Abstract
Mycelium growth rate is a quantitative characteristic that exhibits continuous variation. This trait has applied interest, as growth rate is correlated with production yield and increased advantage against competitors. In this work, we studied growth rate variation in the edible basidiomycete Pleurotus ostreatus growing as monokaryotic or dikaryotic mycelium on Eger medium or on wheat straw. Our analysis resulted in identification of several genomic regions (quantitative trait loci [QTLs]) involved in the control of growth rate that can be mapped on the genetic linkage map of this fungus. In some cases monokaryotic and dikaryotic QTLs clustered at the same map position, indicating that there are principal genomic areas responsible for growth rate control. The availability of this linkage map of growth rate QTLs can help in the design of rational strain breeding programs based on genomic information.
Pleurotus ostreatus is a commercially important edible mushroom commonly known as oyster mushroom. This fungus is industrially produced as human food (4), for the bioconversion of agricultural and industrial lignocellulose wastes (1, 27), and as a source of enzymes and other chemicals for industrial and medical applications (9, 10, 23). Its life cycle alternates between monokaryotic (haploid) and dikaryotic (dihaploid) mycelial phases (8). Two compatible monokaryotic hyphae fuse and produce a dikaryotic mycelium in which the two parental nuclei remain independent (dikaryon, heterokaryon) throughout vegetative growth. Fruiting occurs under appropriate environmental conditions, and true diploidy occurs only at the basidia, where karyogamy takes place immediately before the onset of the meiosis that produces four uninucleate basidiospores.
P. ostreatus can be grown on a wide range of substrates, agricultural subproducts, and industrial wastes (11, 25), although pasteurized straw is the most commonly used substrate. Many pests and diseases can cause important yield losses in P. ostreatus cultures. Currently, one of the most important threats to production of this organism is attack by green molds belonging to the genus Trichoderma (30). Differences in green mold susceptibility among P. ostreatus strains have been related to variations in the growth rate of this basidiomycete (2, 29) Additionally, mycelium growth rate has been found to be positively correlated with fruit body yield (28) and thus is an important characteristic for selection in oyster mushroom breeding programs.
The mycelium growth rate exhibits continuous variation and is presumably under the control of a polygenic genetic system. The monokaryotic growth rates of some filamentous fungi have been studied and have been found to be associated with some chromosomal regions (14, 19, 31). However, no quantitative analyses have been performed to determine exhaustively the number, positions, and effects of the genes involved. Quantitative trait locus (QTL) analysis has been applied widely to commercially important plant crops (22, 32, 33) but is rare in studies of mushrooms. Only for Agaricus bisporus has a QTL analysis of resistance to Pseudomonas tolaasii been described (24). In a previous study a linkage map of P. ostreatus was constructed (21), which provided a basis for performing an efficient QTL analysis. In this paper we describe such an analysis carried out with P. ostreatus to determine mycelium growth rates of monokaryotic and dikaryotic populations. To our knowledge, this is the first QTL analysis of the oyster mushroom, and our results could provide the basis for a marker-assisted selective breeding program.
MATERIALS AND METHODS
Fungal strains and populations.
The P. ostreatus strains and protocols used in this work have been described previously (19, 20). Monokaryotic and dikaryotic cultures were grown at 24°C in the dark and were stored in petri dishes at 4°C in solid Eger medium (SEM) (20 g of malt extract per liter, 15 g of agar per liter [7]).
For analysis of monokaryotic growth rates, a segregating population consisting of 80 monokaryons derived from dikaryotic strain N001 of P. ostreatus var. florida was used. This mapping population (population M1) was the same population that was used for construction of the genetic linkage map of P. ostreatus (21). In order to analyze the growth rate in dikaryons, each monokaryon of population M1 was crossed with five monokaryotic testers (PC21, PC23, ME126, MF129, and MF130) to produce five dikaryotic populations (D21, D23, D126, D129, and D130). Testers PC21 and PC23 are the two protoclones (monokaryons carrying only one of the nuclei present in a dikaryotic strain) of wild dikaryotic strain N003 (P. ostreatus var. ostreatus). ME126 is a monokaryon derived from dikaryotic strain N005 (P. ostreatus var. colombinus). MF129 and MF130 are monokaryons derived from dikaryotic strain N006 (P. ostreatus var. sajor-caju) (18).
Monokaryotic growth rate estimation.
Monokaryotic growth rate segregation in population M1 was studied by using SEM or straw as the culture substrate, while the dikaryotic growth rate was determined exclusively in SEM. To evaluate growth rates on SEM, a 16-mm2 piece of actively growing mycelium was placed in the center of a petri dish. The growth rate was estimated by determining the time that elapsed between the time of inoculation and the time that the mycelium reached the rim of the plate (diameter, 9 cm). To evaluate growth rates on straw, glass tubes (diameter, 16 mm; length, 16 cm) containing 10 g of chopped wheat straw (humidity, 70%) were prepared. The inoculum was placed at the top of each tube, and growth was estimated by determining the distance (in centimeters) colonized by the mycelium during the second and third weeks of culture. Three repetitions were performed for samples grown in SEM, and two repetitions were performed for samples grown in wheat straw. Both kinds of samples were incubated at 24°C in the dark.
Statistical analyses.
Growth data were subjected to normality tests (Kolmogorov-Smirnov), and the effects of the genotypes on growth rate were determined by performing a one-way analysis of variance (ANOVA) with SPSS for Windows V8.0.1S software (SPSS Inc., Chicago, Ill.). In addition, a two-way ANOVA was performed for the dikaryotic populations to evaluate the effects of genotypes and testers and the interaction between these two factors. Correlation coefficients for growth rates of the different populations were determined by using the Pearson procedure (SPSS Inc.).
QTLs were mapped by using least-square interval mapping methods developed for backcross progenies as described by Knapp et al. (15, 16). These procedures were applied to the map intervals defined by two consecutive markers present in the P. ostreatus genetic linkage map (21). SAS software (SAS Institute Inc., Cary, N.C.), in particular the PROC NLIN procedure, was used for computational analysis. Multiple regression was used to calculate the percentage of the variance explained by each QTL, estimated by the coefficient of determination (R2). Two-way ANOVA for pairs of selected intervals with a significance threshold of P < 0.005 was used in order to determine digenic interactions.
QTLs were designated as follows: Q(m/d)gr(e/s)x, where m is the monokaryotic state, d is the dikaryotic state, gr is growth rate, e is SEM, s is the straw-based substrate, and x is a consecutive QTL number.
RESULTS
Analysis of mycelium growth rates in monokaryons.
The growth rates of monokaryons on SEM and straw substrates were normally distributed, and no data transformation was needed. The ANOVA showed that the genotype had a significant effect on the mycelium growth rate (data not shown). Table 1 shows the estimated growth rates in the different experiments (averages based on three repetitions). The estimated variation coefficients were similar for the two substrates, and the correlation coefficient for the two media (SEM and straw) was 0.60 (P < 0.01).
TABLE 1.
Estimated mycelium growth rates for monokaryotic and dikaryotic populations
| Population | Substrate | Growth rate
|
||
|---|---|---|---|---|
| Meana | Range of variation | Coefficient of variation (%) | ||
| M1 | Straw | 5.49 cm | 0.6-8.4 cm | 32.7 |
| M1 | SEM | 15.10 days | 10.0-41.0 days | 35.0 |
| D21 | SEM | 9.61 days | 6.0-18.7 days | 25.0 |
| D23 | SEM | 8.85 days | 7.0-12.0 days | 9.9 |
| D126 | SEM | 12.38 days | 8.0-36.7 days | 40.7 |
| D129 | SEM | 9.39 days | 8.0-15.0 days | 13.6 |
| D130 | SEM | 9.43 days | 7.0-12.3 days | 9.5 |
Values were determined as described in Materials and Methods.
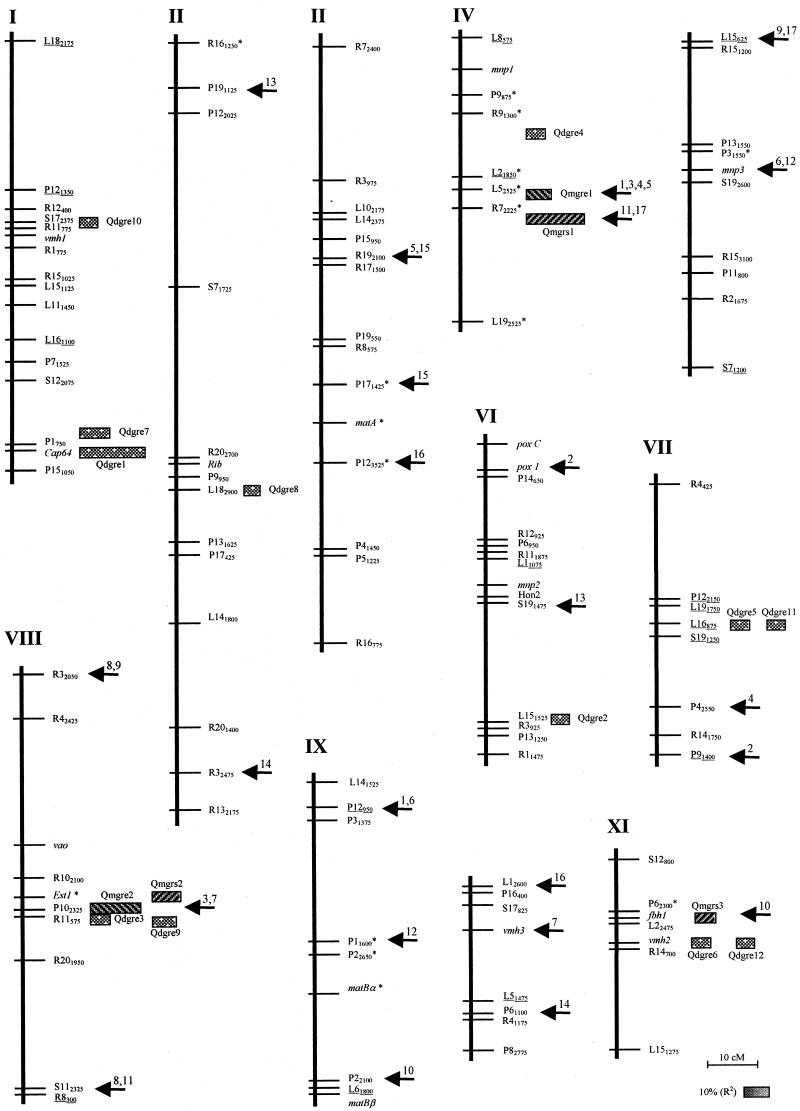
In order to identify the genomic regions involved in growth rate control, the complete genetic linkage map (21) was divided into 109 intervals defined by pairs of consecutive map markers, and each interval was tested for the presence of a QTL effect. For cultures grown on SEM, 11 intervals had a significant effect on growth rate at a confidence level of P < 0.05 and were grouped into two regions (QTLs) located in different linkage groups, linkage groups IV and VIII (Fig. 1). The effects and statistical robustness of the QTLs are shown in Table 2. Taken together, these two QTLs explained 38.46% of the total growth rate variance. This value was higher than the sum of the values for the individual effects, suggesting that there was an epistatic interaction. QTL analyses for the monokaryotic growth rate on the straw-based substrate were performed in a similar way. Three QTLs were detected, which mapped to chromosomes IV, VIII, and XI (Fig. 1) and which accounted for 41.19% of the growth rate variance (Table 2). Two of the straw-based QTLs mapped close to the QTLs found in the experiment performed with SEM. The linkage distance between Qmgre1 and Qmgrs1 (chromosome IV) was 6.1 centimorgans, and the linkage distance between Qmgre2 and Qmgrs2 (chromosome VIII) was 2.6 centimorgans. The relative importance of the members of each QTL pair (R2), however, was inverted in the two substrates.
FIG. 1.
Linkage map of P. ostreatus (21) and locations of QTL and digenic interactions that affect vegetative growth rate in monokaryons and dikaryons. The approximate positions of QTLs are indicated to the right of the markers. The percentages of the phenotypic variation explained by the putative QTLs (R2) are represented by proportional boxes. Significant interactions among QTLs, genes, and other molecular markers are indicated by arrows and are numbered as shown in Table 3. Randomly amplified polymorphic DNA markers on the map are indicated by the primer name and the approximate length of the corresponding amplification product. Randomly amplified polymorphic markers converted to restriction fragment length polymorphism markers are underlined. Genes are in italics. Markers that deviated from the expected 1:1 segregation (P < 0.05) are indicated by asterisks.
TABLE 2.
QTLs for monokaryotic growth rate on SEM and straw
| QTL | Linkage group | Positiona | Probability | Absolute effect | R2 | Total R2 |
|---|---|---|---|---|---|---|
| Qmgre1 | IV | L52525 + 1.4 | 0.017 | 3.83b | 9.90 | |
| Qmgre2 | VIII | P102325 | 0.00011 | 5.03b | 20.27 | 38.46 |
| Qmgrs1 | IV | R72225 + 3.8 | 0.00001 | 2.24c | 23.17 | |
| Qmgrs2 | VIII | R102100 + 3.8 | 0.011 | 1.23c | 11.31 | 41.19 |
| Qmgrs3 | XI | fbh1 | 0.035 | 1.04c | 8.18 |
Map position of QTL at marker locus plus the distance (in centimorgans) towards the bottom of the linkage group (Fig. 1).
Estimated absolute difference between alleles at the QTL, expressed in days.
Estimated absolute difference between alleles at the QTL, expressed in centimeters.
Digenic interactions that significantly (P < 0.005) affected the monokaryotic growth rate in SEM or in straw were studied by analyzing the effects of all possible allelic combinations for each gene pair of genetic markers (monokaryotic QTLs, anonymous markers, and functional genes) present in the genetic linkage map on the estimated monokaryotic growth rates. A total of seven significant digenic interactions that affected the growth rate on the SEM substrate and 10 significant digenic interactions that affected the growth rate on straw were detected (Table 3). A significant digenic interaction (P = 0.00093) between Qmgre1 and Qmgre2 was detected, confirming the epistatic effect between the two QTLs described above. All of the significant digenic interactions that affected the monokaryotic growth rate are shown in Fig. 1.
TABLE 3.
Digenic interactions of monokaryons for mycelium growth rates on SEM and straw substrate
| No. | Interaction | Map location | Probability | Means for the following genotypic classesa:
|
|||
|---|---|---|---|---|---|---|---|
| 00 | 01 | 10 | 11 | ||||
| Interactions on SEM | |||||||
| 1 | Qmgre1 × P12950 | IV-IX | 0.00052 | 14.88 | 23.38 | 14.49 | 13.97 |
| 2 | pox1 × P91400 | VI-VII | 0.00060 | 18.83 | 13.99 | 13.15 | 16.46 |
| 3 | Qmgre1 × Qmgre2 | IV-VIII | 0.00093 | 15.41 | 28.08 | 12.40 | 16.45 |
| 4 | Qmgre1 × P42550 | IV-VII | 0.00286 | 22.48 | 15.71 | 13.48 | 14.64 |
| 5 | Qmgre1 × R192100 | IV-III | 0.00327 | 13.89 | 21.48 | 14.21 | 14.12 |
| 6 | P12950 × mnp3 | IX-V | 0.00422 | 16.16 | 13.64 | 14.04 | 18.07 |
| 7 | vmh3 × Qmgre2 | X-VIII | 0.00457 | 14.42 | 15.02 | 12.96 | 20.05 |
| Interactions on straw-based medium | |||||||
| 8 | R32050 × S112325 | VIII-VIII | 0.00028 | 6.70 | 4.77 | 4.94 | 5.86 |
| 9 | R32050 × L15625 | VIII-V | 0.00057 | 5.41 | 5.91 | 6.14 | 3.90 |
| 10 | P22100 × Qmgrs3 | IX-XI | 0.00058 | 6.32 | 3.61 | 5.70 | 5.70 |
| 11 | Qmgrs1 × S112325 | IV-VIII | 0.00145 | 3.52 | 4.54 | 6.74 | 5.35 |
| 12 | P11600 × mnp3 | IX-V | 0.00161 | 4.19 | 5.76 | 6.06 | 4.90 |
| 13 | S191475 × P191125 | VI-II | 0.00207 | 5.07 | 6.67 | 5.52 | 4.64 |
| 14 | R32475 × P61100 | II-X | 0.00222 | 5.90 | 4.19 | 5.41 | 6.12 |
| 15 | R192100 × P171425 | III-III | 0.00278 | 5.76 | 4.36 | 4.36 | 5.98 |
| 16 | L12600 × P123525 | X-III | 0.00303 | 5.04 | 6.32 | 5.93 | 4.66 |
| 17 | Qmgrs1 × L15625 | IV-V | 0.00397 | 5.02 | 3.00 | 5.95 | 6.18 |
Phenotypic means for genotypic classes 00, 01, 10, and 11 obtained by using the two markers involved in the digenic interaction (0, marker absent; 1, marker present).
Analysis of mycelium growth rates in dikaryons.
The analysis of QTLs controlling the vegetative growth rate in dikaryons was performed by using SEM as described above for identification of monokaryotic QTLs. Five different dikaryotic populations were constructed by crossing individuals belonging to population M1 with each of five compatible testers. The growth rate characteristics of these populations are shown in Table 1. Compared to monokaryons, the dikaryotic populations grew faster and their variation coefficients were more dispersed. All possible pairwise correlation values for growth rates of the different dikaryotic populations were calculated (Table 4). Significant but relatively low correlation coefficients between certain populations were found; however, the growth rates of other population pairs were completely unrelated. The ANOVA revealed highly significant effects of tester strain (between populations) and genotype (within populations) on growth rates and a highly significant interaction between tester and genotype (data not shown), which was reflected in the correlation coefficients calculated (Table 4).
TABLE 4.
Correlation coefficients for dikaryotic growth rates
| Population | Correlation coefficient
|
|||
|---|---|---|---|---|
| D23 | D126 | D129 | D130 | |
| D21 | 0.40 (<0.01)a | 0.32 (0.04) | 0.01 (0.92) | 0.12 (0.29) |
| D23 | 0.19 (0.09) | 0.26 (0.02) | 0.36 (<0.01) | |
| D126 | −0.05 (0.67) | 0.06 (0.57) | ||
| D129 | 0.24 (0.04) | |||
The values in parentheses are P values.
Twelve significant dikaryotic growth rate QTLs were detected (Table 5). Their map positions appeared to be scattered across the P. ostreatus genome (Fig. 1). Only four of the five monokaryotic testers gave rise to populations in which significant QTLs could be identified, and no QTLs were detected in population D129. The QTLs identified could explain between 19.51 and 39.14% (total R2) of the total variation in growth rate (Table 5). The strongest QTL (Qdgre1) was identified in population D1 (tester PC21), accounted for the 25.90% of the growth rate variance, and had the highest confidence level (probability value) of all the QTLs identified in this work. In some cases two dikaryotic QTLs identified by using different monokaryotic testers mapped to closely linked positions (Table 5; Fig. 1). Especially noteworthy are the QTL clusters found on chromosome VIII, which contained Qmgre2, Qmgrs2, Qdgre3, and Qdgre9, and the cluster mapping to the central region of chromosome XI, where Qmgrs3, Qdgre6, and Qdgre12 appeared to be located. When QTL locations in monokaryons and dikaryons were compared, five dikaryotic QTLs mapped very close to monokaryotic QTLs and seven dikaryotic QTLs had no genetically linked relatives in the monokaryons.
TABLE 5.
QTLs for dikaryotic growth rate on SEM when different tester lines were used
| Dikaryotic population | Tester | QTL | Linkage group | Positiona | Probability | Absolute effect (days)b | R2 | Total R2 |
|---|---|---|---|---|---|---|---|---|
| D21 | PC21 | Qdgre1 | I | cap64 | 0.00002 | 2.45 | 25.90 | |
| Qdgre2 | VI | L151525 | 0.026 | 1.43 | 7.11 | 39.14 | ||
| Qdgre3 | VIII | R11575 | 0.048 | 1.34 | 7.58 | |||
| D23 | PC23 | Qdgre4 | IV | R91300 + 4.0 | 0.032 | 0.59 | 7.42 | |
| Qdgre5 | VII | L16875 | 0.034 | 0.52 | 7.24 | 19.51 | ||
| Qdgre6 | XI | vmh2 | 0.033 | 0.51 | 7.48 | |||
| D126 | ME126 | Qdgre7 | I | S122075 + 10.7 | 0.0038 | 3.85 | 11.81 | |
| Qdgre8 | II | L182900 | 0.045 | 2.91 | 6.26 | 25.75 | ||
| Qdgre9 | VIII | R11575 + 1.0 | 0.013 | 3.40 | 9.15 | |||
| D130 | MF130 | Qdgre10 | I | S17575 + 0.3 | 0.041 | 0.51 | 7.04 | |
| Qdgre11 | VII | L16875 | 0.047 | 0.53 | 7.15 | 20.83 | ||
| Qdgre12 | XI | vmh2 | 0.042 | 0.50 | 7.00 |
Map position of QTL at marker locus plus the distance (in centimorgans) towards the bottom of the linkage group (Fig. 1).
Estimated absolute difference between alleles at the QTL.
Candidate gene for the QTL cluster in chromosome VIII.
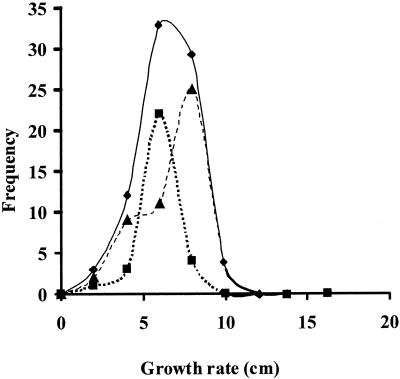
The central region of chromosome VIII is peculiar because it contains four independently identified QTLs, exhibits some statistically significant digenic interactions, and has a functional gene coding for a member of the esterase family (est1). In order to determine if the est1 gene could be involved in growth rate variation, the growth rates of the monokaryons belonging to population M1 were grouped on the basis of their est1 alleles. Figure 2 shows that the two est1 alleles allowed differentiation of two monokaryotic subpopulations whose growth rates differed statistically; monokaryons carrying the est1-1 allele had a significantly lower growth rate than monokaryons harboring the est1-2 allele.
FIG. 2.
Continuous variation of the monokaryotic growth rate in straw-based substrate of the total monokaryotic population (⧫) and of the two populations differing in the est1 allele, est1-1 (▴) and est1-2 (▪).
DISCUSSION
The study of the genetic basis of quantitative characters can be approached by contrasting the phenotypes displayed by different genotypic classes defined either by a single marker locus or by genomic intervals limited by two flanking markers. In the present study, we attempted to map several mycelium growth rate QTLs and assign them to chromosome intervals defined by the molecular markers previously used to construct the genetic linkage map of P. ostreatus var. florida (21). In this study we considered the mycelium growth phenotypic differences in monokaryotic and dikaryotic progeny derived from P. ostreatus strain N001. Despite the small size of the mapping populations (80 individuals), a number of statistically significant QTLs were detected.
The correlation value for the growth rates of monokaryons cultured on SEM and straw substrates was relatively low. This could have been the result of differences in nutrient availability and could have reflected weakly complementary abilities to use simple or complex substrates. The occurrence of QTL pairs mapping to the same position was initially unexpected as previous studies carried out with A. bisporus growing on complete or minimal medium revealed QTLs located on different chromosomes (13). In our study, however, the two substrates used could be considered complex media, although they contained different available carbon sources. The members of the two main monokaryotic QTL clusters (chromosomes IV and VIII) could correspond to different genes or gene families since their map positions are not exactly coincident (Fig. 1; Table 2). It should be considered, however, that the exact position of a QTL is statistically determined by individual growth values and may be influenced by other QTLs, interactions between QTL alleles, and interactions between the environment and specific genotypes. Therefore, these two QTL clusters could correspond to two genetic factors and probably to enzymes having functions in general metabolism.
The relative importance of the QTLs in SEM and straw appeared to be opposite, indicating that growth on the two substrates is influenced by different QTLs or by different interactions between genes and substrates. Furthermore, different digenic interactions were observed with the two substrates (Table 3). On the other hand, a certain concordance between QTLs detected in monokaryons and markers exhibiting distorted segregation in the linkage map (21) was observed, which could reflect how establishment of the mapping population was conditioned by the different growth rates of monokaryons derived from single spores.
Based on the high degree of concordance between the mycelium growth rates in SEM and straw, a preliminary selection test for active substrate colonization by monokaryotic strains can be performed easily in a petri dish before the strains are tested under industrial conditions in breeding programs.
For mapping purposes the use of haploid populations (monokaryons) of this species is convenient. However, evaluation of several important agronomic traits for commercial applications is possible only with the dikaryotic stage. For this reason the monokaryotic mapping population was used to construct different dikaryotic populations with various compatible testers. The use of testers belonging to other P. ostreatus varieties allowed us to determine the general and specific combining abilities of P. ostreatus var. florida monokaryons. Measurement of these genetic abilities is relevant for establishment of efficient breeding programs (3). Since all the dikaryons in each population have the same nucleus (the tester nucleus), the QTLs identified in this work reflect the effect of the segregating nuclei plus their interactions with the constant tester nucleus, which differ depending on this nucleus. Hence, the QTLs identified in the different dikaryotic populations are different.
The 12 QTLs detected in the four dikaryotic populations exhibited moderate correspondence, indicating that the different monokaryotic testers had recessive alleles at the Qdgre loci. This allowed expression of the corresponding dominant alleles present in P. ostreatus var. florida. As discussed above for monokaryons, some of the QTLs detected in the dikaryotic populations probably correspond to the same gene or gene family since they map to identical locations (e.g., Qdgre5 to Qdgre11 and Qdgre6 to Qdgre12). Furthermore, the dikaryotic populations contained QTLs which were not detected in the analysis of monokaryons. The main dikaryotic QTLs detected in this analysis (Qdgre1 and Qdgre7) map to very close or overlapping regions at the end of chromosome I (Fig. 1), and they have the highest significance. cap64 is the only functional gene found at this position (A. G. Pisabarro, unpublished data). This gene codes for a protein that is very similar to Cryptococcus neoformans Cap64, which is required for capsule formation and virulence in this pathogenic fungus (5), although its actual biological function is not known yet. Accordingly, the effect of P. ostreatus Cap64 on the dikaryotic growth rate deserves more thorough study.
QTL analyses involving functional genes may be useful for studying the complexity of the growth rate using a candidate gene approach (17, 26). Understanding the genetic basis of growth rate control is important due to its relationship with yield (28) and tolerance to Trichoderma. Furthermore, disease resistance is a priority in breeding programs for edible mushrooms. In this context, identification of single genes putatively responsible for quantitative effects is very important. The results presented here suggest that the est1 gene plays an important role in the ability of P. ostreatus to colonize a substrate. The relationship between est1 and lipid biosynthesis must be clarified in this fungus, as it has been in other fungal systems (6). The identification of candidate genes and the analysis of the genetic factors controlling the continuous variation of characters provide an opportunity to obtain a new level of understanding of the regulation of gene expression and genome organization, which are factors that can be included in the general concept of genetical genomics (12). The genetic control of growth rate and other traits, such as yield, color, and precocity, is currently being studied in our laboratory.
Acknowledgments
This work was supported by research project BIO99-0278 of the Comisión Nacional de Ciencia y Tecnología, by a research project of the Gobierno de Navarra, and by funds from the Universidad Pública de Navarra (Pamplona, Spain).
REFERENCES
- 1.Ballero, M., E. Mascia, A. Rescigno, and E. S. D. Teulada. 1990. Use of Pleurotus for transformation of polyphenols in waste waters from olive presses into proteins. Micol. Ital. 19:39-41. [Google Scholar]
- 2.Beyer, D. M., P. J. Wuest, and J. J. Kremser. 2000. Evaluation of epidemiological factors and mushroom substrate characteristics influencing the occurrence and development of Trichoderma green mold, p. 633-640. In L. J. L. D. van Griensven (ed.), Science and cultivation of edible fungi, vol. 2. A. A. Balkema, Rotterdam, The Netherlands. [Google Scholar]
- 3.Bos, I., and P. Caligari. 1995. Selection methods in plant breeding, 1st ed. Chapman & Hall, London, United Kingdom.
- 4.Chang, R. 1996. Functional properties of edible mushrooms. Nutr. Rev. 54:91-93. [DOI] [PubMed] [Google Scholar]
- 5.Chang, Y. C., L. A. Penoyer, and K. J. Kwon-Chung. 1996. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect. Immun. 64:1977-1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheng, J., T. S. Park, A. S. Fischl, and X. S. Ye. 2001. Cell cycle progression and cell polarity require sphingolipid biosynthesis in Aspergillus nidulans. Mol. Cell. Biol. 21:6198-6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eger, G. 1976. Pleurotus ostreatus-breeding potential of a new cultivated mushroom. Theor. Appl. Genet. 47:155-163. [DOI] [PubMed] [Google Scholar]
- 8.Eugenio, C. P., and N. A. Anderson. 1968. The genetics and cultivation of Pleurotus ostreatus. Mycology 60:627-634. [Google Scholar]
- 9.Giardina, P., V. Aurilia, R. Cannio, L. Marzullo, A. Amoresano, R. Siciliano, P. Pucci, and G. Sannia. 1996. The gene, protein and glycan structures of laccase from Pleurotus ostreatus. Eur. J. Biochem. 235:508-515. [DOI] [PubMed] [Google Scholar]
- 10.Gunde-Cimerman, N. 1999. Medicinal value of the genus Pleurotus (Fr.) P. Karst. (Agaricales s.l. Basidiomycetes). Int. J. Med. Mushrooms 1:69-80. [Google Scholar]
- 11.Jadhav, A. B., P. K. Bagal, and S. W. Jadhav. 1996. Effects of different substrates on yield of oyster mushroom. J. Maharashtra Agric. Univ. 21:424-426. [Google Scholar]
- 12.Jansen, R. C., and J. Nap. 2001. Genetical genomics: the added value from segregation. Trends Genet 17:388-391. [DOI] [PubMed] [Google Scholar]
- 13.Kerrigan, R. W. 2000. A brief history of marker assisted selection in Agaricus bisporus, p. 183-189. In L. J. L. D. van Griensven (ed.), Science and cultivation of edible fungi, vol. 1. A. A. Balkema, Rotterdam, The Netherlands. [Google Scholar]
- 14.Kerrigan, R. W., J. C. Royer, L. M. Baller, Y. Kohli, P. A. Horgen, and J. B. Anderson. 1993. Meiotic behavior and linkage relationships in the secondarily homothallic fungus Agaricus bisporus. Genetics 133:225-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Knapp, S. J., and W. C. Bridges. 1990. Programs for mapping quantitative trait loci using flanking molecular markers and nonlinear models. J. Hered. 81:234-235. [Google Scholar]
- 16.Knapp, S. J., W. C. Bridges, and D. Birkes. 1990. Mapping quantitative trait loci using molecular marker linkage maps. Theor. Appl. Genet. 79:583-592. [DOI] [PubMed] [Google Scholar]
- 17.Lander, E. S., and N. J. Stork. 1994. Genetic dissection of complex traits. Science 265:2037-2048. [DOI] [PubMed] [Google Scholar]
- 18.Larraya, L., M. M. Peñas, G. Pérez, C. Santos, E. Ritter, A. G. Pisabarro, and L. Ramírez. 1999. Identification of incompatibility alleles and characterisation of molecular markers genetically linked to the A incompatibility locus in the white rot fungus Pleurotus ostreatus. Curr. Genet. 34:486-493. [DOI] [PubMed] [Google Scholar]
- 19.Larraya, L. M., G. Pérez, I. Iribarren, J. A. Blanco, M. Alfonso, A. G. Pisabarro, and L. Ramírez. 2001. Relationship between monokaryotic growth rate and mating type in the edible basidiomycete Pleurotus ostreatus. Appl. Environ. Microbiol. 67:3385-3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larraya, L. M., G. Pérez, M. M. Peñas, J. P. Baars, T. S. P. Mikosch, A. G. Pisabarro, and L. Ramírez. 1999. Molecular karyotype of the white rot fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 65:3413-3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larraya, L. M., G. Pérez, E. Ritter, A. G. Pisabarro, and L. Ramírez. 2000. Genetic linkage map of the edible basidiomycete Pleurotus ostreatus. Appl. Environ. Microbiol. 66:5290-5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin, H., H. Qian, J. Zhuang, J. Lu, S. Min, Z. Xiong, N. Huang, and K. Zheng. 1996. RFLP mapping of QTLs for yield and related characters in rice (Oryza sativa L.). Theor. Appl. Genet. 92:920-927. [DOI] [PubMed] [Google Scholar]
- 23.Marzullo, L., R. Cannio, P. Giardina, M. T. Santini, and G. Sannia. 1995. Veratryl alcohol oxidase from Pleurotus ostreatus participates in lignin biodegradation and prevents polymerization of laccase-oxidized substrates. J. Biol. Chem. 270:3823-3827. [DOI] [PubMed] [Google Scholar]
- 24.Moquet, F., C. Desmerger, M. Mamoun, M. Ramos-Guedes-Lafargue, and J. M. Olivier. 1999. A quantitative trait locus of Agaricus bisporus resistance to Pseudomonas tolaasii is closely linked to natural cap color. Fungal Genet. Biol. 28:34-42. [DOI] [PubMed] [Google Scholar]
- 25.Pani, B. K., S. N. Panda, and S. R. Das. 1997. Utilization of some agricultural bioproducts and other wastes for sporophore production of oyster mushroom. Orissa J. Hortic. 25:36-39. [Google Scholar]
- 26.Prioul, J. P., S. Pelleschi, M. Séne, C. Thévenot, M. Causse, D. de Vienne, and A. Leonardi. 1999. From QTLs for enzyme activity to candidate genes in maize. J. Exp. Bot. 337:1281-1288. [Google Scholar]
- 27.Puniya, A. K., K. G. Shah, S. A. Hire, R. N. Ahire, M. P. Rathod, and R. S. Mali. 1996. Bioreactor for solid-state fermentation of agro-industrial wastes. Indian J. Microbiol. 36:177-178. [Google Scholar]
- 28.Salmones, D., R. Gaitán-Hernández, R. Pérez, and G. Guzmán. 1997. Studies on genus Pleurotus. VIII. Interaction between mycelial growth and yield. Rev. Iberoam. Micol. 14:173-176. [PubMed] [Google Scholar]
- 29.Savoie, J. M., P. Delpech, C. Billette, and G. Mata. 2000. Inoculum adaptation changes the outcome of the competition between Lentinula edodes and Trichoderma spp. during shiitake cultivation on pasteurized wheat straw, p. 667-674. In L. J. L. D. van Griensven (ed.), Science and cultivation of edible fungi, vol. 2. A. A. Balkema, Rotterdam, The Netherlands. [Google Scholar]
- 30.Sharma, S. R., and B. Vijay. 1996. Yield loss in Pleurotus ostreatus spp. caused by Trichoderma viride. Mushroom Res. 5:19-22. [Google Scholar]
- 31.Simchen, G. 1966. Monokaryotic variation and haploid selection in Schizophyllum commune. Heredity 21:241-263. [DOI] [PubMed] [Google Scholar]
- 32.Staub, J. E., F. C. Serquen, and M. Gupta. 1996. Genetic markers, map construction, and their application in plant breeding. HortScience 31:729-738. [Google Scholar]
- 33.Thomas, W., W. Powell, J. Swanston, R. Ellis, K. Chalmers, U. Barua, P. Jack, V. Lea, B. Forster, R. Waugh, and D. Smith. 1996. Quantitative trait loci for germination and malting quality characters in a spring barley cross. Crop Sci. 36:265.. [DOI] [PubMed] [Google Scholar]