Abstract
The development of assays for quantitative analysis of the relative transcript levels of ABC transporter genes by real-time reverse transcription-PCR (RT-PCR) might provide important information about multidrug resistance in filamentous fungi. Here, we evaluate the potential of real-time RT-PCR to quantify the relative transcript levels of ABC transporter Atr genes from Aspergillus nidulans. The AtrA to AtrD genes showed different and higher levels in the presence of structurally unrelated drugs, such as camptothecin, imazalil, itraconazole, hygromycin, and 4-nitroquinoline oxide. We also verified the relative transcript levels of the Atr genes in the A. nidulans imazalil-resistant mutants. These genes displayed a very complex pattern in different ima genetic backgrounds. The imaB mutant has higher basal transcript levels of AtrB and -D than those of the wild-type strain. The levels of these two genes are comparable when the imaB mutant is grown in the presence and absence of imazalil. The imaC, -D, and -H mutants have higher basal levels of AtrA than that of the wild type. The same behavior is observed for the relative transcript levels of AtrB in the imaG mutant background.
The frequency of life-threatening fungal infections is rising worldwide. The need for effective antifungal therapies has been more acute since the emergence of AIDS and AIDS-related complex, which are often associated with opportunistic fungal infections (15). The failure of drugs to treat fungal infections combined with improvements in performance and standardization of antifungal susceptibility testing have drawn attention to the problem of antifungal drug resistance. It is now clear that antifungal agents can create clinical and epidemiological situations that are analogous to those found with antibiotic-resistant bacteria (1, 7, 8, 9, 18, 20, 24, 25). There are several known genetic determinants associated with multidrug resistance in fungi (for reviews, see references 3, 5, 11, 21, 23, and 29). Typical gene products responsible for multiple drug resistance in these organisms are ABC (ATP-binding cassette) transport proteins that are responsible for the efflux of toxic compounds (for a review, see reference 16).
Fungi are becoming very important human pathogens, and there are few drugs that can inhibit fungal growth. The epidemiological combination of these two factors predicts that very soon fungi will acquire new genetic determinants that will provide multidrug resistance. Actually, there are already several reports showing the emergence of multidrug resistance in fungal pathogens (for a review, see reference 29). The evidence so far points to ABC transporter-encoding genes playing an important role in these phenomena. The development of assays for quantitative analysis of the relative transcript levels of ABC transporter genes by real-time reverse transcription-PCR (RT-PCR) might provide important information about multidrug resistance in filamentous fungi. Quantitative nucleic acid sequence analysis has had an important role in many fields of biological research. PCR has proven to be a powerful tool for quantitative nucleic acid analysis. Several amplification methods, such as quantitative PCR, competitive PCR, and more recently 5′ nuclease assays, have been developed to measure target templates quantitatively (for a review, see reference 22). The real-time PCR method offers several advantages. (i) It is a highly sensitive assay. (ii) It is performed in a closed-tube system and requires no post-PCR manipulation of the sample, thereby increasingly the sample throughput. (iii) It supports the use of a normalization gene for quantitative PCR or housekeeping genes for quantitative RT-PCR controls (17).
Aspergillus nidulans is a filamentous fungus of considerable biological interest which provides an excellent model system for investigating different aspects of drug resistance in filamentous fungi. Here, we evaluate the potential of real-time RT-PCR to quantify the relative transcript levels of ABC transporter Atr genes from A. nidulans.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The A. nidulans strains used were R21 (yA1 pabaA1), ima532 (biA1 acrA1 imaB9), ima533 (biA1 acrA1 imaC10), ima534 (biA1 acrA1 imaD13), ima535 (biA1 acriA1 imaG18), and ima537 (biA1 acrA1 imaH19). The ima mutants were obtained from the Fungal Genetics Stock Center (www.fgsc.net). The medium used was the complete YG medium (2% glucose, 0.5% yeast extract, and trace elements). When necessary, agar was added to a final concentration of 2%. Trace elements, vitamins, and nitrate salts were used as described by Kafer (19).
RNA isolation.
Conidia (108 ml−1) were used to inoculate liquid cultures which were incubated in a reciprocal shaker at 37°C for either 8 or 12 h. Mycelia were aseptically transferred to fresh YG medium in the presence or absence of drugs for 8 h, respectively. The following chemicals were used: 25 μM camptothecin, 0.3 μg of imazalil per ml, 0.2 μg of itraconazole per ml, 100 μg of hygromycin per ml, and 2.5 μg of 4-nitroquinoline oxide (4-NQO) per ml. A. nidulans mycelia were harvested by filtration through Whatman no. 1 filter paper, washed thoroughly with sterile water, quickly frozen in liquid nitrogen, and disrupted by grinding, and total RNA was extracted with Trizol (Life Technologies). To verify the RNA integrity, 20 μg of RNA from each treatment was then fractionated in a 2.2 M formaldehyde-1.2% agarose gel, stained with ethidium bromide, and then visualized with UV light. The presence of intact 28S and 18S rRNA bands (the first band twice as intense as the second band) was used as the criterion to verify if the RNA was degraded. About 500 μg of total RNA was obtained for each milligram of mycelia. RNase-free DNase treatment was done in a final volume of 100 μl containing 1× buffer (40 mM Tris-HCl [pH 7.5] and 6 mM MgCl2), 1 μl of RNasin (40 U/μl; Promega), 10 μl of RNase-free DNase (1 U/μl; Promega or Life Technologies), 5 μl of 200 mM dithiothreitol, and 10 μg of total RNA. The reaction mixture was incubated at 37°C for 60 min, and the reaction was stopped by incubating at 70°C for 30 min.
RT-PCRs and PCRs.
PCRs and RT-PCRs were performed using an ABI Prism 7700 Sequence Detection System (Perkin-Elmer Applied Biosystems). Taq-Man EZ RT-PCR kits (Applied Biosystems) were used for RT-PCRs. The RT-PCR thermal cycling conditions were as follows: an initial step at 50°C for 2 min; 30 min at 60°C for RT; 95°C for 5 min; and 40 cycles, with 1 cycle consisting of 20 s at 94°C and 1 min at 60°C. The Taq-Man PCR Reagent kit was used for PCRs. The PCR thermal cycling conditions were as follows: an initial step at 50°C for 2 min; 10 min at 95°C; and 40 cycles, with 1 cycle consisting of 15 s at 95°C and 1 min at 60°C.
In all experiments, appropriate negative controls containing no template DNA or RNA were subjected to the same procedure to exclude or detect any possible contamination or carryover. Each sample was repeated at least three times. All reaction mixtures were analyzed by agarose gel electrophoresis to confirm that only one PCR product was synthesized. The results were normalized using the Cts obtained for the tubC (β-tubulin) RNA amplifications run on the same plate. Ct is defined as the first amplification cycle at which fluorescence indicating PCR products becomes detectable. The tubC RNA is an endogenous control that was used to normalize the samples for differences in the amounts of total RNA added to each cDNA reaction mixture. In real-time PCR analysis, quantification is based on the threshold cycle, which is inversely proportional to the logarithm of the initial copy number (27). Real-time PCR was performed for each primer pair and its Taq-Man probe, using known amounts of target obtained by serial dilutions of genomic DNA as the template over several orders of magnitude. The initial template amount used in the parallel reaction mixtures was estimated, using 30 Mb as the A. nidulans genome size. A linear relationship was obtained by plotting the threshold cycle against the logarithm of known amount of initial template. The equation of the line that best fits the data was determined by regression analysis. The R2 value was calculated for each data set to estimate the accuracy of the real-time PCR with Taq-Man as a quantification method. The amounts of target copies contained in an unknown sample were determined by extrapolation from the linear regression of the standard curve obtained for each primer-Taq-Man set (13). For a description of the primers and Taq-Man fluorescent probes (Applied Biosystems) used in this work, see Table 1.
TABLE 1.
Primers and fluorescent probes used in this work
| Primer or probe | Sequencea | Gene location (accession no.) |
|---|---|---|
| tubCf | 5′-CGGAAACTGGCCGTCAATAT-3′ | 1252-1271 (M17520.1) |
| tubCr | 5′-GGGCAAACCCGACCATAAA-3′ | 1297-1315 (M17520.1) |
| ptubC | 5′-6FAM-TCCCTTCCCGCGGTTGCATTT-TAMRA-3′ | 1275-1295 (M17520.1) |
| Actinf | 5′-TTCCGTGCTTGTCCCTGAAC-3′ | 462-481 (M22869.1) |
| Actinr | 5′-AAGGACGGGTAGGTCGTTGC-3′ | 505-524 (M22869.1) |
| Pactin | 5′-VIC-CTCGCCTGGTCTCGCCGCTGT-TAMRA-3′ | 483-502 (M22869.1) |
| AtrAf | 5′-TCCCTCGCATGACAATCC-3′ | 1542-1560 (Z68904.1) |
| AtrAr | 5′-GAGAGAGAATGAGGCGGAGTTG-3′ | 1596-1618 (Z68904.1) |
| pAtrA | 5′-TET-ACGATGACCTAGAAGACGCAACCCTTGAC-TAMRA-3′ | 1565-1594 (Z68904.1) |
| AtrBf | 5′-CGCTTTGCTATCTAACGAGTTTCAC-3′ | 2736-2761 (Z68905.1) |
| AtrBr | 5′-CGGCGGGCACAAGGT-3′ | 2789-2804 (Z68905.1) |
| patrB | 5′-VIC-AAGATCATTCCCTGCGTGGGCACC-3′ | 2764-2788 (Z68905.1) |
| AtrCf | 5′-TGGCATCCCGCTACATGAT-3′ | 3954-3973 (AF082072.1) |
| AtrCr | 5′-GGGTTGGGTTCTGGGTGAC-3′ | 4009-4028 (AF082072.1) |
| pAtrC | 5′-VIC-CCGTGCCCGGATCGGCCT-TAMRA-3′ | 3990-4008 (AF082072.1) |
| AtrDf | 5′-CATCCGATCTCGACGGAAAA-3′ | 3550-3569 (AF071411.1) |
| AtrDr | 5′-GCGCTGGAAACAGACTTGTG-3′ | 3616-3597 (AF071411.1) |
| pabcD | 5′-TET-CAACCATTGACCGCACGGGCA-TAMRA-3′ | 3574-3594 (AF071411.1) |
Abbreviations: 6FAM, 6-carboxyfluorescein; TAMRA, 6-carboxy-N,N,N′,N′-tetramethylrhodamine; VIC, trademark product from Applied Biosystems; TET, 6-carboxy-4,7,2′,7′-tetrachlorofluorescein.
RESULTS
Quantitative analysis of the relative transcript levels of ABC transporter genes.
The presence of contaminating DNA is always a question of major concern in quantitative RT-PCR experiments. Before initiating our quantitative RT-PCR experiments, we decided to evaluate the DNA contamination in our A. nidulans total RNA by amplifying the tubC template and an intron of the actin gene (for a description of the primers and fluorescent probes as well as their gene location, see Table 1). Both PCR products yielded a single band with the expected size as visualized on an agarose gel (data not shown). The number of A. nidulans tubulin (and later for actin and Atr genes) DNA copies in each reaction mixture was calculated by interpolating the mean Ct for the test sample into the standard curve generated with A. nidulans genomic DNA. Increasing amounts of untreated total RNA provided lower Cts (with 25 ng of RNA, Ct = 33; with 800 ng of RNA, Ct = 28). When RNA is treated with DNase, the Cts increase to 38 to 40 (25 to 800 ng), indicating that the previous values obtained for RNA not treated with DNase were due in part to contamination of DNA. DNase inhibition by heat (70°C for 30 min) was more suitable than EDTA inactivation; EDTA treatment proved to inhibit RT-PCR (data not shown). We were unable to amplify an intron from the A. nidulans actin gene, thus proving the efficiency of the DNase treatment (data not shown). Our results indicate that the treatment of total RNA with RNase-free DNase improves the reliability of the quantitative method considerably. Thus, all the RNAs used in this work were previously RNase-free and treated with DNase before real-time PCRs.
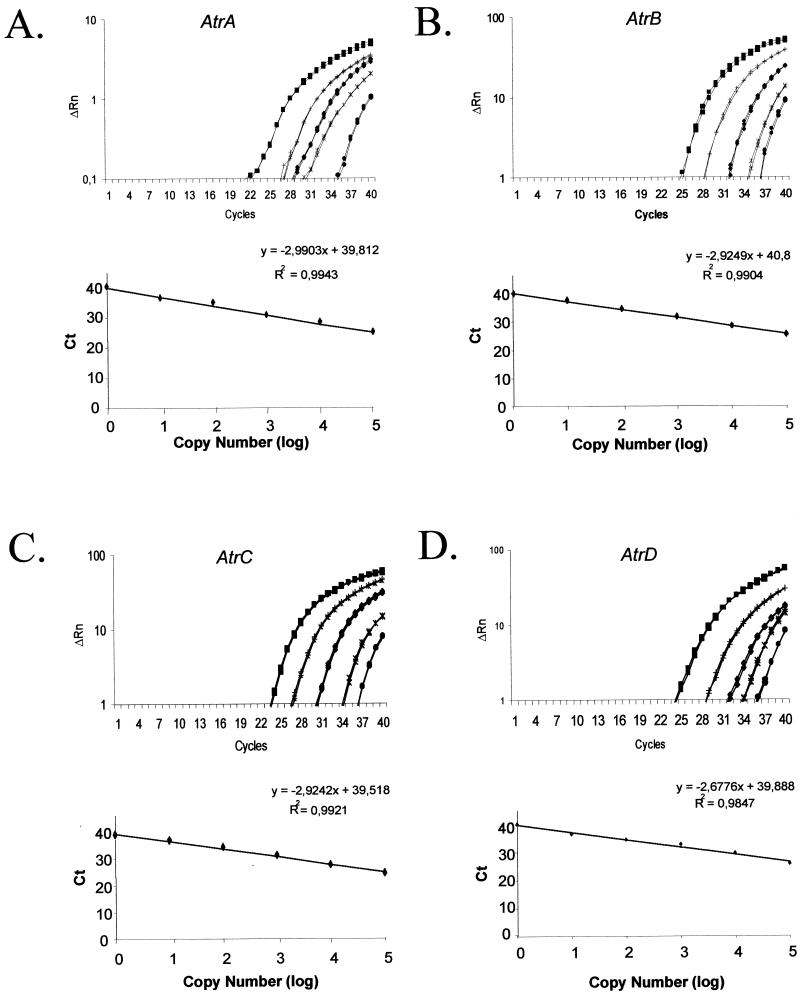
Several ABC transporter-encoding genes have already been reported for A. nidulans (for a review, see reference 5). To perform initial experiments aiming to validate the potential of real-time RT-PCR to quantify the relative transcript levels of ABC transporter genes from A. nidulans, we decided to design primers and Taq-Man fluorescent probes of the AtrA, AtrB (6), AtrC (2a) and AtrD (2) gene products. All the Atr RT-PCR products yielded a single band with the expected size on an agarose gel, and sequencing analysis showed the primers were amplifying the expected products (data not shown). In addition, we have not found in the A. nidulans genome database (http://www.genome.ou.edu/fungal.html) expressed sequences that display high identity with the AtrA to -D probes (data not shown). Figure 1 shows the amplification plots and standard curves for the Atr genes. By using these fluorescent probes, we were able to detect as few as 10 copies of each gene. No PCR products were detected in any of the negative-control samples that contained all reagents except A. nidulans genomic DNA after 40 rounds of amplification (data not shown).
FIG. 1.
Amplification plots and standard curves for the Atr genes. The amplification plot (top panel) and standard curve (bottom panel) for AtrA to -D are shown. The amplification plots correspond to PCRs done with different amounts of genomic DNA (105 ▪, 104 [+], 103 ♦, 102 [×], and 10 • copies) (using 30 Mb as the genome size). ΔRn is the normalized reporter signal minus the baseline level established in the first few cycles of PCR. The data from the amplification plots were used for the construction of the standard curves (i.e., Cts plotted against the logarithm of the DNA copy number). A linear relationship was obtained by plotting the resulting threshold cycle values (Ct) against the logarithm of the initial template amount. The best-fit line was determined through linear regression analysis. The results are averages of four repetitions.
Wild-type A. nidulans was grown in the absence of any drug, transferred to a subinhibitory concentration of camptothecin, imazalil, itraconazole, hygromycin, or 4-NQO for 8 h, and then the RNA was isolated and analyzed for the relative transcript level of each gene. Table 2 shows the quantitation results of the Atr genes (for a description of the methodology used, see Materials and Methods). The AtrA gene was induced about 3- and 14-fold when A. nidulans was grown in the presence of hygromycin and 4-NQO, respectively. The AtrB gene was induced about 30-, 39-, and 4,700-fold in the presence of imazalil, itraconazole, and 4-NQO, respectively. The AtrC gene was induced about 13-, 5-, 62-, and 2-fold when the fungus was grown in the presence of imazalil, itraconazole, hygromycin, and 4-NQO, respectively. The AtrD gene was induced about 4-, 9-, 23-, 279-, and 4-fold in the presence of camptothecin, imazalil, itraconazole, hygromycin, and 4-NQO, respectively (Table 2). Curiously, the relative transcript levels of the AtrA gene were repressed in the presence of camptothecin, imazalil, and itraconazole (Table 2).
TABLE 2.
Quantitation of the relative transcript levels of ABC transporter Atr genesa
| Drug | Relative transcript level
|
|||
|---|---|---|---|---|
| AtrA | AtrB | AtrC | AtrD | |
| None (control) | 1.00 | 1.00 | 1.00 | 1.00 |
| Camptothecin | 0.09 | ND | 0.52 | 4.25 |
| Imazalil | 0.04 | 30.06 | 13.68 | 9.83 |
| Itraconazole | 0.05 | 39.05 | 5.45 | 23.31 |
| Hygromycin | 2.81 | ND | 62.77 | 279.04 |
| 4-NQO | 14.21 | 4,760.57 | 2.30 | 3.94 |
Quantitation of the relative transcript levels of Atr genes from the A. nidulans wild-type strain grown in the presence of several unrelated drugs compared to the wild type grown in the absence of any drug. The measured quantity of each ABC transporter gene mRNA in each of the treated samples was normalized using the Cts obtained for the tubC RNA amplifications run on the same plate. The relative levels of ABC transporter and tubulin transcripts were determined by a standard curve (i.e., Cts plotted against logarithm of the DNA copy number). The results are averages of three repetitions. The values represent the number of times the genes are expressed compared to the wild-type control grown without any drug (set at 1.00). ND, not determined.
Relative transcript levels of Atr genes in A. nidulans imazalil-resistant mutants.
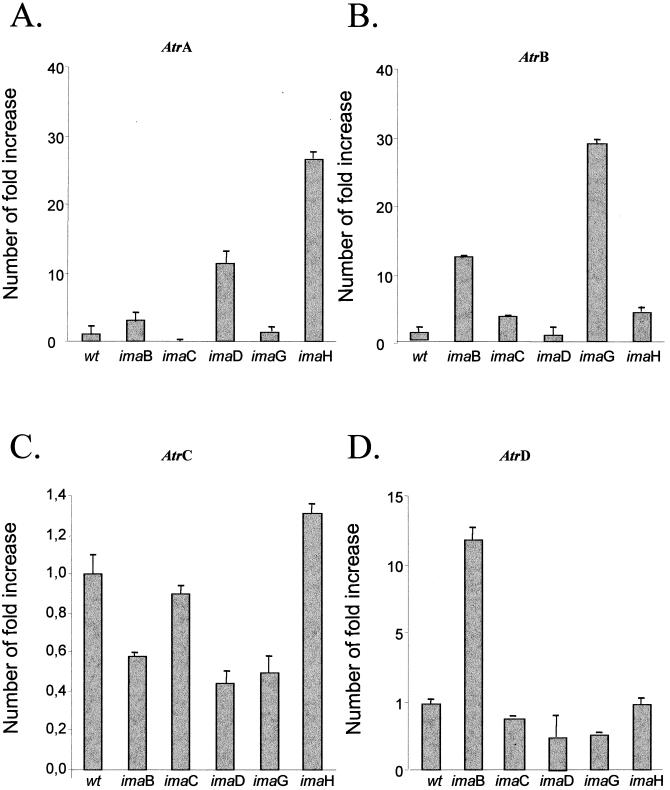
Genetic determinants displaying pleiotropic effects have been identified in A. nidulans (10, 28). Resistance to the azole fungicide imazalil can be determined by mutations in at least eight different genes (28). The mechanism of drug resistance in the imazalil mutants depends on decreased accumulation caused by energy-dependent efflux of the azole fungicides from the mycelium (12). We performed real-time RT-PCR assays to verify if any of the ABC transporter genes used in this work were overexpressed in the imazalil-resistant mutants (Fig. 2 and 3). First, we compared the relative transcript levels of ABC transporter genes when the ima mutants were grown in the absence of imazalil with those for the wild-type strain grown in the absence of the drug (Fig. 2). The AtrA gene had high levels in the imaD and -H mutants, while the AtrB gene had high levels in the imaB and -G mutants (Fig. 2A and B). In addition, the relative transcript levels of the AtrD gene were high in the imaB mutant (Fig. 2D). These results indicate the relative transcript levels of the Atr genes are high in the absence of the inducer drug imazalil.
FIG. 2.
Relative quantitation of the imazalil-resistant mutants of A. nidulans grown in the absence of imazalil compared to the wild type grown in the absence of the drug. The measured quantity of each ABC transporter gene mRNA in each of the treated samples was normalized using the Cts obtained for the tubC RNA amplifications run in the same plate. The relative amounts of ABC transporter genes and relative levels of tubulin transcripts were determined by a standard curve (i.e., Cts plotted against logarithm of the DNA copy number). The values represent the numbers of times the genes are expressed compared to that of the wild-type (wt) control grown without any drug (set at 1.00). The results are averages of three repetitions.
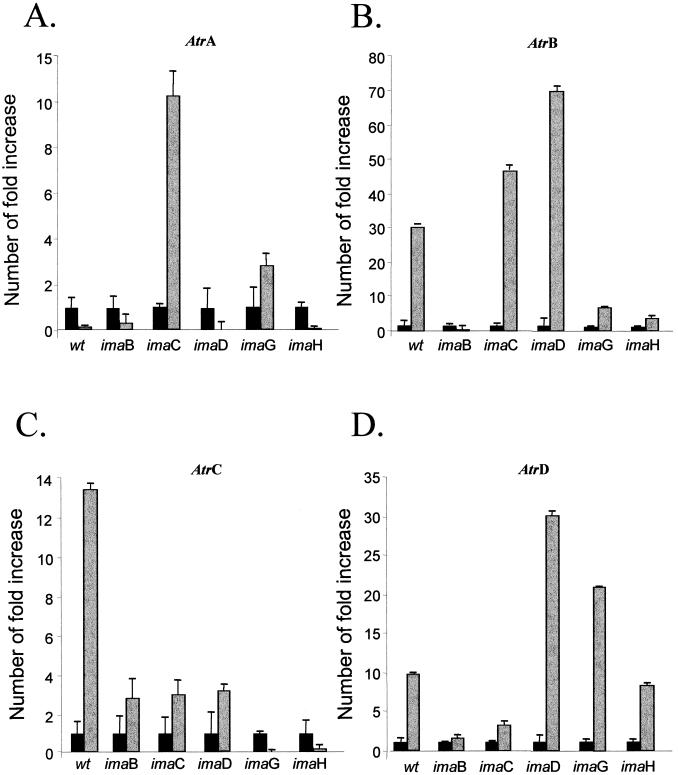
FIG. 3.
Relative quantitation of the imazalil-resistant mutants of A. nidulans grown in the presence and absence of imazalil. The same methodology described in the legend to Fig. 2 was used to quantify the relative transcript levels of the AtrA, AtrB, AtrC, and AtrD genes in the imazalil-resistant mutants grown in the presence of 0.3 μg of imazalil per ml. The values represent the numbers of times the genes are expressed (shaded bars) compared to that of the same strain grown without any drug (set at 1.00; solid black bars). The results are averages of three repetitions.
Second, we compared the relative transcript levels of the ABC transporter genes when the ima mutants were grown in the presence and absence of imazalil (Fig. 3). As shown in Table 2, the AtrB, -C, and -D genes were induced by imazalil in high levels (Fig. 3B to D). Surprisingly, although imazalil did not increase the relative AtrA transcript levels (Table 2), the imaC mutant displayed high levels of this gene when grown in the presence of imazalil (Fig. 3A). Curiously, the relative AtrC transcript levels were repressed in imaG and -H mutants and moderately increased in the imaB, -C, and -D mutants but highly increased in the wild-type strain (Fig. 3C). The same behavior can be observed in the imaH and -C mutants, where the relative AtrB and -D transcript levels, respectively, were moderately increased compared to that of the wild-type strain.
DISCUSSION
Gene quantification has been hampered by the lack of fast, reliable, and accurate methods (13). Current methods to determine gene expression are time-consuming and labor-intensive. We have evaluated the potential of real-time PCR to quantify the relative transcript levels of ABC transporter genes from the wild-type strain and imazalil-resistant A. nidulans strains. The Atr genes were shown to be induced by several structurally different drugs (for instance, AtrB and -D, a hallmark of the multiple drug resistance phenotype) (3). We were able to clearly distinguish quantitative differences in the relative transcript levels of these genes. This method was shown to be suitable for the identification and quantitative comparison of the relative transcript levels of different ABC transporter-encoding genes.
We have studied the transcript levels of the Atr genes in the A. nidulans imazalil-resistant mutants. These genes displayed a very complex pattern in different ima genetic backgrounds. The imaB mutant has higher basal levels of AtrB and -D than those of the wild-type strain. These two genes have comparable levels when the imaB mutant is grown in the presence and absence of imazalil. A possible explanation is that the gene encoded by imaB could be acting as a factor that regulates the expression of AtrB and -D genes. The imaC, -D, and -H mutants have higher basal levels of AtrA than that of the wild type. These mutations could correspond to genes that affect either AtrA gene expression or a mutation in the AtrA gene itself. The same behavior is observed for the AtrB levels in the imaG mutant background. Curiously, the relative Atr transcript levels were repressed in some of the ima mutants; for instance, the AtrC levels were repressed when the imaI, -G, and -H mutants were grown in the presence of imazalil. The same was also observed for the AtrA gene in the wild-type strain, where the levels were repressed in the presence of several drugs. This could suggest that regulatory factors important for AtrA or -C gene expression were being sequestered due to the overexpression of other transporters. According to White et al. (29), only increased mRNA levels have demonstrated the involvement of the efflux pumps in drug resistance of clinical isolates, but the causes of the increase have not been determined. Possible mechanisms for increased mRNA levels are the following: (i) gene amplification; (ii) increased transcription from endogenous copies of the gene, which can be the result of mutations in the promoter of the gene or of alterations in the levels of activity of transcriptional activators that interact with the gene promoter; and (iii) enhanced half-life of the mRNA, which could be the result of a mutation in the 3′ untranslated region. Our results showed a complex net of interactions among expression levels of the different ABC transporter genes in the ima mutant background. Additional information about these interactions will be provided when the genes that complement the ima mutants are cloned and characterized.
The investigation of the quantitative relative transcript levels of the ABC transporter genes in clinical isolates could provide an important contribution to antifungal therapy. The quantitative real-time RT-PCR described here is sensitive and accurate, and it could be used to quantitate large numbers of samples in a relatively short time. Antifungal therapy would be benefited by cataloguing the existing ABC transporter genes in a pathogenic species, developing specific fluorescent probes for each of them, and performing the quantitative analysis of the relative transcript levels of these genes in multidrug-resistant clinical isolates. The lack of structural similarity to mammalian P-glycoproteins may be exploited in designing specific inhibitors of the fungal efflux pumps (14). In addition, combined therapy of an antifungal drug associated with a drug that blocks the drug efflux pump (for instance, a multiple drug resistance reversal agent or modulator [4, 26]) might decrease the deleterious effects of these important pathogens.
Acknowledgments
This work was funded in part by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Brasília, Brazil.
We thank the two anonymous referees for their suggestions and comments.
REFERENCES
- 1.Albertson, G. D., M. Niimi, R. D. Cannon, and H. F. Jenkinson. 1996. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob. Agents Chemother. 40:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andrade, A. C., J. G. M. Van Nistelrooy, R. B. Peery, P. I. Skatrud, and M. A. De Waard. 2000. The role of ABC transporters from Aspergillus nidulans in protection against cytotoxic agents and in antibiotic production. Mol. Gen. Genet. 263:966-977. [DOI] [PubMed] [Google Scholar]
- 2a.Angermayr, K., W. Parson, G. Stoffler, and H. Haas. 1999. Expression of atrC—encoding a novel member of the ATP binding cassette transporter family in Aspergillus nidulans—is sensitive to cycloheximide. Biochim. Biophys. Acta 1453:304-310. [DOI] [PubMed] [Google Scholar]
- 3.Balzi, E., and A. Goffeau. 1994. Genetics and biochemistry of yeast multidrug resistance. Biochim. Biophys. Acta 1187:152-162. [DOI] [PubMed] [Google Scholar]
- 4.Berger, D., R. Citarella, M. Dutia, L. Greenberger, W. Hallett, and P. R. Powell. 1999. Novel multidrug resistance reversal agents. J. Med. Chem. 17:2145-2161. [DOI] [PubMed] [Google Scholar]
- 5.Del Sorbo, G., H. Schoonbeek, and M. A. De Waard. 2000. Fungal transporters involved in efflux of natural toxic compounds and fungicides. Fungal Genet. Biol. 30:1-15. [DOI] [PubMed] [Google Scholar]
- 6.Del Sorbo, G., A. C. Andrade, J. G. M. Van Nistelroy, J. A. L. Van Kan, E. Balzi, and M. A. De Waard. 1997. Multidrug resistance in Aspergillus nidulans involves novel ATP-binding cassette transporters. Mol. Gen. Genet. 254:417-426. [DOI] [PubMed] [Google Scholar]
- 7.Denning, D. W., and D. A. Stevens. 1990. Antifungal and surgical treatment of invasive aspergillosis: review of 2,121 published cases. Rev. Infect. Dis. 12:1147-1201. [DOI] [PubMed] [Google Scholar]
- 8.Denning, D. W., K. Venkateswarlu, K. L. Oakley, M. J. Anderson, N. J. Manning, D. A. Stevens, D. W. Warnock, and S. L. Kelly. 1997. Itraconazole resistance in Aspergillus fumigatus. J. Clin. Microbiol. 41:1364-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denning, D. W., S. A. Radford, K. L. Oakley, L. Hall, E. M. Johnson, and D. W. Warnock. 1997. Correlation between in-vitro susceptibility testing to itraconazole and in-vivo outcome of Aspergillus fumigatus infection. J. Antimicrob. Chemother. 40:401-414. [DOI] [PubMed] [Google Scholar]
- 10.de Souza, C. C., C. H. Pellizzon, M. Hiraishi, M. H. S. Goldman, and G. H. Goldman. 1998. Isolation of cycloheximide-sensitive mutants of Aspergillus nidulans. Curr. Genet. 33:60-69. [DOI] [PubMed] [Google Scholar]
- 11.de Souza, C. C., M. H. S. Goldman, and G. H. Goldman. 2000. Tagging of genes involved in multidrug resistance in Aspergillus nidulans. Mol. Gen. Genet. 263:702-711. [DOI] [PubMed] [Google Scholar]
- 12.De Waard, M. A., and J. G. M. Van Nistelrooy. 1980. An energy-dependent efflux mechanism for fenarimol in a wild-type strain and fenarimol-resistant mutants of Aspergillus nidulans. Pestic. Biochem. Physiol. 13:255-266. [Google Scholar]
- 13.Freeman, W. M., S. J. Walker, and K. E. Vrana. 1999. Quantitative RT-PCR: pitfalls and potential. BioTechniques 26:112-125. [DOI] [PubMed] [Google Scholar]
- 14.Georgopapadakou, N. H. 1998. Antifungals: mechanism of action and resistance, established and novel drugs. Curr. Opin. Microbiol. 1:547-557. [DOI] [PubMed] [Google Scholar]
- 15.Georgopapadakou, N. H., and T. J. Walsh. 1996. Antifungal agents: chemotherapeutic targets and immunologic strategies. Antimicrob. Agents Chemother. 40:279-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottesman, M. M., C. A. Hrycyna, P. V. Schoenlein, U. A. Germann, and I. Pastan. 1995. Genetic analysis of the multidrug transporter. Annu. Rev. Genet. 29:607-649. [DOI] [PubMed] [Google Scholar]
- 17.Heid, C. A., J. Stevens, K. J. Livak, and P. M. Williams. 1996. Real time quantitative PCR. Genome Res. 6:986-994. [DOI] [PubMed] [Google Scholar]
- 18.Joseph-Horne, T., and D. W. Hollomon. 1997. Molecular mechanisms of azole resistance in fungi. FEMS Microbiol. Lett. 149:141-149. [DOI] [PubMed] [Google Scholar]
- 19.Kafer, E. 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberrations. Adv. Genet. 19:33-131. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, S. L., D. C. Lamb, D. E. Kelly, J. Loeffler, and H. Einsele. 1996. Resistance to fluconazole and amphotericin in Candida albicans from AIDS patients. Lancet 348:1523-1524. [DOI] [PubMed] [Google Scholar]
- 21.Kolaczkowski, M., and A. Goffeau. 1997. Active efflux by multidrug transporters as one of the strategies to evade chemotherapy and novel practical implications of yeast pleiotropic drug resistance. Pharmacol. Ther. 76:219-242. [DOI] [PubMed] [Google Scholar]
- 22.Lie, Y. S., and C. J. Petropoulos. 1998. Advances in quantitative PCR technology: 5′ nuclease assays. Curr. Opin. Biotechnol. 9:43-48. [DOI] [PubMed] [Google Scholar]
- 23.Nakaune, R., K. Adachi, O. Nawata, M. Tomiyama, K. Akutsu, and T. Hibi. 1998. A novel ATP-binding cassette transporter involved in multidrug resistance in the phytopathogenic fungus Penicillium digitatum. Appl. Environ. Microbiol. 64:3983-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nolte, F. S., T. Parkinson, D. J. Falconer, S. Dix, J. Williams, C. Gilmore, C. Geller, R. Geller, and J. R. Wingard. 1997. Isolation and characterization of fluconazole- and amphotericin B-resistant Candida albicans from blood of two patients with leukemia. Antimicrob. Agents Chemother. 41:196-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rex, J. H., M. G. Rinaldi, and M. A. Pfaller. 1995. Resistance of Candida species to fluconazole. Antimicrob. Agents Chemother. 39:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan, B., D. Piwnica-Worms, and L. Ratner. 2000. Multidrug-resistance transporters and modulation. Curr. Opin. Oncol. 12:450-458. [DOI] [PubMed] [Google Scholar]
- 27.Tyagi, S., D. P. Bratu, and F. R. Kramer. 1998. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16:49-53. [DOI] [PubMed] [Google Scholar]
- 28.van Tuyl, J. M. 1977. Genetics of fungal resistance to systemic fungicides. Meded. Landbouwhogesch. Wageningen 77:1-136. [Google Scholar]
- 29.White, T. C., K. A. Marr, and R. A. Bowden. 1998. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin. Microbiol. Rev. 11:382-402. [DOI] [PMC free article] [PubMed] [Google Scholar]