Abstract
Ganoderma lucidum is a rapidly growing mushroom, wood-colonizing fungus belonging to the phylum Basidiomycota. G. lucidum exhibits a wide range of pharmacological properties such as antimicrobial and anticancer activities. However, with its potentials, the antibacterial and antifungal effects of its purified compounds have been relatively understudied. This study aims to investigate the phytochemical analysis and antimicrobial effects of Ganoderma lucidum against selected bacteria and fungi of medical importance. Molecular Identification was done by targeting the internal transcribed spacer (ITS) region of the rDNA gene. The phytochemical composition of the extract was determined using gas chromatography-mass spectrometry (GC–MS). Antimicrobial testing was performed using the agar well diffusion method to evaluate the inhibitory effects of the extract against select microorganisms. Gas chromatography-mass spectrometry (GC–MS) analysis identified 44 compounds in the methanol and 29 compounds in the aqueous extracts of the mushroom. Antimicrobial analysis demonstrated significant inhibitory effects against Staphylococcus aureus (MIC: 10.5 mm), Escherichia coli (MIC: 33.5 mm), and Candida albicans (MIC: 16 mm), as well as Pseudomonas aeruginosa (MIC: 15 mm). The methanol extract showed moderate effects against Rhizopus sp. at concentrations of 5 and 10 mg/ml with (MIC: 16 mm and 18 mm) but showed resistance to the chloramphenicol. However, Aspergillus flavus, Aspergillus niger and Penicillium showed resistance against Ganoderma lucidum. Overall, this findings show the extracts has the potential for combating the growing threat of antimicrobial resistance.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-025-06068-9.
Keywords: Ganoderma lucidum, Phytochemical analysis, Antimicrobial activity, Medical importance, GC–MS, PCR authentication
Subject terms: Medicinal chemistry, Microbiology
Introduction
Ganoderma species, particularly Ganoderma lucidum, are well known for their medicinal and bioactive properties, including antimicrobial, antioxidant, and immunomodulatory effects. These fungi contain diverse secondary metabolites such as polysaccharides, triterpenoids, and phenolic compounds, which contribute to their therapeutic potential1. With the rising global concern over antimicrobial resistance, there is an increasing need to explore natural sources for novel bioactive compounds, and medicinal mushrooms have emerged as promising candidates. While Ganoderma lucidum has been extensively studied for its medicinal properties, the use of Gas Chromatography-Mass Spectrometry (GC–MS) to characterize its phytochemical composition remains underutilized, particularly in African countries. Comparative studies from other regions demonstrate the value of this technique in revealing the mushroom complex metabolite profile2. For example, a study conducted in China used high-resolution mass spectrometry and multivariate analysis to differentiate G. lucidum samples from three provinces Jilin, Henan, and also Shandong identifying 84 unique compounds and biogeographical herbalism markers such as ganoderenic acid A and ganodernoid D, which varied significantly by region3. Similarly, Turkish researchers analyzed volatile compounds in fresh G. lucidum using headspace GC–MS, identifying key aroma compounds like 1-octen-3-ol and 3-methyl butanal, which contribute not only to flavor but potentially to bioactivity as well4. The antimicrobial activity of Ganoderma lucidum has been studied and several studies have highlighted it potency, particularly against both Gram-positive and Gram-negative bacteria, including multidrug-resistant (MDR) strains. In a study carried out by Ahmed (2024), prepared the extracts of G. lucidum using methanol, acetone, chloroform, and water exhibited broad-spectrum antibacterial activity, with methanol and acetone extracts showing the highest inhibition zones. Gram-positive bacteria, such as Staphylococcus aureus and Bacillus subtilis, were more susceptible compared to Gram-negative species like E. coli and Pseudomonas aeruginosa5. Complementing this, a recent in vitro study found ethanolic extracts of G. lucidum to be particularly effective, significantly inhibiting E. coli and S. aureus. Notably, the study employed SEM to reveal cellular damage and deformation in treated bacteria, suggesting a direct action of the extracts on bacterial membranes6. Ganoderma lucidum exhibits physical characteristics with other closely related species, molecular characterization has become a important technique for reliably identifying this species. At an International DNA Barcod of Life Conference, the internal transcribed spacer (ITS) region was suggested as a worldwide DNA barcode sequence for fungal identification. Based on 6600 samples representing 4800 species, Chen in 2010 suggested using the nuclear ribosomal DNA second internal transcribed spacer (ITS2) region as a unique universal DNA barcode to identify herbs. After comparing the ITS and ITS2 sections7. In a research carried out, it was concluded that the ITS2 region was better suited for species identification due to its short length and excellent PCR amplification efficiency. Furthermore, ITS2 secondary structures and sequences may be regarded as molecular morphological traits for species identification8.
Despite several studies highlighting the antimicrobial and bioactive properties of G. lucidum, data on its effectiveness against specific bacterial and fungal strains endemic, to Nigeria remain scarce. Additionally, there has been minimal exploration of the phytochemical composition of G. lucidum from different ecological zones in Nigeria, and limited comparisons between the efficacy of aqueous and methanolic extracts9. Furthermore, the molecular characterization of indigenous Ganoderma species remains underexplored, leading to limited understanding of their genetic diversity and pharmaceutical potential. These gaps should be addresses for enhancing local fungal biodiversity in antimicrobial drug discovery.
The increasing prevalence of antimicrobial-resistant pathogens necessitates the search for alternative treatment strategies. G. lucidum, a widely recognized medicinal mushroom, represents a valuable natural resource for bioactive compound discovery10.
The molecular identification of Nigerian G. lucidum, combined with phytochemical profiling using GC–MS isolates ensures accurate species characterization, evaluating its antimicrobial activity against clinically relevant bacterial and fungal strains provides insights into its bioactive compounds, highlighting its potential as a natural antimicrobial agent also for pharmaceutical applications.
This study aims to identify the phytochemical constituents and evaluate the antimicrobial activity of Ganoderma lucidum against clinically important bacterial and fungal pathogens from ofadeto Osogbo regions in Nigeria.
Materials and methods
Sample collection
Mushroom samples were collected during the rainy season in June 2024 from Ofatedo Osogbo (Osun State) ), Nigeria. Two specimens, exhibiting morphological characteristics consistent with Ganoderma sp. were confirmed through molecular identification. The samples were harvested using sterile tools carefully cleaned to remove surface debris, and preserved by drying in a hot air oven at 50 °C for 50 min refrigerated at 4 °C until further analysis. This controlled drying process was employed to prevent microbial contamination and preserve the integrity of bioactive compounds for subsequent analysis. These locations were selected due to their favorable ecological conditions for fungal growth, with the samples intended for antimicrobial and bioactive compound studies (Fig. 1).
Fig. 1.
Geographical distribution of Ganoderma lucidum collection sitess in Nigeria.
Sample preparation
Fresh mushrooms were sliced and air-dried in an oven (Selecta, Madrid, Spain) at 50 °C until a constant weight was achieved. The dried material (60 g) was ground into a fine powder using a hammer stirrer (IKA, Staufen, Germany). A 30 g portion was extracted in 500 mL of hot water and methanol for 96 h to dissolve phytochemicals. The extract was filtered using Whatman No. 1 filter paper11.
Molecular identification
DNA extraction
DNA was extracted using the ZYMO DNA extraction kit. A 50 mg wet sample was resuspended in 200 μL of water in a ZR BashingBead™ Lysis Tube, with 750 μL BashingBead™ Buffer added. The mixture was processed in a bead beater for 5 min, centrifuged at 10,000 × g for 1 min, and the 400 μL supernatant was transferred to a Zymo-Spin™ III-F Filter. Following sequential filtrations and centrifugations at 10,000 × g, 100 μL of DNA Elution Buffer was added, yielding 2.80 ng/μL DNA.
Primer design and PCR amplification
PCR was performed with 12.5 μL PCR master mix, 1.25 μL of each 10 mM primer, and 10 μL of diluted DNA template, adjusted to 25 μL with sterile deionized water12. Thermal cycling conditions included initial denaturation at 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 50 °C for 30 s, and a final extension at 72 °C for 1 min. PCR products were purified using Montage™ PCR Centrifugal Filter Devices (Millipore Corp., USA)13. The ITS1 and ITS4 primers14 were used to amplify the ITS ribosomal region. The ITS region was chosen because it is widely regarded as the most reliable genetic marker for species-level identification in fungi. ITS provides greater variability than Large subunit (LSU) and other regions, making it more useful for differentiating closely related species. Also, ITS region has been recommended as the universal DNA barcode marker for fungi.
Genome sequencing and analysis
Genome sequencing was conducted using PacBio Sequel (long-read) and Illumina NovaSeq (short-read) platforms. De novo assembly was performed with SMARTdenovo1 and polished using Pilon15. Assembly integrity was assessed with BUSCO16. The ITS1-5.8S-ITS2 region (588 bp) was analyzed using BLAST with 99.55% identity, 99.55% query coverage, and an E-value ≤ 1.00E-135. Sequences were matched to Ganoderma lucidum (taxid:5315). Phylogenetic relationships were inferred using Maximum Likelihood (ML) analysis in MEGA 717.
Extract concentration
Aqueous extracts of G. lucidum were concentrated using a freeze dryer (Model LGJ-10) at 50 °C. Methanolic extracts were concentrated via evaporation in a water bath at 50 °C.
Test organisms
Bacterial isolates (E. coli, Staphylococcus aureus, Enterobacter aerogenes, Pseudomonas aeruginosa) were obtained from the Microbiology Laboratory, Ladoke Akintola University, Ogbomosho, Nigeria. Fungal strains (Aspergillus niger, Rhizopus stolonifer, Penicillium sp., A. flavus, candida sp.) were sourced from Bowen University, Iwo, Nigeria. Cultures were maintained on agar slants at 4 °C to prevent contamination11.
Phytochemical analysis (GC–MS)
GC–MS analysis was performed using an Agilent Technologies Model 7000 GC–MS Triple Quad (TQQQ) system, equipped with Hunter workstation software (version B.04.00) [https://www.agilent.com/]. The software was used for data acquisition and analysis. The electron ionization potential was 70 eV. Compounds were separated using an OPTIMA-5 column (30 m × 250 μm × 0.25 μm) at 360 °C. Helium was the carrier gas (flow rate: 1.129 mL/min; split ratio: 5:1). A 2.5 μL sample was injected via an automatic liquid sampler. Compounds were identified by comparing mass spectra with the NIST database, using retention indices calculated via Kovat’s formula with n-alkanes (C9–C33) as standards18.
Inoculum preparation
Test organisms were sub-cultured on nutrient agar and potato dextrose agar, then incubated at 37 °C for bacteria and 24 °C for fungi for 24–36 h. Bacterial suspensions were adjusted to a 0.5 McFarland standard (~ 1.5 × 108 CFU/mL).
Antimicrobial test
The agar well diffusion method was used. A 24-h broth culture was swabbed onto sterile Mueller–Hinton agar in Petri dishes. Three equidistant wells (6 mm) were created using a sterile cork borer and filled with 0.1 mL of extract. A positive control (chloramphenicol, 50 μg/mL) and a negative control (solvent) were included. Plates were incubated at 37 °C for 24 h, and inhibition zones were measured in millimetres11.
Statistical analysis
Data were analyzed using SPSS (Version 25.0) and expressed as mean ± standard deviation. Statistical significance was determined using one-way ANOVA, Student’s t-test, and chi-square analysis (p < 0.05). Bar charts with error bars were generated using Microsoft Excel with Pearson correlation analysis.
Results
Molecular identification
The ITS sequence (AS_ITS1_C06_09) showed high-confidence alignments with several Ganoderma species, especially Ganoderma lucidum, according to the BLAST analysis. The top matches displayed Elevated Maximum Scores of 663, Percent Identities above 99%, 75% of queries were covered, E-values of 0.0 signify matches that are highly significant. G. lucidum isolates (e.g., OQ701698.1, MF755277.1) and allied species such as G. resinaceum and G. sp. were among the notable sequences (supplementary Table S1). These findings support the query sequence’s identity within the Ganoderma lucidum complex by confirming a strong taxonomic relationship between it and its components.
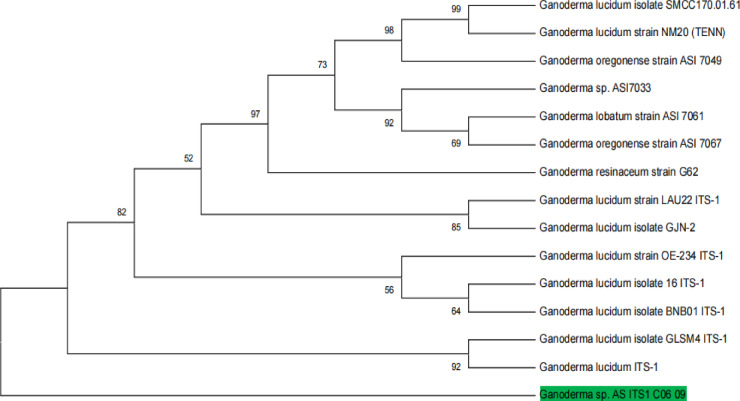
The phylogenetic analysis of the sample AS_ITS1_C06_09 (Fig. 2) provided key insights into its evolutionary relationships within the Ganoderma genus. Using the Maximum Likelihood method with 100 bootstrap replicates, the analysis showed a strong clustering of AS_ITS1_C06_09 with Ganoderma sessile (KF605634.1), indicating significant genetic similarity and evolutionary relatedness. High bootstrap values further validated this placement. The phylogenetic tree also revealed distinct clades for other Ganoderma species, illustrating the genus’s genetic diversity. Notably, Ganoderma lucidum (ON394695.1), isolated from Ogbomoso, Nigeria, formed a separate, well-supported clade, underscoring its evolutionary divergence and unique genetic identity (Table 1).
Fig. 2.
Evolutionary relationship of AS_ITS1_C06_09 with Other Ganoderma isolates based on ITS Region.
Table 1.
GC–MS profiling of bioactive compounds in Ganoderma lucidum methanol extract.
| Compounds peak | Retention time | Area (%) | Molecular weight |
|---|---|---|---|
| Decane,2,3,7-trimethyl3-ethyl-3-methylheptaneundecane,3-ethyl- | 6.944 | 0.82 | 116.16 |
| Hexadecane, tridecane, sulfurous acid | 7.499 | 2.01 | 158.24 |
| Nonane acid | 10.497 | 3.74 | 242.40 |
| D-Arabinitol | 10.772 | 5.32 | 652.9 |
| Pentadecanoic acid, 14-methyl-este | 19.46 | 12.87 | 270.45 |
| Hexadecanoic acid, ethyl ester | 19.907 | 7.45 | 284.55 |
| Deca-6,9-diene-2,8-dione | 13..324 | 22.11 | 294.47 |
| Heptadecane, 2, 6, 10, 15 Tetramethy | 20.95 | 1.66 | 296.12 |
| (E)-9-Octadecanoic acid, methyl ester | 21.631 | 1.29 | 296.48 |
| Octadecanoic acid | 22.737 | 33.24 | 284.47 |
| Carbamic acid, 2-(dimethylamino)ethyl ester | 27.391 | 1.21 | 132.16 |
| Oleic acid | 15.367 | 1.88 | 281.48 |
| Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester | 19.325 | 0.21 | 330.50 |
| 6,9-Octadecadienoic acid, methyl ester | 30.876 | 0.41 | 294.50 |
| 4H-1,2,4-triazol-3-ol, 5- (phenylmethyl) | 18.199 | 1.55 | 56.50 |
GC–MS profiling of bioactive compounds in ganoderma lucidum
The Gas Chromatography-Mass Spectrometry (GC–MS) analysis of Ganoderma lucidum Methanol Extract (GLME) and Aqueous Extract (GLAE) revealed a diverse range of bioactive compounds (Fig. 3), each identified by its retention time (RT). The GC–MS analysis of Ganoderma lucidum extracts (Table 2) identified various bioactive compounds with significant therapeutic potential. The most abundant was octadecanoic acid (RT: 22.737 min, 33.24%), known for its antimicrobial, anti-inflammatory, and lipid metabolism-regulating effects. Another major compound, deca-6,9-diene-2,8-dione (RT: 13.324 min, 22.11%), is also bioactive. Various saturated and unsaturated fatty acids, including hexadecanoic acid, pentadecanoic acid methyl ester, and oleic acid, exhibited cardioprotective, antimicrobial, and antioxidant properties.
Fig. 3.
GC–MS analysis identified the bioactive compounds present in Ganoderma lucidum methanol extract (GLME) and aqueous extract (GLAE), along with their corresponding retention times.
Table 2.
GC–MS profiling of bioactive compounds in Ganoderma lucidum aqeouse extract.
| Compound peak | Retention time | Area (%) | Molecular weight |
|---|---|---|---|
| 1,2-propanediol, 3-benzyloxy-1-2-diol | 18.211 | 0.82 | 116.16 |
| Hexadecane, tridecane, sulfurous acid | 7.499 | 2.01 | 158.24 |
| Methyl stearate | 15.069 | 3.74 | 242.40 |
| Oxacyclododecan-2-one | 19.212 | 5.32 | 652.9 |
| pentadecanoic acid, 14-methyl-ester | 13.765 | 12.87 | 270.45 |
| Hexadecanoic acid | 13,456 | 7.45 | 284.55 |
| deca-6,9-diene-2,8-dione | 14.869 | 22.11 | 294.47 |
| Heptadecane2, 6, 10, 15 Tetramethyl | 11.722 | 1.66 | 296.12 |
| Octadecanoic acid | 12.609 | 33.24 | 284.47 |
| Oleic acid | 15.367 | 1.88 | 282.47 |
Antimicrobial test
The aqueous extract across most tested microorganisms. Escherichia coli showed the highest susceptibility, with inhibition zones of 35.33 ± 0.58 mm at 10 mg/mL, decreasing slightly with lower concentrations. This was followed by Candida albicans (16.67 ± 0.58 mm) and Rhizopus stolonifer (17.67 ± 0.58 mm), which also showed strong susceptibility to the methanol extract at 10 mg/mL. Notably, Rhizopus stolonifer demonstrated dose-dependent inhibition, confirming its sensitivity to methanol-extracted bioactive compounds while Aspergillus flavus, A. niger, and Penicillium exhibited minimal inhibition zones (~ 5 mm) with both extracts at all concentrations, indicating resistance. The bacteria test organism; Pseudomonas aeruginosa and Staphylococcus aureus were moderately inhibited by the methanol extract, while the aqueous extract had a limited effect (Table 3).
Table 3.
Antimicrobial activity of methanol and aqueous extracts of Ganoderma lucidum against selected bacterial and fungal strains (mean inhibition zone in mm ± SD).
| Microorganism | Methanol extract (10 mg) | Methanol extract (5 mg) | Methanol extract (2 mg) | Control | Water extract (10 mg) | Water extract (5 mg) | Water extract (2 mg) | Control |
|---|---|---|---|---|---|---|---|---|
| Aspergillus flavus | 5.33 ± 0.58 | 5.33 ± 0.58 | 5.33 ± 0.58 | 0.00 ± 0.00 | 5.00 ± 0.00 | 5.00 ± 0.00 | 5.33 ± 0.58 | 0.00 ± 0.00 |
| Aspergillus niger | 5.33 ± 0.58 | 5.33 ± 0.58 | 5.33 ± 0.58 | 0.00 ± 0.00 | 5.00 ± 0.00 | 5.00 ± 0.00 | 5.33 ± 0.58 | 0.00 ± 0.00 |
| Candida albicans | 16.67 ± 0.58 | 17.33 ± 0.58 | 5.33 ± 0.58 | 34.33 ± 3.06 | 10.33 ± 0.58 | 10.00 ± 0.00 | 5.33 ± 0.58 | 34.33 ± 3.06 |
| Escherichia coli | 35.33 ± 0.58 | 32.33 ± 0.58 | 31.67 ± 0.58 | 43.33 ± 6.66 | 19.67 ± 0.58 | 17.33 ± 0.58 | 5.33 ± 0.58 | 43.33 ± 6.66 |
| Penicillium | 5.00 ± 0.00 | 5.33 ± 0.58 | 5.00 ± 0.00 | 0.00 ± 0.00 | 5.00 ± 0.00 | 5.00 ± 0.00 | 5.33 ± 0.58 | 0.00 ± 0.00 |
| Pseudomonas aeruginosa | 14.67 ± 0.58 | 5.33 ± 0.58 | 5.67 ± 0.58 | 56.33 ± 1.53 | 5.33 ± 0.58 | 5.33 ± 0.58 | 5.33 ± 0.58 | 56.33 ± 1.53 |
| Rhizopus stolonifer | 17.67 ± 0.58 | 16.33 ± 0.58 | 5.67 ± 0.58 | 0.00 ± 0.00 | 5.00 ± 0.00 | 5.00 ± 0.00 | 5.33 ± 0.58 | 0.00 ± 0.00 |
| Staphylococcus aureus | 11.00 ± 0.00 | 9.67 ± 0.58 | 5.00 ± 0.00 | 48.67 ± 4.04 | 10.67 ± 0.58 | 5.33 ± 0.58 | 5.33 ± 0.58 | 48.67 ± 4.04 |
Figure 4 present the zones of inhibition for different concentrations of Ganoderma lucidum methanolic and aqueous extracts with control against selected test organisms.
Fig. 4.
Antibacterial activity of methanol and water extracts at varying concentrations (10 mg/mL, 5 mg/mL, and 2 mg/mL) compared to control against selected microbial isolates. The zone of inhibition (mm) was measured for each extract against Aspergillus flavus, Aspergillus niger, Candida albican, Escherichia coli, Penicillium, Pseudomonas aeruginosa, Rhizopus stolinofer, and Staphylococcus aureus.
The methanolic extract demonstrated moderate antibacterial activity against Escherichia coli, Staphylococcus aureus, Candida albicans, Pseudomonas aeruginosa, and Rhizopus stolonifer. The inhibition zones suggest that E. coli was the most sensitive organism, with concentration values at 2, 5, and 10 mg/mL, confirming its greater susceptibility compared to the other test strains. The aqueous extract also exhibited moderate antimicrobial activity, particularly against E. coli, S. aureus, and C. albicans, with concentration values recorded at 10 and 5 mg/mL.
Discussion
Phylogenetic and biochemical characterization of ganoderma species
The phylogenetic analysis of Ganoderma species revealed strong genetic relationships, with G. lucidum, Ganoderma sp., G. oregonense and G. lucidum forming a well-supported clade. The high bootstrap values reinforce the genetic integrity of this classification. Notably, G. resinaceum exhibited clear genetic distinctiveness, corroborating its status as a unique species within the G. lucidum complex19. The placement of AS_ITS1_C06_09 alongside G. resinaceum suggests close evolutionary ties, further supported by robust bootstrap values. This study emphasizes the diversity within Ganoderma, where G. lucidum and G. resinaceum demonstrate distinct evolutionary paths. The ITS-based taxonomic approach proved reliable in delineating these relationships.
Bioactive compounds and functional potential
GC–MS analysis identified several bioactive compounds with significant pharmacological properties. Oxaspiro (C17H24O), exhibits antioxidant and metabolic regulatory functions20, affirming its potential in mitigating oxidative stress-related diseases. Notably, hexadecenoic acid (palmitoleic acid), detected at RTs of 13.782 min (GLME) and 13.456 min (GLAE), has been previously reported for its anti-inflammatory, cardioprotective, and antimicrobial effects. This aligns with findings by Randeni et al.21 who identified similar fatty acids in G. lucidum with potential in managing cardiovascular and inflammatory disorders22.
Also, in line with earlier studies such as23 and24, this study also identified octadecanoic acid (stearic acid) as the most abundant compound (RT: 22.737 min, 33.24%). This compound is well-documented for its lipid metabolism-regulating, antimicrobial, and immunomodulatory properties, reinforcing its relevance in nutraceutical and therapeutic applications. Another significant compound, deca-6,9-diene-2,8-dione (RT: 13.324 min, 22.11%), is less commonly reported but shows strong potential as a novel bioactive molecule, warranting further functional evaluation. Similarly, the detection of oleic acid, pentadecanoic acid methyl ester, and nonanoic acid is consistent with the findings of25,26, who reported similar fatty acid derivatives with antioxidant and antimicrobial activities in G. lucidum extracts. Of particular interest is the detection of D-arabinitol (RT: 10.772 min, 5.32%), a sugar alcohol with known antifungal effects, also reported by27 in their GC–MS profiling of medicinal mushrooms. Its presence suggests a potential mechanism for G. lucidum's antifungal activity. In addition, minor compounds such as carbamic acid derivatives and 4H-1,2,4-triazol-3-ol were identified, adding to the extract’s neuroprotective and antifungal potential, respectively in agreement with secondary metabolite profiles from other studies. The presence of tetradecanoic acid in Ganoderma lucidum corroborates findings from earlier studies which demonstrated its anti-virulence and antimicrobial activities, particularly against Pseudomonas aeruginosaal. This supports its potential as a bioactive agent in managing resistant pathogens. Similarly, previous research on G. lucidum has consistently highlighted its richness in bioactive polysaccharides and triterpenoids, which are widely acknowledged for their immunomodulatory, anticancer, and antimicrobial properties28. These compounds have long been used in traditional medicine and are now being explored as components of nutraceuticals and functional foods.
The compound phenylmethyl exhibited strong antimicrobial activity, inhibiting carbonic anhydrase enzymes and showing broad-spectrum efficacy against bacterial and fungal pathogens2. Additionally, 1,2-propanediol, 3-benzyloxy-1,2-diacetyl, identified in the aqueous extract, demonstrated antibacterial and antiviral properties, suggesting its potential as a biopharmaceutical agent.
GLME contained a higher concentration of bioactive compounds and exhibited a stronger antimicrobial effect compared to GLAE. This suggests solvent-specific extraction efficiency, emphasizing the role of polarity in optimizing bioactive compound recovery. Future studies should explore alternative extraction techniques, such as supercritical fluid extraction, to enhance yield and efficacy.
Antimicrobial efficacy and clinical relevance
The antimicrobial test demonstrated that G. lucidum extracts exhibit concentration-dependent activity against clinical pathogens. The methanol extract exhibited no inhibitory effect against Pseudomonas aeruginosa at concentrations of 2 mg/mL and 5 mg/mL. However, a notable zone of inhibition (15 mm) was observed at 10 mg/mL, indicating concentration-dependent efficacy. This finding aligns with previous studies reporting the dose-dependent antimicrobial activity of G. lucidum extracts29,30.
In contrast, the aqueous extract showed resistance across all tested concentrations (2–10 mg/mL), with zones of inhibition consistently below 9 mm. This suggests that antimicrobial efficacy against P. aeruginosa may depend on specific bioactive constituents extracted by the solvent. Kumar et al. similarly reported enhanced antimicrobial activity in methanol extracts compared to water extracts31. Staphylococcus aureus was susceptible to the methanol extract at all concentrations (2–10 mg/mL), consistent with earlier findings5. In contrast, the aqueous extract showed limited activity, with resistance at 2 mg/mL and 5 mg/mL, and a moderate inhibition zone (10 mm) at 10 mg/mL. These results underscore the importance of solvent selection in antimicrobial screening. Heleno et al. also observed stronger antibacterial activity in methanol extracts of G. lucidum against S. aureus and Bacillus cereus compared to conventional antibiotics. Escherichia coli exhibited susceptibility to both methanol and aqueous extracts. However, the aqueous extract showed resistance at 2 mg/mL. These findings align with previous reports on G. lucidum's antimicrobial activity against E. coli32,33. A broader regional study from India, Nepal, Pakistan, and Northern Nigeria also documented efficacy of methanol extracts against E. coli at 20–200 mg/mL, although minimum bactericidal concentrations (MBCs) were not determined33,34. The notable susceptibility of Rhizopus spp. to the methanol extract of G. lucidum, with inhibition zones of 18 mm at 10 mg/mL and 16 mm at 5 mg/mL, demonstrates a clear dose-dependent antifungal effect and positions G. lucidum as a promising source of antifungal agents against Rhizopusrelated to food spoilage and infections. In contrast, Aspergillus flavus, A. niger, and Penicilliumspp. exhibited resistance not only to G. lucidum extracts but also to the conventional antibiotic (control) chloramphenicol, indicating a broader resistance profile among these molds35 This selective efficacy aligns with previous findings that reported minimal antifungal activity of G. lucidum against filamentous fungi such as Aspergillus and Candida species36,37. These results underscore the potential of G. lucidum methanol extract in targeting susceptible molds like Rhizopus, and highlight the critical role of extraction solvents in enhancing the bioavailability of antifungal compounds38,39. However, Interestingly, Candida albicans showed susceptibility to both extracts at 10 mg/mL but resistance at lower concentrations. This observation supports the concentration-dependent antifungal activity of G. lucidum against Candida species, as previously reported.
Differences in susceptibility between Gram-negative and Gram-positive bacteria can be attributed to their cell wall structures. Gram-negative bacteria’s outer membrane and lipopolysaccharide (LPS) layer restrict compound diffusion, whereas the porous structure of Gram-positive bacteria facilitates greater permeability40. Understanding these mechanisms could aid in optimizing Ganoderma-derived antimicrobials for targeted applications.
Implications for food and pharmaceutical industries
The findings highlight the vast potential of G. lucidum as a functional food and pharmaceutical resource. With an annual global market exceeding $1.5 billion, G. lucidum’s bioactive compounds, including triterpenoids and polysaccharides, offer diverse health benefits. Their roles in immune modulation, anticancer therapy, and metabolic regulation position Ganoderma as a promising natural alternative to synthetic pharmaceuticals.
The structural diversity of bioactive compounds, such as oxaspiro and methyl esters, underscores their potential applications in nutraceuticals and functional foods. The ability of these compounds to regulate hormonal balance, relieve pain, and inhibit cancer cell proliferation further enhances their therapeutic relevance.
Moreover, the antimicrobial properties of G. lucidum suggest its application in food preservation and alternative medicine. The strain isolated from Osogbo exhibits promising potential for commercial exploitation, particularly in developing countries where affordable, natural antimicrobial agents are needed. Our findings demonstrate a concentration-dependent antimicrobial effect of G. lucidum methanol extract, particularly evident in the inhibition of Staphylococcus aureus only at 10 mg/mL, but not at lower concentrations—providing a clearer threshold than previously reported studies. Furthermore, the antifungal response of Rhizopus at 10 mg/ml and 5 mg/mland Candida albicans to both extracts at higher concentrations is noteworthy, especially given prior reports of resistance. The GC–MS analysis also revealed bioactive compounds such as deca-6,9-diene-2,8-dione and sulfurous acid derivatives, which are not commonly emphasized in earlier work. These differences may account for the observed variations in bioactivity, potentially influenced by strain specificity and regional factors.
Conclusion
Despite the promising bioactivities demonstrated by Ganoderma lucidum extracts, this study has several limitations that should be acknowledged. The antimicrobial assays were performed against a limited number of bacterial strains, which may not capture the full spectrum of clinically relevant or multidrug-resistant pathogens. Although both aqueous and methanol extracts were evaluated, the range of extraction techniques and solvent systems was relatively narrow. Additionally, the extracts were not further purified or standardized, which may have influenced the consistency and specificity of the observed bio-active compounds. The absence of in vivo validation further limits the transnational relevance of the findings. Future research should incorporate a broader microbial panel, employ more diverse and standardized extraction and purification strategies, and include in vivo studies to fully assess therapeutic potential.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Author contributions
S.S and A.O conceived and designed the research work. S.S., A.O.O, I.O, A.A and F.D.O. prepared the manuscript text. O.B.A and I.A performed the molecular analysis. I.O and A.A provided the isolates. O.O performed the Gas Chromatography Mass Spectrometry analysis. A.O.O performed the statistical analysis and prepared the figures. O.O.O, O.O., O.A.O supervised the work. All authors reviewed the manuscript.
Data availability
The nucleotide sequence data generated in this study have been deposited in GenBank under the accession number PV444608. The data will be publicly available on April 9, 2025, or upon the publication of this manuscript, whichever occurs first. The submission can be accessed via the GenBank Submission Portal at https://submit.ncbi.nlm.nih.gov/subs/?search=SUB15234193.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sharma, C., Kumar, N., Pandey, R., Meis, J. F. & Chowdhary, A. Whole genome sequencing of emerging multidrug resistant Candida auris isolates in India demonstrates low genetic variation. New Microb. New Infect.1(13), 77–82 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Erbiai, E. H. et al. Chemical characterization and evaluation of antimicrobial properties of the wild medicinal mushroom Ganoderma lucidum growing in northern Moroccan forests. Life13(5), 1217 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu, J. et al. Deciphering the chemical composition of Ganoderma lucidum from different geographical origins by mass spectrometry molecular networking coupled with multivariate analysis. Biomed. Chromatogr. BMC37(1), e5506 (2023). [DOI] [PubMed] [Google Scholar]
- 4.Oyetayo, V. O. & Ogidi, C. O. Potential of mycochemicals in the prevention and control of microbial diseases. In Eco-Friendly Biobased Products Used in Microbial Diseases. (CRC Press, 2022).
- 5.Ahmad, M. F. et al. Ganoderma lucidum: Insight into antimicrobial and antioxidant properties with development of secondary metabolites. Heliyon10(3), e25607 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ewunkem, A. et al. In vitro antimicrobial efficacy assessment of ethanolic, aqueous, and dual solvent extracts of mushroom Ganoderma lucidum: genomic and morphological analysis. Antibiotics13(12), 1109 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, S. et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE5(1), e8613 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liao, B. et al. Identification of commercial Ganoderma (Lingzhi) species by ITS2 sequences. Chin. Med.19(10), 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oke, M. A. et al. Ganoderma lucidum: Unutilized natural medicine and promising future solution to emerging diseases in Africa. Front. Pharmacol.13, 952027 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vallavan, V., Krishnasamy, G., Zin, N. M. & Abdul, L. M. A Review on antistaphylococcal secondary metabolites from basidiomycetes. Molecules25(24), 5848 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adeoyo, O. R. & Oluborode, O. O. Antimicrobial properties of some Nigerian edible mushrooms. J. Phytopharmacol.9(2), 110–114. 10.31254/phyto.2020.9206 (2020). [Google Scholar]
- 12.Aime, M. C., Vilgalys, R. & Miller, O. K. Jr. The Crepidotaceae (Basidiomycota, Agaricales): Phylogeny and taxonomy of the genera and revision of the family based on molecular evidence. Am. J. Bot.92(1), 74–82 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Adotey et al. Mangrove carbon stock assessment. ResearchGate (2022). Available from: https://www.researchgate.net/publication/364309816_Adotey_et_al_2022_Mangrove_carbon_stock_assessment
- 14.Manter, D. K. & Vivanco, J. M. Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis. J. Microbiol. Methods71(1), 7–14 (2007). [DOI] [PubMed] [Google Scholar]
- 15.Walker, D. J., Lutts, S., Sánchez-García, M. & Correal, E. Atriplex halimus L.: Its biology and uses. J. Arid. Environ.100–101, 111–121 (2014). [Google Scholar]
- 16.Waterhouse, A. et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res.46(W1), W296-303 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar, S., Stecher, G., Li, M., Knyaz, C. & Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms Battistuzzi FU, ediitor. Mol. Biol. Evol.35(6), 1547–1549 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farooqi, W. et al. Comparison of efficacy and safety of device-based interventions versus pharmacological therapy in the management of patients with advanced Parkinson’s disease: A literature review. Cureus16(12), e76044 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keith Vlasak. Available from: https://www.goodreads.com/author/show/8451611.Keith_Vlasak
- 20.National Center for Biotechnology Information. Available from: https://www.ncbi.nlm.nih.gov/
- 21.Randeni, N. & Xu, B. New insights into signaling pathways of cancer prevention effects of polysaccharides from edible and medicinal mushrooms. Phytomedicine1(132), 155875 (2024). [DOI] [PubMed] [Google Scholar]
- 22.Filbay, S. R. et al. Healing of acute anterior cruciate ligament rupture on MRI and outcomes following non-surgical management with the cross bracing protocol. Br. J. Sports Med.57(23), 1490–1497 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oke, M. A. et al. Ganoderma lucidum: Unutilized natural medicine and promising future solution to emerging diseases in Africa. Front. Pharmacol.22(13), 952027 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ekiz, E. et al. Exploring the potential medicinal benefits of Ganoderma lucidum: From metabolic disorders to coronavirus infections. Foods12(7), 1512 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wachtel-Galor, S., Yuen, J., Buswell, J. A. & Benzie, I. F. F. Ganoderma lucidum (Lingzhi or Reishi): A medicinal mushroom. In Herbal Medicine: Biomolecular and Clinical Aspects 2nd edn (eds Benzie, I. F. F. & Wachtel-Galor, S.) (CRC Press, Boca Raton, 2011). [PubMed] [Google Scholar]
- 26.Piccinin, E., Villani, G. & Moschetta, A. Metabolic aspects in NAFLD, NASH and hepatocellular carcinoma: The role of PGC1 coactivators. Nat. Rev. Gastroenterol. Hepatol.16(3), 160–174 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Zhou, D. X. et al. Breeding a new Ganoderma lucidum strain with increased contents of individual ganoderic acids by mono–mono crossing of genetically modified monokaryons. Front. Microbiol.6, 15. 10.3389/fmicb.2024.1410368/full (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toral-Juárez, M. A. et al. Drought-tolerant coffee plants display increased tolerance to waterlogging and post-waterlogging reoxygenation. Environ. Exp. Bot.1(182), 104311 (2021). [Google Scholar]
- 29.Naveenkumar, K. L. Morphological diversity analysis of novel inbred lines of maize (Zea mays L.) for development of single cross hybrids. Int. J. Pure Appl. Biosci.5(1), 489–497 (2017). [Google Scholar]
- 30.Yoon, S. Y., Eo, S. K., Kim, Y. S., Lee, C. K. & Han, S. S. Antimicrobial activity of Ganoderma lucidum extract alone and in combination with some antibiotics. Arch. Pharm. Res.17(6), 438–442 (1994). [DOI] [PubMed] [Google Scholar]
- 31.Wang, G., Chang, L., Zhang, R., Wang, S., Wei, X., Rickert, E., et al. Supplement to: Wang, G et al. (2019): Can targeted defense elicitation improve seaweed aquaculture? Journal of Applied Phycology, 31(3), 1845–1854, 10.1007/s10811-018-1709-6. PANGAEA; 2018 [cited 2025 Mar 20]. Experiment on the effects of oligo-alginate elicitation on juvenile and adult sporophytes of Saccharina japonica and Saccharina latissima. Available from: https://doi.pangaea.de/10.1594/PANGAEA.896664
- 32.Cör Andrejč, D., Knez, Ž & Knez, M. M. Antioxidant, antibacterial, antitumor, antifungal, antiviral, anti-inflammatory, and nevro-protective activity of Ganoderma lucidum: An overview. Front. Pharmacol.22(13), 934982 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anusiya, G. et al. A review of the therapeutic and biological effects of edible and wild mushrooms. Bioengineered12(2), 11239–11268 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.OSF | Lee et al. (2022). Available from: https://osf.io/vjpw4/
- 35.Al-Qursh, F. H. & Ali, I. A. Antifungal activity of ethanol extracts of Ganoderma resinaceum against vaginal fungal isolates. Trop. J. Nat. Prod. Res. TJNPR9(4), 1610–1615 (2025). [Google Scholar]
- 36.Gizaw, A. et al. Phytochemical screening and in vitro antifungal activity of selected medicinal plants against Candida albicans and Aspergillus niger in West Shewa Zone, Ethiopia. Adv. Pharmacol. Pharm. Sci.28(2022), 3299146 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ajith, A. & Janardhanan, K. Indian medicinal mushrooms as a source of antioxidant and antitumor agents. J. Clin. Biochem. Nutr.40(3), 157–162 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanodiya, B. S., Thakur, G. S., Baghel, R. K., Prasad, G. B. K. S. & Bisen, P. S. Ganoderma lucidum: A potent pharmacological macrofungus. Curr. Pharm. Biotechnol.10(8), 717–742 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Kei Kumakura’s research works | Takasaki University of Health and Welfare and other places. Available from: https://www.researchgate.net/scientific-contributions/Kei-Kumakura-2125777496
- 40.Mustafin, C. et al. Diagnostics of ovarian tumors in postmenopausal patients. Diagn. Basel Switz.12(11), 2619 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The nucleotide sequence data generated in this study have been deposited in GenBank under the accession number PV444608. The data will be publicly available on April 9, 2025, or upon the publication of this manuscript, whichever occurs first. The submission can be accessed via the GenBank Submission Portal at https://submit.ncbi.nlm.nih.gov/subs/?search=SUB15234193.