemoautotrophic Beggiatoa sp. in oxygen-sulfide microgradients. Appl. Environ. Microbiol. 52:225-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Norsker, N. H., P. H. Nielsen, and T. Hvitved-Jacobsen. 1995. Influence of oxygen on biofilm growth and potential sulfate reduction in gravity sewer biofilm. Water Sci. Technol. 31:159-167. [Google Scholar]
- 36.Okabe, S., T. Itoh, H. Satoh, and Y. Watanabe. 1999. Analyses of spatial distributions of sulfate-reducing bacteria and their activity in aerobic wastewater biofilms. Appl. Environ. Microbiol. 65:5107-5116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postgate, J. R. 1984. The sulphate-reducing bacteria, 2nd ed. Cambridge University Press, Cambridge, United Kingdom.
- 38.Ramsing, N. B., M. Kühl, and B. B. Jorgensen. 1993. Distribution of sulfate-reducing bacteria, O2, and H2S in photosynthetic biofilms determined by oligonucleotide probes and microelectrodes. Appl. Environ. Microbiol. 59:3840-3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Revsbech, N. P. 1989. An oxygen microelectrode with a guard cathode. Limnol. Oceanogr. 55:1907-1910. [Google Scholar]
- 40.Revsbech, N. P., and B. B. Jorgensen. 1986. Microelectrodes: their use in microbial ecology. Adv. Microb. Ecol. 9:293-352. [Google Scholar]
- 41.Risatti, J. B., W. C. Capman, and D. A. Stahl. 1994. Community structure of a microbial mat: the phylogenetic dimension. Proc. Natl. Acad. Sci. USA 91:10173-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saito, N., and M. Nei. 1987. The neighbor-joining method: a new method for constructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 43.Santegoeds, C. M., L. R. Damgaard, G. Hesselink, J. Zopfi, P. Lens, G. Muyzer, and D. de Beer. 1999. Distribution of sulfate-reducing and methanogenic bacteria in anaerobic aggregates determined by microsensor and molecular analyses. Appl. Environ. Microbiol. 65:4618-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Santegoeds, C. M., T. G. Ferdelman, G. Muyzer, and D. de Beer. 1998. Structural and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl. Environ. Microbiol. 64:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sass, H., H. Cypionka, and H.-D. Babenzien. 1997. Vertical distribution of sulfate-reducing bacteria at the oxic-anoxic interface in sediments of the oligotrophic Lake Stechlin. FEMS Microbiol. Ecol. 22:245-255. [Google Scholar]
- 46.Schramm, A., C. M. Santegoeds, H. K. Nielsen, H. Ploug, M. Wagner, M. Pribyl, J. Wanner, R. Amann, and D. de Beer. 1999. On the occurrence of anoxic microniches, denitrification, and sulfate reduction in aerated activated sludge. Appl. Environ. Microbiol. 65:4189-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teske, A., C. Wawer, G. Muyzer, and N. B. Ramsing. 1996. Distribution of sulfate-reducing bacteria in a stratified fjord (Mariager Fjord, Denmark) as evaluated by most-probable-number counts and denaturing gradient gel electrophoresis of PCR-amplified ribosomal DNA fragments. Appl. Environ. Microbiol. 62:1405-1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Widdel, F. 1988. Microbiology and ecology of sulfate-and sulfur-reducing bacteria, p. 469-585. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. A. John Wiley & Sons, Inc., New York, N.Y.
- 50.Widdel, F., and N. Pfennig. 1982. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. II. Incomplete oxidation of propionate by Desulfobulbus propionicus gen. nov., sp. nov. Arch. Microbiol. 131:360-365. [DOI] [PubMed] [Google Scholar]