Abstract
Streptococcus mutans, the primary etiological agent of human dental caries, is an obligate biofilm-forming bacterium. The goals of this study were to identify the gene(s) required for biofilm formation by this organism and to elucidate the role(s) that some of the known global regulators of gene expression play in controlling biofilm formation. In S. mutans UA159, the brpA gene (for biofilm regulatory protein) was found to encode a novel protein of 406 amino acid residues. A strain carrying an insertionally inactivated copy of brpA formed longer chains than did the parental strain, aggregated in liquid culture, and was unable to form biofilms as shown by an in vitro biofilm assay. A putative homologue of the enzyme responsible for synthesis of autoinducer II (AI-2) of the bacterial quorum-sensing system was also identified in S. mutans UA159, but insertional inactivation of the gene (luxSSm) did not alter colony or cell morphology or diminish the capacity of S. mutans to form biofilms. We also examined the role of the homologue of the Bacillus subtilis catabolite control protein CcpA in S. mutans in biofilm formation, and the results showed that loss of CcpA resulted in about a 60% decrease in the ability to form biofilms on an abiotic surface. From these data, we conclude that CcpA and BrpA may regulate genes that are required for stable biofilm formation by S. mutans.
Streptococcus mutans, an “obligate” biofilm-forming bacterium (5) and the primary etiological agent of human dental caries, has developed a variety of mechanisms to colonize the tooth surfaces and, under certain conditions, to become a numerically significant species in cariogenic biofilms (4). Moreover, this bacterium possesses the ability to scavenge multiple fermentable sugars at micromolar concentrations and to grow and carry out glycolysis at the low plaque pH values attained in the oral cavity. The production of acids by S. mutans causes dissolution of minerals in tooth enamel and formation of dental caries.
Despite the complexity of molecular mechanisms implicated in the process, biofilm formation is generally considered to be a two-step sequential process requiring attachment of the bacterial cells to a surface (early adherence), followed by growth-dependent accumulation of bacteria in multilayered cell clusters, involving intra- and intergeneric adhesion events. Considerable effort has been devoted to identifying factors of S. mutans and other oral streptococci that participate in biofilm initiation and development. Surface-associated proteins, such as SpaP and Fap1, have been found to function as high-affinity adhesins and play an important role in the initiation of biofilm formation by oral streptococci (2, 14). Extracellular glucans, especially the insoluble glucans synthesized from sucrose by the glucosyltransferases, are of central importance in adhesive interactions by mutans streptococci and are key factors in the accumulation of mutans streptococci on smooth surfaces (22). Reduction in the synthesis of water-insoluble glucan through directed mutagenesis was found to result in decreased biofilm formation by S. mutans and reduced cariogenicity in rat caries models (8, 18, 37). S. mutans also synthesizes three glucan-binding proteins: GbpA, GbpB, and GbpC. GbpC has similarities to members of the Spa family of proteins of oral streptococci and is involved in rapid, dextran-dependent aggregation (31). Immunization with GbpB has been reported to be protective against caries (33), although Hazlett et al. (18) have reported that a gbpA-deficient strain is hypercariogenic in a rat model. It was later found that inactivation of gbpA alters the structural and functional aspects of plaque biofilm that could be compensated by recombination between the gtfB and gtfC genes (17). Thus, there are numerous cell surface and extracellular proteins that work in concert to successfully establish S. mutans in tooth biofilms.
While much is now known about initial adhesion of S. mutans to the tooth, very little is known about maturation of oral biofilms formed by this organism or about how the organism controls expression of virulence when it is growing as adherent populations. Importantly, it is generally believed that the regulation of gene expression in biofilm populations is different from that in planktonic cells (10, 29) and that the bacteria must coordinate the expression of sets of genes to form stable biofilms (9, 28). Here, we report the identification and characterization of a gene that was found to encode a novel protein that appears to be involved in regulation of biofilm development by S. mutans, and we explore the impact of inactivation of genes potentially involved in quorum sensing (QS) and in the control of carbohydrate metabolism on the ability of this bacterium to form biofilms.
MATERIALS AND METHODS
Bacterial cultivation.
Escherichia coli DH10B strains were grown in Luria-Bertani medium and, when necessary, ampicillin (100 μg/ml) or erythromycin (500 μg/ml) was included. S. mutans UA159 and its derivatives were maintained on brain heart infusion (BHI) medium, with or without addition of erythromycin (10 μg/ml). Preparation of competent cells and transformation of S. mutans were done as previously described (6). For growth studies and biofilm formation assays, S. mutans strains were grown in the semidefined medium BM (25). All agar media were prepared similarly, but agar (Difco Laboratories, Detroit, Mich.) was added at a concentration of 1.5% (wt/vol).
DNA manipulations.
Unless otherwise stated, standard recombinant DNA techniques were performed as described by Sambrook et al. (30). All restriction and modifying enzymes and reagents were purchased from Gibco-BRL (Life Technologies, Inc., Gaithersburg, Md.), New England Biolabs (Beverly, Mass.), or MBI Fermentas (Amherst, N.Y.) and used as described by the suppliers. For Southern hybridizations, 12 μg of genomic DNAs was digested with various restriction enzymes and transferred to NewBond Nylon membranes (NEN Life Science Products, Inc.). Probes were labeled with [α-32P]dATP by uing a random primer DNA labeling system from Gibco-BRL.
Biofilm formation assay.
Biofilm assays were done by using the protocol of Loo et al. (25) with minor modifications. Briefly, overnight cultures of S. mutans UA159 and its derivatives were transferred to prewarmed BM medium and grown at 37°C, in a 5% CO2 aerobic atmosphere to an optical density at 600 nm (OD600) of ∼0.5. The cultures were diluted 1:100 in fresh BM, and then 200 μl of the cell suspension was inoculated into the wells of a 96-well (flat bottom) cell culture cluster (Costar 3595; Corning, Inc., Corning, N.Y.). Wells containing uninoculated growth medium were used as negative controls. Plates were incubated at 37°C in a 5% CO2 aerobic atmosphere for 16 to 24 h. Before biofilm quantification, growth was assessed by measuring the absorbance of cultures in the wells at 600 nm by using an enzyme-linked immunosorbent assay (ELISA) reader (EIA Reader 2550; Bio-Rad Laboratories, Hercules, Calif.). Media and unattached bacterial cells were decanted from the wells after 5 min of agitation at 200 rpm on a shaker (Shaker 20E; Labnet International, Edison, N.J.), and the remaining planktonic or loosely bound cells were removed by gentle rinsing with 200 μl of sterile distilled water. The plates were then blotted on paper towels and air dried, and adherent bacteria were stained with 50 μl of 0.1% crystal violet for 15 min at room temperature. After two rinses with 200 μl of water each time, the bound dye was extracted from the stained cells by using 200 μl of 99% ethanol, and the plates were set on a shaker to allow full release of the dye. Biofilm formation was then quantified by measuring the absorbance of the solution at 600 nm with an ELISA reader. In this quantitative biofilm assay, both primary attachment and accumulation in multilayered cell clusters, which together lead to biofilm formation, can be measured by altering the incubation time of bacterial cultures.
Phase-contrast microscopy.
Examination of the cell morphology of the wild-type and mutant S. mutans was done by using phase-contrast microscopy. Mid-log and overnight cultures grown in BM were examined by using a Nikon Eclipse E800 microscope, and digitized images were captured by using a Spot RT monochrome camera driven by Spot v3.0.4 software (Diagnostic Instruments, Inc.).
Autolysis assay.
Autolysis assays were performed as described by Garcia et al. (16) with minor modifications. Cell samples (50 ml) were collected from late-exponential-phase cultures grown in BHI medium (OD600 ≅ 0.7). Cell pellets, obtained after centrifugation at 3,800 × g, at 4°C for 10 min, were washed twice with 50 ml of ice-cold water and resuspended in 50 ml of 0.05 M Tris-HCl (pH 8.0) containing 0.2% Triton X-100 (Sigma). The cells were then incubated at 37°C with agitation, and the absorbance at 600 nm was measured at 30-min intervals.
RESULTS AND DISCUSSION
Characterization of the brpA gene.
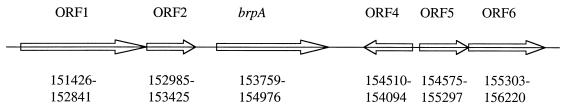
During a search of the genome of S. mutans UA159 for various types of transcriptional regulators, a gene of 1,218 bp was identified and designated brpA (for biofilm regulatory protein A). The brpA gene was located at nucleotides (nt) 153759 to 154976 in the S. mutans UA159 chromosome and was found to encode a polypeptide of 406 amino acid residues with a high degree of similarity to a putative transcriptional regulator of Streptococcus pyogenes (spy1733, accession number AAK34477; 65% identity) and a putative transcriptional regulator of Lactococcus lactis subsp. lactis (yeeG, accession number AAK04533; 34% identity) (Fig. 1 and 2). BrpA was predicted by using computer algorithms to be a membrane-associated protein with a serine-rich tail extending from the cell wall. The brpA-encoded protein also showed some homology to the transcriptional regulator LytR of Bacillus subtilis (26% identity) and the putative regulator of capsule production, CpsX, of Streptococcus agalactiae (12% identity) (21, 23). LytR of B. subtilis functions as an attenuator of the LytABC and LytR operons: LytC is an N-acetylmuramoyl-l-alanine amidase, one of the two major autolysins in the walls of vegetative cells; LytB is a modifier of the amidase; and LytA is a lipoprotein. However, the brpA-encoded protein was ca. 100 residues larger than LytR due to the presence of the serine-rich C-terminal domain that was absent in LytR. Further, no apparent homologues of LytABC could be identified in the S. mutans genome database. CpsX, a predicted sensor kinase, in concert with CpsY, a response regulator of the putative bacterial two-component system CpsXY, is thought to function in the regulation of capsule expression in group B streptococci (21). A gene that codes for a protein virtually identical to CpsY was identified at nt 935327 to 936229 in the S. mutans chromosome, but it was not linked to brpA and S. mutans is not known to produce a capsule by traditional means. Moreover, BrpA does not contain domains that are highly conserved in known two-component regulatory systems. Analysis of the regions flanking brpA (Fig. 1) revealed that no genes were apparently cotranscribed with brpA. Therefore, brpA constitutes a locus that has some similarity to, but also some key differences with, lytR of B. subtilis and cpsX of S. agalactiae, and which is highly conserved with putative transcriptional regulators in some other gram-positive cocci.
FIG. 1.
Schematic diagram of the brpA locus and its flanking region. ORF1 and -2 encode hypothetical proteins; ORF4 codes for a protein of the histidine triad family; and ORF5 and ORF6 encode an ATP-binding protein and a permease of the ABC transporter system, respectively. Arrows represent the orientation of each individual ORF, and the numbers underneath indicate their respective positions in the chromosome.
FIG. 2.
Alignment of predicted amino acid sequence of BrpA of S. mutans (Sm) with the transcriptional regulator LytR of B. subtilis (Bs), and putative regulatory proteins CpsX of S. agalactiae (Sa), YeeG of L. lactis subsp. lactis (Lc), and Spy1733 of S. pyogenes (Sp). Identical residues are highlighted. Conserved regions of the protein are in boxes, and a consensus sequence is shown below these alignments. See the text for more detail.
Isolation and characterization of a brpA mutant.
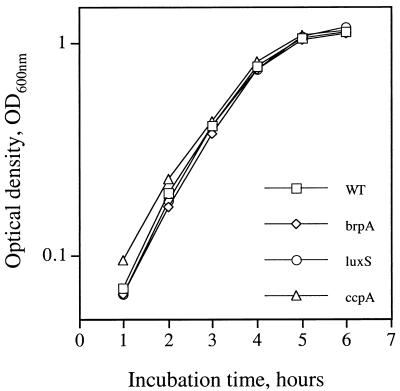
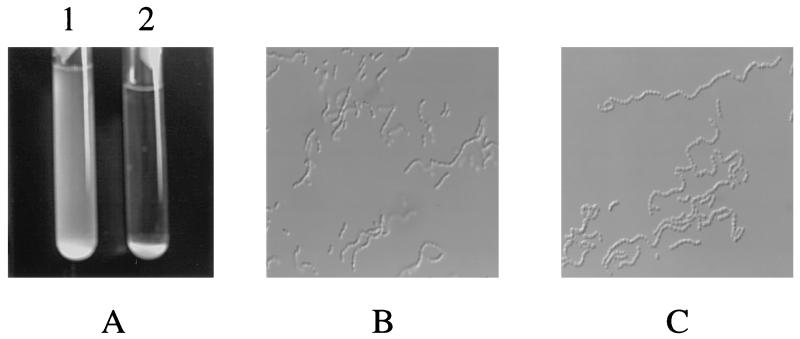
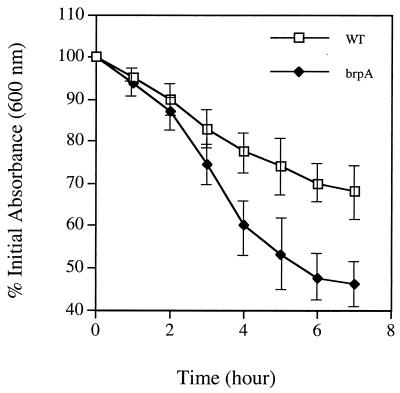
The brpA gene was amplified from S. mutans UA159 by PCR as a 1.8-kbp DNA fragment and directly cloned into pUC19 by EcoRI/XbaI fusions. To inactivate the gene, which was positioned at nt 247 to 1464 in the 1.8-kb fragment, a 270-bp EcoRV/SmaI fragment (nt 311 to 581, relative to the start codon) was deleted and replaced by an erythromycin resistance (Emr) marker. The resulting construct was then used to transform S. mutans UA159. Mutants with an inactivated brpA gene, resulting from double-crossover recombination between the chromosomal brpA and the incoming disrupted copy in the plasmid, were isolated on BHI-erythromycin plates and confirmed by Southern blot analysis (data not shown). Compared to the parental strain, the brpA mutant (strain TW14) grew similarly well in both BHI and BM media (Fig. 3). No differences in colony morphology were noted between the brpA-deficient and parental strains. However, one striking feature of the brpA mutant was its marked propensity to form longer chains (average number of cells per chain, 27.9 ± 14.1 for the mutant versus 11.6 ± 3.7 for the wild type) and to settle to the bottom of the test tube, leaving an almost clear liquid phase after overnight growth (Fig. 4). Increased chaining and aggregation were primarily associated with late-exponential-phase to stationary-phase cultures. An autolysis assay, described in Materials and Methods, also showed that the brpA mutant underwent autolysis at a higher rate than the parental strain (Fig. 5).
FIG. 3.
Growth curves of S. mutans UA159 (WT) and its derivatives. brpA, luxS, and ccpA refer to brpA, luxSSm, and ccpA mutant strains, respectively. Cultures were grown in BM medium at 37°C in 5% CO2, and growth was monitored by measuring the absorbance of the cultures at 600 nm as described in Materials and Methods.
FIG. 4.
Morphological characteristics in BM medium. (A) Twenty-four-hour cultures of the brpA-deficient strain TW14 (tube 2) and its parent S. mutans UA159 (tube 1). (B and C) Phase-contrast microscopy of the 24-h cultures of UA159 and the brpA mutant, respectively (magnification, ×900). Data are representative of no fewer than three separate experiments.
FIG. 5.
Autolysis of the whole cells of S. mutans UA159 (WT) and the brpA-deficient strain (brpA) grown in BHI medium. Mid-log-phase cultures (OD600 ≅ 0.7) were harvested by centrifugation, washed twice with ice-cold water, resuspended in 0.05 M Tris-HCl (pH 8.0) containing 0.2% Triton X-100, and incubated at 37°C with agitation (200 rpm). The changes of absorbance at 600 nm were determined as described in Materials and Methods. Data represent the average of three separate experiments, and the error bars indicate standard deviations.
Biofilm formation impairment in the brpA strain.
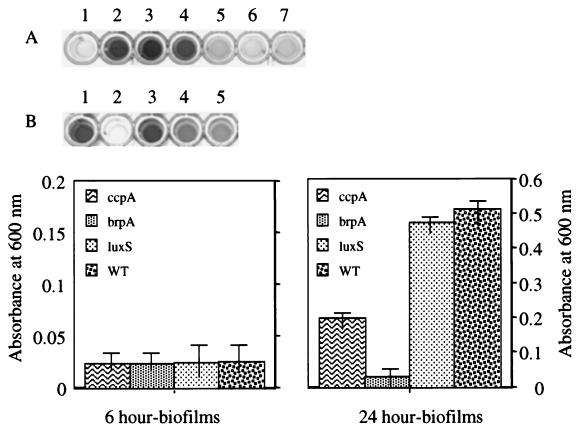
Similar to previous reports on biofilm formation by oral streptococci, S. mutans formed better biofilms when grown on BM medium than when grown on BHI medium (25). After 16 to 24 h of growth in the wells of 96-well microtiter plates, strain TW14 looked different than the parental strain. In contrast to the wild type, which formed an adhesive film of bacteria on the polystyrene surface, the brpA mutant formed large clumps and almost all of the cells were removed during washing of the plates. The results achieved by staining of the biofilms with crystal violet were in agreement with visual observations (Fig. 6). However, measurement of the biofilm-forming capacity after only a 6-h incubation, which primarily measures initial attachment of cells to the polystyrene surface (25), showed that the amounts of biofilm formed by TW14 and by the parent were not different (Fig. 6), indicating that inactivation of brpA resulted mainly in defects in biofilm development rather than in adherence to the polystyrene surface.
FIG. 6.
Biofilm formation of S. mutans UA159 and its derivatives in BM medium. Crystal violet-stained 24-h biofilms of brpA (wells A5 to A7), ccpA (wells B4 and B5), and luxSSm (well B3) mutants and their parental strain UA159 (wells A2 and A4 and well B1). Wells A1 and B2 are uninoculated BM medium as negative control. The graphs show quantitation of the biofilms formed after 6 h (left) and 24 h (right) by ccpA (ccpA), brpA (brpA), and luxSsm (luxS) mutants and the wild-type (WT) strains. See the text for more details. Data are representative of no fewer than three separate experiments. The error bars represent standard deviations.
It is known that autolytic activity regulates the chain-length of chain-forming bacteria. The similarity of BrpA to LytR of B. subtilis could lead one to infer that the increased chain-forming tendencies of S. mutans TW14 were a result of decreased autolytic activity due to inactivation of brpA. Interestingly, Heilmann et al. (19) have recently shown that the Staphylococcus epidermidis autolysin AltE is important for initial adherence to hydrophobic, abiotic surfaces. Also, Loo et al. (25) recently reported that in Streptococcus gordonii many genes that are required for biofilm formation are also involved in peptidoglucan biosynthesis. However, unlike altE-deficient Staphylococcus epidermidis strains, S. mutans TW14 demonstrated an increased rate of autolysis, which could be a result of increased susceptibility of cell walls to autolysins and/or of an increase in an autolytic activity. It is also conceivable that changes in surface protein expression or envelope biogenesis as a result of brpA mutation lead to the autolytic phenotype. Therefore, it is not clear if the basis for diminished biofilm formation is related to alterations in the expression of streptococcal autolysins or changes in expression of cell surface proteins by TW14.
It is noteworthy that, in some ways, the brpA-deficient strain resembles S. aureus LytS-deficient mutants (3). LytS is the sensor/kinase of a two-component signal transduction system, LytSR, and is involved in regulation of autolysis and cell separation. Mutation of lytS resulted in increased autolysis and formation of aggregates in liquid culture, although no data were presented in that study on the impact of inactivation of lytS on biofilm formation. We have identified apparent homologues of S. aureus LytSR in the S. mutans genome database (nt 310037 to 308298 and nt 308401 to 307583, respectively). The predicated S. mutans LytR and LytS proteins were 34 and 42% identical, respectively, to the S. aureus proteins, although the genes are not tightly linked to brpA. Analysis of the relationship of S. mutans lytSR with brpA is currently under way and will provide additional information on the regulation of autolytic activity and biofilm formation.
Biofilm formation by a luxS mutant.
QS plays an important role in initial adherence, as well as in biofilm maturation. Davies et al. (11) showed that Pseudomonas aeruginosa mutants defective in the production of acylated homoserine lactone, an autoinducer of the QS system, formed a thin, flat, unstructured biofilm. Such mutants were later found to be significantly impaired in the ability to attach to glass coverslips compared to the wild-type (12). The S. aureus QS system Agr, which is known to regulate virulence factors, is also involved in the regulation of autolysis, murein hydrolase activity, adherence, and biofilm formation (15, 36). In S. gordonii and S. mutans, the cell-cell signaling Com system controls genetic competence and functions optimally when cells are living in actively growing biofilms (24, 25). A recent study showed that the Com system of S. mutans also affects initial adherence and biofilm architecture (Y. Li, Y. N. Tang, W. Y. Chen, and D. Cvitkovitch, Abstr. 101st Gen. Meet. Am. Soc. Microbiol., abstr. J-8, p. 442, 2001). Therefore, different types of QS systems function in various capacities in the development of bacterial biofilms.
A search of the S. mutans genome was undertaken with a variety of genes of known QS pathways. Using this strategy, we were able to identify a gene predicted to encode a protein with significant homology to LuxS of Vibrio harveyi. V. harveyi possesses two autoinducer-response systems that function in parallel to control the density-dependent expression of bioluminescence (lux) (1). Despite a lack of definition of its exact function, the luxS gene for autoinducer of QS system 2 (AI-2) has been shown to be functional in stimulating expression of the V. harveyi lux operon and has now been shown to be present in several gram-positive and gram-negative bacteria (7, 13, 20, 34). An open reading frame (ORF) of 480 bp in S. mutans, designated luxSSm and located at nt 443111 to 442632 in the chromosome, was identified and predicted to encode a polypeptide of 160 residues that was 33, 31, and 33% identical to the LuxS proteins of E. coli, Salmonella enterica serovar Typhimurium, or V. harveyi, respectively (34).
To determine whether LuxSSm was required for biofilm formation, luxSSm was amplified by PCR from S. mutans UA159 and cloned into pUC19 by EcoRI/XbaI fusion, resulting in pUC:luxS. A luxS mutant was generated by allelic exchange by using an erythromycin resistance determinant (Emr). Briefly, a 1.5-kb DNA fragment containing the Emr gene was released from plasmid pUC:Emr, which is pUC19 carrying an Emr determinant that functions in E. coli and oral streptococci (Z. T. Wen and R. A. Burne, unpublished data), by SstI/HindIII digestions and treated with T4 DNA polymerase to create blunt ends. The determinant was then inserted into pUC:luxS at a unique PstI site, located at nt 161 with respect to the start codon, after PstI digestion and T4 DNA polymerase treatment. The resulting plasmid pUC:luxS:Emr was then used to transform S. mutans UA159, and mutants with disruptions of the luxSSm gene from insertion of the Emr marker by double-crossover recombination were isolated from BHI-erythromycin plates and confirmed by Southern blot analysis (data not shown). When BHI and BM were used to support growth, the luxSSm mutant (strain TW26) grew as well as the wild-type (Fig. 3). To assess the role played by luxSSm in biofilm formation, S. mutans UA159 and TW26 were grown in 96-well microtiter plates on BM medium. As shown in Fig. 6, inactivation of luxSSm did not significantly (<7%) impair biofilm formation after 16 to 24 h of growth.
To demonstrate the expression of luxSSm, we amplified the luxSSm promoter region and fused it with a promoterless chloramphenicol acetyltransferase (CAT) gene (cat). Expression of cat under the direction of luxSSm promoter was analyzed by measuring CAT activity as a function of growth phase and cell density. CAT activity was detectable in the early log phase and was optimal when the cells were in the mid-exponential phase of growth (OD600 ≅ 0.3) (data not shown).
Production of AI-2 has also been reported in Prevotella intermedia, Fusobacterium nucleatum, and Porphyromonas gingivalis, three common isolates of subgingival dental biofilms (13). In Porphyromonas gingivalis, loss of LuxS resulted in upregulation of a putative hemin-acquisition protein and the arginine-specific protease; however, LuxS-deficient strains did not show any impairment of biofilm formation by P. gingivalis on abiotic surface (7), which is similar to what we observed with a luxSSm mutant of S. mutans UA159 (Fig. 6). The luxSSm-encoded protein of S. mutans belongs to the family of proteins that are responsible for production of AI-2 and are highly conserved among gram-positive and gram-negative organisms (34). Interestingly, however, similar to Porphyromonas gingivalis and S. pyogenes, homologues of other V. harveyi QS system 2 components could not be identified in the S. mutans genome, perhaps indicating that there are mechanistic differences in cell-cell signaling between these bacteria or that the functional equivalents of QS system 2 components of S. mutans are too dissimilar to display significant matches in BLAST searches with V. harveyi constituents. Further characterization of luxSSm gene expression in relation to cell density and the role of LuxS in virulence is under way in an effort to determine the potential function of LuxS in S. mutans.
The contribution of CcpA to biofilm formation.
In Bacillus and other gram-positive bacteria, carbon catabolite control protein CcpA is a global regulator that plays a central role in coordinating the metabolism of carbon and energy sources and in regulating other metabolic pathways (26). Given the importance of coordination of carbon source utilization and regulation of metabolic pathways by oral streptococci in the human mouth, we reasoned that CcpA could have an important role in regulation of genes that were important for biofilm formation by S. mutans. We previously isolated an S. mutans ccpA-deficient mutant (strain TW1) and analyzed the impact of ccpA deficiency on regulation of fruA expression (Wen and Burne, unpublished). The ccpA mutant grew as well as the wild-type strain on both BHI and BM media (Fig. 3), and no other morphologic defects were apparent. To elucidate the role that CcpA may play in biofilm formation, the ccpA mutant was grown in BM medium and evaluated by the in vitro biofilm assay. As can be seen in Fig. 6, the loss of CcpA resulted in a 60% decrease of biofilms formed after 26 h on polystyrene surfaces compared to the parental strain. However, no significant difference in biofilms formed after 6 h was noted between UA159 and TW1 (Fig. 6).
The ccpA gene is flanked by pepQ and amyA (32). The upstream gene, pepQ, encodes a dipeptidase and is transcribed in the opposite orientation. The amyA gene encodes an α-amylase with its own promoter. As such, deletional and/or replacement inactivation of ccpA would not have polar effects on expression of the linked genes. Notably, we also have data that demonstrated that single-crossover, nonpolar mutants have the same phenotype as the double-crossover mutants in terms of growth on a variety of carbohydrates tested.
It is becoming clear that the capacity of bacteria to form biofilms varies in response to environmental factors, including carbon source and availability. In Pseudomonas aeruginosa, the catabolite repression control protein Crc has been shown to be a component of a signal transduction pathway that is required for biofilm development (27). Mutation of crc resulted in a decrease in pilA (pilin) transcription, which led to diminished type IV pilus-mediated twitching motility and a markedly reduced capacity to form biofilms. It is well documented that in low-G+C gram-positive bacteria, CcpA binds to a cis-element CRE (for carbon responsive element) near the promoter regions of catabolite repression-sensitive genes, resulting in repression of genes for utilization of secondary carbon source when the cells are also provided with glucose or some other readily metabolizable carbohydrate source. Besides the role in carbon catabolite repression, the B. subtilis CcpA is also required for the activation of many other metabolic operons, including those for acetoin biosynthesis and acetate production by cells grown on glucose (35). Inactivation of ccpA also has other important effects on cellular physiology. For example, a B. subtilis ccpA null mutant displays defects in both the timing and magnitude of the pH decrease when cultured with glucose (35). CcpA also appears to be involved in regulation of the oxidoreduction state and in general stress responses (26). Although the actual function of CcpA in S. mutans remains unclear, sequences highly similar to CREs of B. subtilis CcpA are widely distributed in S. mutans (data not shown). Inactivation of ccpA could cause disruption of the normal flow of carbohydrate, lead to an imbalance of intra- and extra-cellular pH, or alter the ability of the organisms to respond to environmental stresses, all of which could contribute, directly or indirectly, to the observed impairment in biofilm formation by strain TW1.
Summary.
As is the case for many other bacteria inhabiting the oral cavity, S. mutans, the primary etiological agent of dental caries, has evolved as an obligate biofilm-forming organism. The findings generated in this study reinforce that biofilm formation is a process that is linked to several aspects of cellular physiology and biochemistry that function cooperatively in response to multiple environmental cues, such as pH, oxygen, osmolarity, carbon source and availability, cell density, and the presence of solid surfaces, to allow the organisms to adhere to surfaces and to form stable biofilms (9, 12, 14, 26, 28). Although much remains to be learned about the molecular basis for the behavior of the BrpA-deficient strain and how CcpA impacts biofilm formation, BrpA and CcpA appear to be essential for efficient biofilm formation by S. mutans. Because of its unique molecular architecture, it is also reasonable to suggest that BrpA could be considered as a potential target to attenuate the virulence of S. mutans and perhaps other pathogenic streptococci. Since BrpA appears to be part of the cell envelope, BrpA may also be a reasonable candidate for development of a vaccine against S. mutans and dental caries, or chemical agents may be developed that specifically subvert the function of BrpA.
Acknowledgments
We thank J. Lemos and S. Bhagwat for critical suggestions during the preparation of the manuscript. We also thank F. Hagen for help with the phase-contrast microscopy.
This work was supported by NIDCR grant DE12236.
REFERENCES
- 1.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signaling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 2.Bowen, W. H., K. Schilling, E. Giertsen, S. Pearson, S. F. Lee, A. Bleiweis, and D. Beeman. 1991. Role of a cell surface-associated protein in adherence and dental caries. Infect. Immun. 59:4604-4609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunskill, E. W., and K. W. Bayles. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611-618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burne, R. A. 1998. Oral streptococci… products of their environment. J. Dent. Res. 77:445-452. [DOI] [PubMed] [Google Scholar]
- 5.Burne, R. A. 1998. Regulation of gene expression in adherent population of oral streptococci, p. 55-71. In D. J. LeBlanc, M. S. Lantz, and L. M. Switalski (ed.), Microbial pathogenesis: current and emerging issues. Proceedings of the 2nd Annual Indiana Conference. Indiana University Press, Indianapolis, Ind.
- 6.Burne, R. A., Z. T. Wen, Y. M. Chen, and J. E. C. Penders. 1999. Regulation of expression of the fructan hydrolase gene of Streptococcus mutans GS-5 by induction and carbon catabolite repression. J. Bacteriol. 181:2863-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, W. O., Y. Park, R. J. Lamont, R. McNab, B. Barbieri, and D. R. Demuth. 2001. Signaling system in Porphyromonas gingivalis based on a LuxS protein. J. Bacteriol. 183:3903-3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Colby, S. M., R. E. McLaughlin, J. J. Ferretti, and R. R. Russell. 1999. Effect of inactivation of gtf genes on adherence of Streptococcus downei. Oral Microbiol. Immunol. 14:27-32. [DOI] [PubMed] [Google Scholar]
- 9.Costerton, J. W., Z. Lewandowski, D. E. Caldwell, D. R. Korber, and H. M. Lappin-Scott. 1995. Microbial biofilms. Annu. Rev. Microbiol. 49:711-745. [DOI] [PubMed] [Google Scholar]
- 10.Davies, D. G., A. M. Chakrabarty, and G. G. Geesey. 1993. Exopolysaccharide production in biofilms: substratum activation of alginate gene expression by Pseudomonas aeruginosa. Appl. Environ. Microbiol. 59:1181-1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 12.De Kievit, T. R., R. Gillis, S. Marx, C. Brown, and B. H. Iglewski. 2001. Quorum-sensing genes in Pseudomonas aeruginosa biofilms: their role and expression patterns. Appl. Environ. Microbiol. 67:1865-1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frias, J., E. Olle, and M. Alsina. 2001. Periodontal pathogens produce quorum sensing signal molecules. Infect. Immun. 69:3431-3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Froeliger, E. H., and P. Fives-Taylor. 2001. Streptococcus parasanguis fimbria-associated adhesin Fap1 is required for biofilm formation. Infect. Immun. 69:2512-2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto, D. F., and K. W. Bayles. 1998. Opposing roles of the Staphylococcus aureus virulence regulators, Agr and Sar, in Triton X-100- and penicillin-induced autolysis. J. Bacteriol. 180:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia, P., E. Garcia, C. Ronda, R. Lopez, R. Z. Jiang, and A. Tomasz. 1986. Mutants of Streptococcus pneumoniae that contain a temperature-sensitive autolysin. J. Gen. Microbiol. 132:1401-1405. [DOI] [PubMed] [Google Scholar]
- 17.Hazlett, K. R., J. E. Mazurkiewicz, and J. A. Banas. 1999. Inactivation of the gbpA gene of Streptococcus mutans alters structural and functional aspects of plaque biofilm which are compensated by recombination of the gtfB and gtfC genes. Infect. Immun. 67:3909-3914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hazlett, K. R., S. M. Michalek, and J. A. Banas. 1998. Inactivation of the gbpA gene of Streptococcus mutans increases virulence and promotes in vivo accumulation of recombinations between the glucosyltransferase B and C genes. Infect. Immun. 66:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 20.Joyce, E. A., B. L. Bassler, and A. Wright. 2000. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J. Bacteriol. 182:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koskiniemi, S., M. Sellin, and M. Norgren. 1998. Identification of two genes, cpsX and cpsY, with putative regulatory function on capsule expression in group B streptococci. FEMS Immunol. Med. Microbiol. 21:159-168. [DOI] [PubMed] [Google Scholar]
- 22.Kuramitsu, H. K. 1993. Virulence factors of mutans streptococci: role of molecular genetics. Crit. Rev. Oral Biol. Med. 4:159-176. [DOI] [PubMed] [Google Scholar]
- 23.Lazarevic, V., P. Margot, B. Soldo, and D. Karamata. 1992. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J. Gen. Microbiol. 138:1949-1961. [DOI] [PubMed] [Google Scholar]
- 24.Li, Y. H., P. C. Lau, J. H. Lee, R. P. Ellen, and D. G. Cvitkovitch. 2001. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 183:897-908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Loo, C. Y., D. A. Corliss, and N. Ganeshkumar. 2000. Streptococcus gordonii biofilm formation: identification of genes that code for biofilm phenotypes. J. Bacteriol. 182:1374-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moreno, M. S., B. L. Schneider, R. R. Maile, W. Weyler, and M. H. Saier. 2001. Catabolite repression mediated by the CcpA protein in Bacillus subtilis: novel modes of regulation revealed by whole-genome analyses. Mol. Microbiol. 39:1366-1381. [DOI] [PubMed] [Google Scholar]
- 27.O'Toole, G. A., K. A. Gibbs, P. W. Hager, P. V. Phibbs, Jr., and R. Kolter. 2000. The global carbon metabolism regulator Crc is a component of a signal transduction pathway required for biofilm development by Pseudomonas aeruginosa. J. Bacteriol. 182:425-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 29.Prigent-Combaret, C., O. Vidal, C. Dorel, and P. Lejeune. 1999. Abiotic surface sensing and biofilm-dependent regulation of gene expression in Escherichia coli. J. Bacteriol. 181:5993-6002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 31.Sato, Y., Y. Yamamoto, and H. Kizaki. 1997. Cloning and sequence analysis of the gbpC gene encoding a novel glucan-binding protein of Streptococcus mutans. Infect. Immun. 65:668-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Simpson, C. L., and R. R. Russell. 1998. Identification of a homolog of CcpA catabolite repressor protein in Streptococcus mutans. Infect. Immun. 66:2085-2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith, D. J., and M. A. Taubman. 1996. Experimental immunization of rats with a Streptococcus mutans 59-kilodalton glucan-binding protein protects against dental caries. Infect. Immun. 64:3069-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Surette, M. G., M. B. Miller, and B. L. Bassler. 1999. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc. Natl. Acad. Sci. USA 96:1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Turinsky, A. J., T. R. Moir-Blais, F. J. Grundy, and T. M. Henkin. 2000. Bacillus subtilis ccpA gene mutants specifically defective in activation of acetoin biosynthesis. J. Bacteriol. 182:5611-5614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuong, C., H. L. Saenz, and F. Gotz. 2000. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J. Infect. Dis. 182:1688-1693. [DOI] [PubMed] [Google Scholar]
- 37.Yamashita, Y., W. H. Bowen, R. A. Burne, and H. K. Kuramitsu. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]