Abstract
To quantify target genes in biological samples using DNA microarrays, we employed reference DNA to normalize variations in spot size and hybridization. This method was tested using nitrate reductase (nirS), naphthalene dioxygenase (nahA), and Escherichia coli O157 O-antigen biosynthesis genes as model genes and lambda DNA as the reference DNA. We observed a good linearity between the log signal ratio and log DNA concentration ratio at DNA concentrations above the method's detection limit, which was approximately 10 pg. This approach for designing quantitative microarrays and the inferred equation from this study provide a simple and convenient way to estimate the target gene concentration from the hybridization signal ratio.
DNA microarrays provide a powerful tool for the parallel analysis of many genes. Most DNA microarray studies conducted so far evaluate gene expression (cf. references 13, 22, and 23) by competitive hybridizations between different populations of mRNA expressed under different culture conditions. The relative extents of hybridizations of target genes to probes on the microarray provide information on the degree of expression of genes of interest. Recently several research groups applied the DNA microarray-based approach to other fields, such as single nucleotide polymorphism and mutation detection (9, 12), sequencing (2, 6), genetic linkage analysis and population genetics (5, 6, 8), comparative genomics (2, 17), and identification of bacterial species (7). However, the quantitative characteristics of microarray hybridization, other than competitive hybridization, have not been investigated. Studies on the quantitative properties and kinetics of microarray hybridization are needed to fully evaluate the potential uses of the DNA microarray method.
DNA microarray technology holds promise for microbial ecology (20), for example, detecting and quantifying different gene families involved in biogeochemical cycling, biodegradation, and pathogenesis in a high-throughput manner. However, the approaches used for gene expression analysis or other previously reported applications are inappropriate for the titration of genes or DNA sequences in environmental samples, because the probe sizes (printed spots) vary and evenness of hybridization cannot be assured. Hence, interpretation of hybridization profiles obtained from one-color hybridization (similar to conventional Southern hybridization) is not accurate. On the other hand, gene expression analysis, which uses two-color competitive hybridization, is not affected by the above two factors, since it uses the ratio values from the competitive hybridizations of each probe. However, DNA microarray hybridization to quantify the amount of a gene requires direct binding of the target sequence to the probe DNA rather than competitive hybridizations, and the extent of this direct binding should be normalized against the concentration variations in the probe DNA and the spatial variation in the extent of hybridization. An alternative approach with a different microarray design and hybridization scheme is required to quantify target genes in biological samples. This need led us to develop the new format described below.
Test genes.
We used three model genes: heme-containing nitrite reductase (nirS), naphthalene dioxygenase (nahA), and Escherichia coli O-antigen biosynthesis gene, as representatives of genes involved in biogeochemical cycling, biodegradation, and pathogen detection, respectively. nirS (900 bp), nahA (700 bp), and O-antigen gene (500 bp) were amplified from Pseudomonas stutzeri ATCC 14405, Pseudomonas putida G7 DSM 4476, and Escherichia coli O157:H7 ATCC 33150 by PCR according to the methods of Braker et al. (3), Herrick et al. (14), and Maurer et al. (16), respectively.
A fragment (500 bp) of lambda DNA (GenBank accession no. J02459) encoding capsid component (6135 to 7160), DNA packaging (7202 to 7600), and head-tail joining (7612 to 7965) was PCR amplified with primer pairs F (GAT GAG TTC GTG TCC GTA CAA CT) and R (GGT TAT CGA AAT CAG CCA CAG CG). Vector pSP72 (Promega, Madison, Wis.) was digested with the restriction enzyme EcoRI. The yeast gene ACT (for actin; GenBank accession no. L00026) was PCR amplified with primer pairs ACT1F1 (GAT GGA GCC AAA GCG GTG A) and ACT1R1 (GCG CTT GCA CCA TCC CAT T). The yeast gene STE (for pheromone receptor; GenBank accession no. M12239) was PCR amplified with primer pairs STE3F1 (CCC CTT CAA AAT TGG AGC TTG C) and STE3R1 (CCC CCT TTA GCA TGG CAT TCA). PCR products and restriction enzyme digests were purified with the QIAquick PCR purification kit (Qiagen, Valencia, Calif.) and quantified with PicoGreen (Molecular Probes, Eugene, Ore.). Other details were done according to the method of Cho and Tiedje (7).
Microarray fabrication.
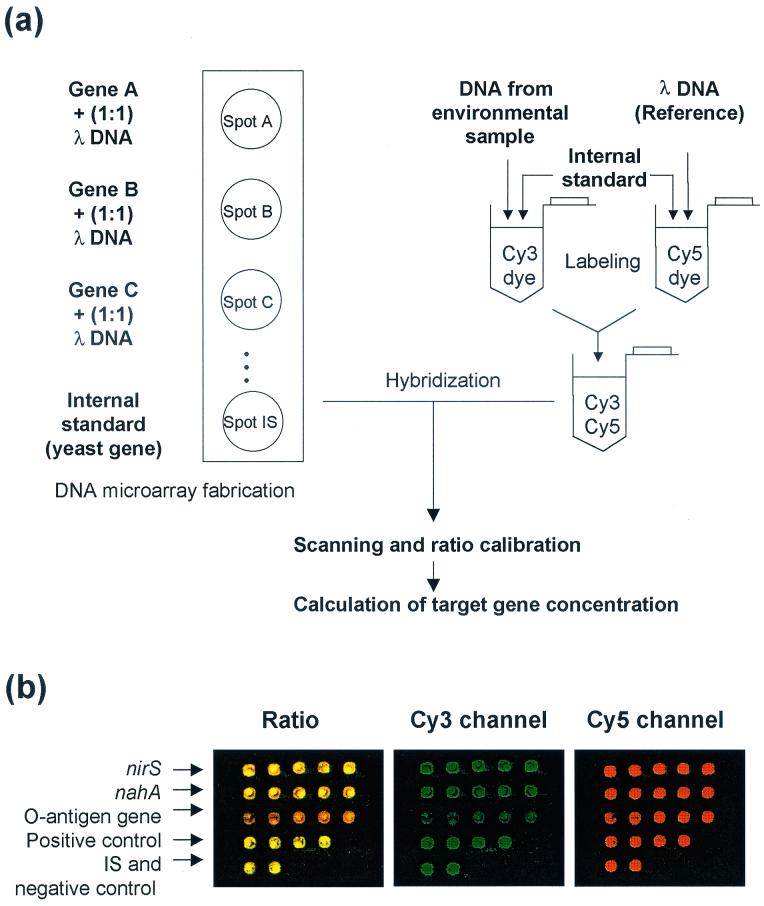
Each of the amplified model genes was mixed with an equal amount of the lambda DNA fragment (Fig. 1a). The mixtures were then resuspended (200 ng/μl) in 3× SSC buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and printed (ca. 1 nl/spot, five replicate spots/each model gene) on CMT-GAPS amino silane coated slides (Corning Inc., Corning, N.Y.) with microspotting pins (TeleChem, Sunnyvale, Calif.). pSP72 vector and the yeast gene ACT were spotted as positive and negative control spots, respectively, and the yeast gene STE was spotted as the internal standard (IS). After drying, the slides were processed using the succinic anhydride blocking method according to the manufacturer's protocol and stored at room temperature until use.
FIG. 1.
(a) Schematic diagram showing microarray design and hybridization procedure. (b) Representative scanning image of microarray. A sample containing 10 ng of each model gene was used for the microarray hybridization. Pseudo-colors indicate the ratio (R) between Cy3 channel and Cy5 channel (green, R > 1; yellow, R = 1; red, R < 1).
DNA labeling and hybridization.
Each model gene (100 ng, 50 ng, 10 ng, 5 ng, 2 ng, 1 ng, 500 pg, 200 pg, 40 pg, 4 pg, 800 fg, 400 fg, or 80 fg) was diluted (total, 1 μg) in mouse genomic DNA (Sigma, St. Louis, Mo.). The dilution series of model genes (test DNAs) was labeled with FluoroLink Cy3-dCTP (Amersham Pharmacia, Piscataway, N.J.) by random priming (High Prime; Roche, Indianapolis, Ind.) and used as sample DNAs. pSP72 vector (10 ng) was included in Cy3-labeling reaction as a positive control. Lambda DNA (1 μg) was labeled with FluoroLink Cy5-dCTP (Amersham Pharmacia, Piscataway, N.J.) by random priming (High Prime; Roche) and used as reference DNA for hybridization signal ratio calculation (Cy3-Test/Cy5-Ref). The yeast gene STE (10 ng) was included in each labeling reaction as an internal standard (Cy3-IS and Cy5-IS) for labeling efficiency correction.
The arrays were prehybridized in prehybridization buffer (3.5× SSC, 0.1% sodium dodecyl sulfate (SDS), 10 mg of bovine serum albumin/ml) for 20 min at 65°C, hybridized with approximately 1 μg of Cy3- and Cy5-labeled DNA mixture (1:1) in hybridization buffer (3× SSC, 0.1% SDS, 0.5 mg of yeast tRNA/ml) at 65°C overnight and then washed once with primary wash buffer (0.1× SSC, 0.1% SDS) at room temperature for 5 min and twice with secondary wash buffer (0.1× SSC) for 5 min.
Scanning.
Hybridized arrays were scanned with a GenePix 4000 laser scanner (Axon, Foster City, Calif.). Laser lights of wavelengths at 532 and 635 nm were used to excite Cy3 dye and Cy5 dye, respectively. Fluorescent images were captured in multi-image tagged image file format and analyzed with GenePix Pro 3.0 software (Axon). Microsoft EXEL, SYSTAT (SPSS, Chicago, Ill.), and SigmaPlot (SPSS) were used for statistical analysis and data presentation. Other details on microarray hybridization, scanning, and data processing were carried out as described previously (7).
All experiments were performed in duplicate. Since each array contained five replicate spots for each model gene, hybridization signal ratios were obtained from 10 replicate spots (Fig. 1b). Hybridization signals from the Cy5 channel enabled us to examine the quality of microarray fabrication and hybridization, since all spots contain λ DNA. Almost all spots satisfied our quality control criterion, i.e., that >90% of pixels of the spot should show >2 × standard deviation [SD] + background signal in the Cy5 channel. Negative control spots showed < 2 × SD + background signal in both the Cy5 and Cy3 channels. The standard deviation of Cy5 channel signal was ca. 25% of mean signal (within a slide) and ca. 50% of mean signal (slide-to-slide variation). By using our normalization approach, the standard deviation was <10% of the mean value (within a slide as well as between slides).
Quantification by hybridization signal ratio.
Hybridization signal ratios (R) between model DNAs and reference DNAs were calculated (Cy3-Test/Cy5-Ref) and corrected with the correction factor (c = Cy5-IS/Cy3-IS) from the internal standard (yeast gene STE) (corrected signal ratio R′ = c × [Cy3-Test/Cy5-Ref]). The ratio of Cy5 incorporation to Cy3 incorporation (Cy5-IS/Cy3-IS) during the DNA labeling was 1.01 ± 0.05 for all experiments, and it was consistent with our previous result (7). All positive control spots gave significantly high hybridization signal ratios (1.22 ± 0.26), and negative control spots showed no hybridization signals in both Cy3 and Cy5 channels.
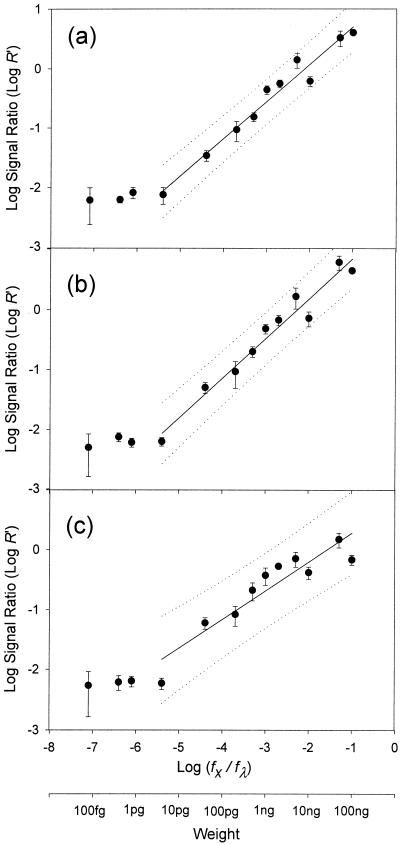
Corrected hybridization signal ratios (R′) were plotted against the weight ratios of model genes to reference DNA (λ DNA) in the Cy dye-labeled DNA mixtures (sample DNA) (Fig. 2). The weight ratio of model gene to reference DNA [(fx/fλ) × 100] also reflects the weight percentage of the model gene in the sample DNA. Since it is unlikely that >10% of total environmental DNA is comprised of a single gene, model gene weight ratios of >0.1 were not examined. The lower detection limit was in the range of 1 to 10 pg for the three model genes. Good linearity between log (hybridization signal ratio, R′) and log (model gene weight ratio, fx/fλ) was observed above the detection limit. Regression analyses resulted in an equation, log (R′) = α log (fx/fλ) + β, describing the linearity (Table 1). This inferred equation allows calculation of the amount of target gene (fx) from the hybridization signal ratio (R′), when α and β values are obtained from a standard curve. α and β values should be a function of a hybridization constant, which is affected by genome complexity, base composition, ionic strength, etc. (4).
FIG. 2.
Relationship between hybridization signal ratios and weight fraction of model genes, nirS (a), nahA (b), and E. coli O157 O-antigen biosynthesis gene (c). The solid and dotted lines indicate the regression curve and 95% prediction interval, respectively. Error bars indicate SDs.
TABLE 1.
Regression analysis of model gene concentrations versus hybridization signal ratios from microarray hybridization [log (R′) = α log (fx/fλ) + β]a
| Model gene | Coefficient of determination (r2) | α, regression coefficient (slope) | β, constant (y-intercept) |
|---|---|---|---|
| nirS | 0.97 | 0.63** | 1.33** |
| nahA | 0.96 | 0.66** | 1.50** |
| O-antigen biosynthesis | 0.86 | 0.48** | 0.76* |
Asterisk, P < 0.05; double asterisk, P < 0.001.
Quantification by hybridization kinetics.
We considered an alternative way to estimate the target gene concentration using hybridization kinetics. Southern-type hybridization kinetics can be described by H = C (1 − e−k F t), where H is the amount (fraction) of DNA hybridized; F is the amount of target DNA to be measured; C is the amount of DNA preimmobilized on the spot (F ≪ C); k is the hybridization constant; and t is the hybridization time (15). In the case of microarray hybridization, we used the extent of hybridization of the model gene x, Hx (Cy3 channel), as Hx = Cx (1 − e−kxFxt), and the extent of hybridization of λ DNA, Hλ (Cy5 channel), as Hλ = Cλ (1 − e−kλFλt). Under the same hybridization conditions, kFt is reduced to k′F. Since the amount of model gene in the spot i is the same as the amount of λ DNA in the spot i, Cx = Cλ. The hybridization signal ratio (R′) equals Hx/Hλ = (Cx/Cλ) [(1 − e−kxFxt)/(1 − e−kλFλt)] = (1 − e−kx′Fx)/[(1 − e−kλ′Fλ)]. Hence, the hybridization signal ratio in the microarray hybridization is a function of Fx (amount of target gene in the sample), since the amount of λ DNA (Fλ) is known and constant. Hybridization constants for nirS and nahA, which were calculated by regression analysis using a two-parameter exponential model, y = a(1 − e−bx), where a = 1/(1 − e−kγ′Fλ); b = kx′, were 0.004 (r2 = 0.85) and 0.004 (r2 = 0.68), respectively. The hybridization constant, k′, for the O-antigen synthesis gene could not be calculated due to the low r2 value (0.09). Using the hybridization constants for each model, the concentration of model genes in the sample DNA can be estimated by interpolating the hybridization signal ratio, R′.
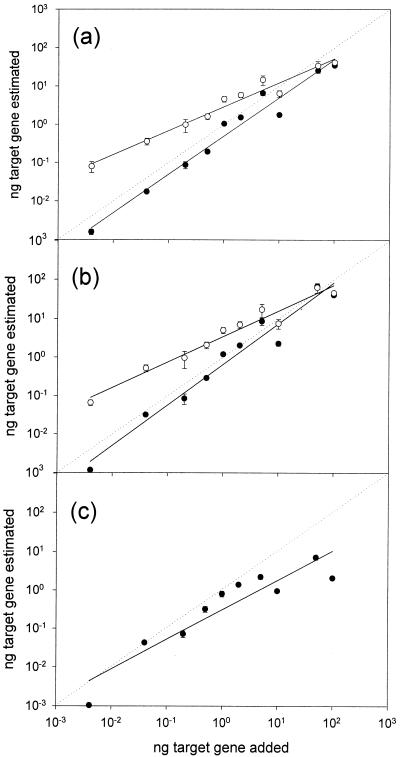
We compared the estimated model gene concentrations by using two different approaches, our inferred equation and the hybridization kinetics. In Fig. 3, a diagonal line (dotted) indicates perfect estimation. Both methods resulted in good approximations to true values, but the estimates by hybridization kinetics tended to diverge more with decreasing target gene concentration than those by our equation. This could be due to two possible reasons. The regression analyses might not give accurate values for the hybridization constants due to the low r2 values (0.68 to 0.85). Secondly, the existing hybridization kinetic model may not correctly explain the microarray hybridization. All hybridization kinetics are based on solution hybridizations, but adjustments even for membrane hybridization kinetics are not established. Studies on the kinetics of DNA hybridization on microarrays are needed to provide the background for quantification as well as gene expression.
FIG. 3.
Relationship between estimated model gene concentration and actual concentration of model genes, nirS (a), nahA (b), and E. coli O157 O-antigen biosynthesis gene (c). Closed circle, calculated by log (R′) = α log (fx/fλ) + β, in this study; open circle, estimation by hybridization kinetics. The solid line indicates the regression curve, and the diagonal dotted line indicates a 1:1 relationship. Error bars indicate standard deviations. No error bars are shown when smaller than symbols.
Since a microarray can contain thousands of genes for quantification, it is not practical to do regression analyses for each gene by using hybridization kinetics. However, the high linearity of our inferred equation suggests that two or three concentration standards may give reasonable estimates for α and β values, enabling massively parallel quantification.
Environmental detection limit.
To test microarray quantification with environmental DNA, we extracted DNAs from sediment (Red Cedar River, East Lansing, Mich.) and soil (Kellogg Biological Station, Hickory Corners, Mich.) using a soil DNA isolation kit (MoBio, Solana Beach, Calif.) and labeled the DNA as described above. Assuming that one cell has a single copy of nirS and that PCR can detect one copy of nirS in the environmental DNA, PCR amplification of nirS with the most-probable-number approach showed that both the sediment and soil samples contain approximately 105 cells (and copies of nirS)/g (wet), which is consistent with real-time PCR data (11). The microarray, however, did not detect nirS sequences in either the sediment or soil samples. Even when 105 cells of denitrifying P. stutzeri were spiked into the soil, the hybridization signal was not detected. This high detection limit agrees with what is to be expected. Considering the genome size of Pseudomonas (5 Mbp) (10, 19), 10 pg of nirS (the detection limit) (Fig. 2) roughly corresponds to 50 ng of denitrifying P. stutzeri genome. This amount is 1/20 of the total DNA load (1 μg/40 μl) per slide for microarray hybridization. Hence, only when genomes with P. stutzeri-like nirS sequences contribute more than 1/20 of the environmental DNA extracted can they be detected. Since soil DNA (or environmental DNA) contains DNA originating from a huge diversity of bacteria plus fungi, archaea, protozoa, plant debris, and microinvertebrates, the quantitative detection of particular genes by microarray using current technology is problematic.
The detection limit of conventional membrane hybridization (dot blotting) using either radioactive labeling or fluorescent dye labeling is generally femtogram quantities. This discrepancy in the detection limits of membrane hybridization versus microarray hybridization can be explained by the differences in the amount of DNA (probe) immobilized on the substrata (membrane, >1 μg/dot; glass slide, <10 to 20 pg/spot) and the corresponding limit in loading capacity. Voordouw et al. (21) showed that the detection limit in the Southern-type DNA hybridization is dependent on and proportional to the amount of DNA immobilized on the substrata. The 105-fold difference in the amount of DNA immobilized on the two substrata likely explains the 105-fold difference in the detection limit between conventional membrane hybridization and microarray hybridization.
There are three possible solutions for improving the detection limit of microarray hybridization. First, increase the amount of DNA (probe) immobilized on the microarray substratum. This requires the development of better chemistry for microarray fabrication or novel substrata to bind larger amounts of probes on the microarray. Second, develop a microarray signal detection system with higher sensitivity. Third, develop a procedure that can selectively enrich bacterial genomes or genes of interest from the environmental samples, for example the use of magnetic or biotinylated probes (1, 18). Once the detection limit issue is solved, the microarray-based method should be a powerful tool for massively parallel analyses of important genes and microbes in the environment.
Use of the test gene-λ DNA mixture as probes on the microarray provided the standardization that allowed the microarray to quantify target genes. Cy5 channel signals from each spot, which were generated by hybridizations of λ DNAs in the probe spots to the Cy5-labeled λ DNA in the sample DNA-reference DNA mixture, standardize Cy3-channel signals as hybridization signal ratios. Regardless of the variations in the amount of DNA spotted on the microarray and spatial variations in the extent of microarray hybridization, these standardized Cy3 channel signals were then comparable to each other, providing quantitative measures of the extent of hybridization between target genes in probes and samples. This standardization is necessary to avoid errors originating from the probe concentration and the spatial variations. Another advantage of using this reference DNA approach is that gene expression profiles under many different conditions can be compared and analyzed simultaneously. Currently, only pairwise comparison of expression profiles under two conditions and using two different dyes is possible. However, using our approach, each mRNA population from many culture conditions is labeled with one dye and hybridized to the microarray after being mixed with reference DNA labeled with another dye. The resulting hybridization signal profile (titration of specific mRNAs) then can be compared to many other profiles generated in the same manner.
This study shows that by using reference DNA, microarrays can accurately quantify genes in DNA samples, but the detection limit with current technology limits environmental applications.
Acknowledgments
We thank Alison Murray and Jizhong Zhou for helpful discussion and Syed Hashsham for use of microarray facilities.
This research was supported by NSF grant no. DEB-0075564 and the Center for Microbial Ecology.
REFERENCES
- 1.Albretsen, C., K. H. Kalland, B. I. Haukanes, L. S. Havarstein, and K. Kleppe. 1990. Applications of magnetic beads with covalently attached oligonucleotides in hybridization: isolation and detection of specific measles virus mRNA from a crude cell lysate. Anal. Biochem. 189:40-50. [DOI] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Braker, G., A. Fesefeldt, and K. P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Britten, R. J., and E. H. Davidson. 1988. Hybridization strategy, p. 3-14. In B. D. Hames and S. J. Higgins (ed.), Nucleic acid hybridization, a practical approach. IRL Press, Oxford, England.
- 5.Chakravarti, A. 1999. Population genetics—making sense out of sequence. Nat. Genet. 21:56-60. [DOI] [PubMed] [Google Scholar]
- 6.Cheung, V. G., J. P. Gregg, K. J. Gogolin-Ewens, J. Bandong, C. A. Stanley, L. Baker, M. J. Higgins, N. J. Nowak, T. B. Shows, W. J. Ewens, S. F. Nelson, and R. S. Spielman. 1998. Linkage-disequilibrium mapping without genotyping. Nat. Genet. 18:225-230. [DOI] [PubMed] [Google Scholar]
- 7.Cho, J.-C., and J. M. Tiedje. 2001. Bacterial species determination from DNA-DNA hybridization using genome fragments and DNA microarrays. Appl. Environ. Microbiol. 67:3677-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gentalen, E., and M. Chee. 1999. A novel method for determining linkage between DNA sequences: hybridization to paired probe arrays. Nucleic Acids Res. 27:1485-1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerry, N. P., N. E. Witowski, J. Day, R. P. Hammer, G. Barany, and F. Barany. 1999. Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol. 292:251-262. [DOI] [PubMed] [Google Scholar]
- 10.Ginard, M., J. Lalucat, B. Tummler, and U. Romling. 1997. Genome organization of Pseudomonas stutzeri and resulting taxonomic and evolutionary considerations. Int. J. Syst. Bacteriol. 47:132-143. [DOI] [PubMed] [Google Scholar]
- 11.Grüntzig, V., S. C. Nold, J. Zhou, and J. M. Tiedje. 2001. Pseudomonas stutzeri nitrite reductase gene abundance in environmental samples measured by real-time PCR. Appl. Environ. Microbiol. 67:760-768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hacia, J. G., J. B. Fan, O. Ryder, L. Jin, K. Edgemon, G. Ghandour, R. A. Mayer, B. Sun, L. Hsie, C. M. Robbins, L. C. Brody, D. Wang, E. S. Lander, R. Lipshutz, S. P. Fodor, and F. S. Collins. 1999. Determination of ancestral alleles for human single-nucleotide polymorphisms using high-density oligonucleotide arrays. Nat. Genet. 22:164-167. [DOI] [PubMed] [Google Scholar]
- 13.Harrington, C. A., C. Rosenow, and J. Retief. 2000. Monitoring gene expression using DNA microarrays. Curr. Opin. Microbiol. 3:285-291. [DOI] [PubMed] [Google Scholar]
- 14.Herrick, J. B., E. L. Madsen, C. A. Batt, and W. C. Ghiorse. 1993. Polymerase chain reaction amplification of naphthalene-catabolic and 16S rRNA gene sequences from indigenous sediment bacteria. Appl. Environ. Microbiol. 59:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kafatos, F. C., C. W. Jones, and A. Efstratiadis. 1979. Determination of nucleic acid sequence homologies and relative concentrations by a dot hybridization procedure. Nucleic Acids Res. 7:1541-1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maurer, J. J., D. Schmidt, P. Petrosko, S. Sanchez, L. Bolton, and M. D. Lee. 1999. Development of primers to O-antigen biosynthesis genes for specific detection of Escherichia coli O157 by PCR. Appl. Environ. Microbiol. 65:2954-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murray, A. E., D. Lies, G. Li, K. Nealson, J. Zhou, and J. M. Tiedje. 2001. DNA/DNA hybridization to microarrays reveals gene-specific differences between closely related microbial genomes. Proc. Natl. Acad. Sci. USA 98:9853-9858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oelmüller, U., N. Krüger, A. Steinbüchel, and C. G. Friedrich. 1990. Isolation of prokaryotic RNA and detection of specific mRNA with biotinylated probes. J. Microbiol. Methods 11:73-84. [Google Scholar]
- 19.Schmidt, K. D., B. Tummler, and U. Romling. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiedje, J. M., J. C. Cho, A. Murray, D. Treves, B. Xia, and J. Zhou. 2000. Soil teeming with life: new frontiers for soil science, p. 393-412. In B. M. Rees, B. C. Ball, C. D. Campbell, and C. A. Watson (ed.), Sustainable management of soil organic matter. CAB International, Oxon, United Kingdom.
- 21.Voordouw, G., Y. Shen, C. S. Harrington, A. J. Telang, T. R. Jack, and D. W. S. Westlake. 1993. Quantitative reverse sample genome probing of microbial communities and its application to oil field production waters. Appl. Environ. Microbiol. 59:4101-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wei, Y., J. M. Lee, C. Richmond, F. R. Blattner, J. A. Rafalski, and R. A. LaRossa. 2001. High-density microarray-mediated gene expression profiling of Escherichia coli. J. Bacteriol. 183:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye, R. W., W. Tao, L. Bedzyk, T. Young, M. Chen, and L. Li. 2000. Global gene expression profiles of Bacillus subtilis grown under anaerobic conditions. J. Bacteriol. 182:4458-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]