Abstract
Soil bacteria are important contributors to primary productivity and nutrient cycling in arid land ecosystems, and their populations may be greatly affected by changes in environmental conditions. In parallel studies, the composition of the total bacterial community and of members of the Acidobacterium division were assessed in arid grassland soils using terminal restriction fragment length polymorphism (TRF, also known as T-RFLP) analysis of 16S rRNA genes amplified from soil DNA. Bacterial communities associated with the rhizospheres of the native bunchgrasses Stipa hymenoides and Hilaria jamesii, the invading annual grass Bromus tectorum, and the interspaces colonized by cyanobacterial soil crusts were compared at three depths. When used in a replicated field-scale study, TRF analysis was useful for identifying broad-scale, consistent differences in the bacterial communities in different soil locations, over the natural microscale heterogeneity of the soil. The compositions of the total bacterial community and Acidobacterium division in the soil crust interspaces were significantly different from those of the plant rhizospheres. Major differences were also observed in the rhizospheres of the three plant species and were most apparent with analysis of the Acidobacterium division. The total bacterial community and the Acidobacterium division bacteria were affected by soil depth in both the interspaces and plant rhizospheres. This study provides a baseline for monitoring bacterial community structure and dynamics with changes in plant cover and environmental conditions in the arid grasslands.
The arid grasslands of the Colorado Plateau region are productive ecosystems that are very sensitive to environmental changes (16). These expansive grasslands have been severely impacted by human activities over the past 200 years. Changing land practices, extensive animal grazing, and invasion of exogenous plant species have dramatically altered and continue to change the composition and productivity of the native plant communities (6, 10, 16, 25, 31). The grasslands of the Colorado Plateau are dominated by two native perennial bunchgrasses, Stipa hymenoides R.& S. (= Oryzopsis hymenoides [R.& S.] Ricker; Indian ricegrass, hereafter called Stipa) and Hilaria jamesii L. (galleta grass, hereafter called Hilaria) (21, 43), and a nonnative species, Bromus tectorum L. (cheatgrass, hereafter called Bromus) that has invaded the native grassland (6, 31). The bunchgrasses are widely spaced, generally covering 30% or less of the land surface, and the between-plant interspaces are colonized by cyanobacterial and lichen crusts (16, 22).
Nitrogen and water are the two most limiting resources in arid grasslands of the Colorado Plateau. Soil bacteria are responsible for all nitrogen fixation in the soil, and they play essential roles in nitrogen cycling in soil crusts and plant rhizospheres (3, 5, 15, 30, 39). Soil bacteria are also central to cycling carbon and other nutrients, degrading organic matter, and soil development processes that promote moisture retention and fertility (16, 30). Although processes mediated by soil bacteria are essential to arid ecosystem productivity and stability, virtually nothing is known about the composition of natural soil bacterial communities in arid grasslands or the response of these communities to changes in environmental conditions.
DNA-based techniques can provide a comprehensive measure of the diversity and composition of soil bacterial communities, since they survey both the cultured and often-predominant noncultured members of the community. Surveys of soil bacteria by PCR amplification of 16S rRNA genes followed by cloning and sequencing have shown that soil bacterial communities are extremely diverse and may contain abundant noncultured representatives of novel, undescribed bacterial divisions (20). However, due to the complexity of soil communities and the effort required for this type of analysis, cloning and sequencing efforts have been restricted to analysis of a single or a few samples in an environment. No study has yet provided a complete survey of the bacterial community in a single soil sample, or even an adequate set of surveys for field-scale community comparisons. Thus, the spatial heterogeneity in the soil bacterial community and community changes in response to different environmental conditions along gradients remain largely unknown. Information on the changes in the diversity and composition of the soil bacterial community associated with changes in the soil environment is important to understanding and predicting plant productivity and ecosystem health in arid grasslands.
Bacterial community fingerprinting techniques, such as terminal restriction fragment analysis (TRF; reviewed in references 23 and 32), have been used to describe natural bacterial communities in soil, marine, and freshwater environments using the 16S rRNA gene or functional genes. TRF analysis is based on detection of a single restriction fragment in each sequence amplified directly from the environmental sample DNA and is capable of surveying dominant members comprising at least 1% of the total community (12). While the TRF fingerprinting method provides a much lower resolution survey of the diversity and composition of bacterial communities than clone and sequence analysis, it enables analysis of many samples and is thus useful in replicated, field-scale studies. TRF analysis to assess spatial or temporal heterogeneity of microbial communities at the field scale has been used only for a marine environment (36). In the present study, we assessed the use of TRF analysis for field-scale comparisons of soil bacterial communities at two levels of resolution. TRF profiles were generated from 16S rRNA genes representing the total bacterial community and a single bacterial division (the Acidobacterium division) (26, 29). The Acidobacterium division was chosen for this study because it is phylogenetically very diverse and its members have been detected in many soil types and a variety of environments worldwide (1, 20, 26). Our previous studies suggest that they may be abundant in arid land soils (11), yet the vast majority of Acidobacterium division bacteria are not yet cultured and their ecological functions in soils remain unknown.
We used field sample replication to estimate the natural variability expected between individual soil samples and TRF profiles to compare the bacterial communities associated with Stipa, Hilaria, and Bromus rhizospheres and with the interspace areas colonized by cyanobacterial crusts. The bacterial communities present at soil depths of 0 to 10, 10 to 20, and 20 to 30 cm were examined for each rhizosphere and interspace environment. We report broad-scale differences in the soil bacterial communities inhabiting different locations in the soil of this arid grassland.
MATERIALS AND METHODS
Field site and grassland plant species.
The experimental field site was located in the Needles District, Canyonlands National Park, Utah, and was typical of high-elevation arid grasslands in the Colorado Plateau region. The site is sparsely covered with the native bunchgrasses Stipa and Hilaria and has been invaded by the annual grass Bromus. Vegetative cover on this site was approximately 5% Stipa, 5% Hilaria, and 10% Bromus (34). Between the bunchgrasses are open areas that are colonized by cyanobacteria (cyanobacterial soil crusts). The soil on this site is a calcareous loamy sand formed from materials weathered from Permian-aged Cedar Mesa Sandstone, classified as a mesic ustollic camborthid of the Begay series (42). For soil from a depth of 0 to 30 cm, the bulk density averaged 1.52, and pH averaged 7.46 (34). Moisture in this region is deposited as snow and rain in winter and spring and as monsoon storms in the late summer or early fall. Average annual precipitation at the study site was 214 mm, with approximately 45% of this total occurring in July to October.
Soil sample collection.
Soil samples were collected in May 1998, near the end of the spring season of moisture availability and at the end of the growing season for Bromus. Soil samples were collected from four locations in the grassland: the rhizospheres of mature Hilaria and Stipa bunchgrasses, the rhizospheres of Bromus plants where plants had formed a continuous lawn between the bunchgrasses, and the interspaces. Soil samples were taken using a soil corer (3 by 30 cm) that was hammered into the soil to a depth of 30 cm. For each bunchgrass plant, three replicate soil cores were collected from different compass points at the base of the plant into the rhizosphere. For each Bromus or interspace plot, three replicate soil cores were collected approximately 20 cm apart in an area of 0.5 m2. Soil samples were stratified into layers at soil depths of 0 to 10, 10 to 20, and 20 to 30 cm and collected separately. The three replicate soil cores were pooled by stratified layer and were mixed well by shaking in a plastic bag. Each set of three replicate soil cores comprised one field replicate sample. Three replicate field samples were collected from each of three plants per bunchgrass species and each of three Bromus or interspace plots (total 12 field sample locations with three layers of soil collected at each location). The soil coring device was cleaned between samples. Soil samples for DNA analysis were immediately frozen on dry ice. A portion of each sample for bacterial plating assays was stored in ice.
Soil characteristics.
The concentrations of organic matter, nutrients, and cations were measured for soil samples from the Stipa and Bromus rhizospheres and interspaces. A composite sample representing the soil layer at a depth of 0 to 30 cm was collected from three plants or interspace locations, sieved through a 2-mm mesh screen, and sent to the Soil Water and Air Testing Laboratory at New Mexico State University (protocols at http://swatlab.nmsu.edu). Nitrogen concentration and soil moisture were measured in the interspace areas. Soil ammonium and nitrate concentrations were extracted at two sample depths, 0 to 10 and 20 to 30 cm, from water-saturated soil pastes with ion-exchange resin capsules (37, 38, 45), following methods previously described by Miller (34). Over a 4-month period prior to collecting samples for bacterial community analysis, soil moisture at depths of 0 to 10, 10 to 20, and 20 to 30 cm was measured gravimetrically at biweekly intervals and converted to volumetric water content using bulk density values (34).
Soil DNA preparation.
DNA was extracted from two 0.5-cm3 subsamples for each soil sample using a small-scale procedure that included incubation in sodium dodecyl sulfate detergent lysis buffer at 70°C, followed by bead mill homogenization and ethanol precipitation (27). Extracted DNA was visualized under UV light after electrophoresis in ethidium bromide-stained agarose gels. Extracted DNA was predominantly high molecular weight (≥24 kb). DNA was quantified from the gels using a calibrated set of standard DNAs of known concentrations, and duplicate samples were pooled. Soil DNA was purified for PCR by passage through Sephadex G-200 minicolumns prepared in 96-well plates as previously described (27). DNA in the column eluate was precipitated in sodium acetate and ethanol to concentrate for storage.
PCR of 16S rRNA genes.
Two primer sets were used to compare the bacterial communities in the soil samples. Primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′, Escherichia coli positions 8 to 27) (14) and 1492R (5′-TACCTTGTTACGACTT-3′, E. coli positions 1507 to 1492) (44) were used to amplify representatives of the total bacterial community. Acidobacterium division-specific primer 31F (5′-GATCCTGGCTCAGAATC-3′, E. coli positions 15 to 31) (1) was used with bacterial primer 1492R to amplify rRNA genes from members of the Acidobacterium division. The forward primers pA and 31F were labeled with 6-carboxyfluorescein during synthesis. Each 50-μl reaction mixture contained 30 mM Tris (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 50 μM (each) deoxynucleoside triphosphate, 50 pmol (each) primer, 0.75 U of Taq polymerase (AmpliTaq LD; Perkin-Elmer, Foster City, Calif.), and 200 pg of soil DNA. The cycling conditions for the pA-1492R reactions follow: (i) 2 min of denaturation at 94°C, (ii) 35 cycles, with 1 cycle consisting of 30 s at 50°C, 1 min at 72°C, and10 s at 94°C, and (iii) a final cycle of annealing at 55°C for 1 min and extension at 72°C for 5 min. Cycling conditions for the 31F-1492R reactions were the same as described above except that an annealing temperature of 42°C was used. Three independent PCRs on 50-μl reaction mixtures were performed for each soil sample and combined. PCR products were purified with a QIAquick PCR cleanup kit (Qiagen, Inc., Chatsworth, Calif.). Amplified products were separated by electrophoresis on agarose gels to quantify the amplicon and to verify that the correct size product was produced.
TRF profiles.
Purified amplicons from the PCRs were digested with two restriction enzymes in separate reactions. Four different enzymes (RsaI, MspI, HaeIII, and HhaI) were tested in preliminary trials and selected for the ability to generate many fragments that were well separated in acrylamide gels. The RsaI and MspI enzymes were used for analysis of the total bacteria, and the RsaI and HaeIII enzymes were used for analysis of the Acidobacterium division (12). One microliter (∼5.0 ng) of each amplicon was dried, suspended in 1.75 μl of loading buffer containing 0.25 μl of Genescan 2500 tetrachloro-6-carboxyfluorescein (TAMRA)-labeled size standard (Applied Biosystems, Inc., Foster City, Calif.) and a 5:1 mixture of deionized formamide-dextran blue and 25 mM EDTA, and then denatured at 94°C for 2 min. DNA fragments were separated by electrophoresis in denaturing 4% polyacrylamide gels (reagents from Bio-Rad, Hercules, Calif.) using an ABI 377 DNA sequencer. The sizes of fluorescently labeled TRFs between 90 and 827 bp in length were determined using Genescan version 2.02 analytical software (Applied Biosystems, Inc.). To maximize the number of TRFs retained in comparisons between locations, we kept the profile baseline low, at 25 absorbance units. This increased the observed variability between replicate samples by retaining small variable peaks but also retained some small peaks that were consistently present (13). For each sample, an aliquot of each digest was applied to three separate gels to obtain three replicate profiles (technical replicates). In addition, one sample was run on all gels as an additional internal control to detect between-gel variability (12). All samples were treated uniformly during DNA extraction, PCR, and TRF analysis, using master mixes for all reactions.
Analysis of TRF profiles.
The TRF profiles derived from each one of the two restriction digests for each technical replicate were combined in tandem array. Technical replicates for each sample were compared to identify the fragment sizes that appeared in at least two of the three technical replicates using clustering algorithms for profile alignment (41). Peak heights ranged from 26 to several thousand absorbance units. The average height of each peak was calculated from the technical replicates for an individual sample. The averaged profile for each set of three technical replicates comprised one field replicate sample and was used in all subsequent analyses. TRF profiles from different samples must be standardized to control for variability arising from slightly different DNA concentrations loaded on gels (13) and to enable statistical comparison based on distance measures that require equal likelihood of a fragment appearing if it is present in a sample (i.e., equal number of peaks in each profile or equal total fluorescence value). Two techniques were compared for standardization of the soil sample TRF profiles.
The peak height standardization method restricted comparisons to an equal number of the most-abundant fragments in each sample (largest peaks in the profile). The number of fragments used in the analysis was set to the number of fragments in the profile having the fewest fragments (about 100 fragments for the total community comparisons and about 20 fragments for the Acidobacterium division comparisons). The underlying assumption in this standardization is that if the samples are identical, the most-abundant fragments (largest peaks) should be identical. Thus, comparing an equal number of the largest peaks gives a measure of similarity between TRF profiles. By confining analysis to the largest peaks in the profile, this method compared the most-abundant TRFs in the community.
The fluorescence standardization method used the total fluorescence value of each TRF profile as a relative measure of the total DNA in the sample. This method controlled for slight differences in sample DNA concentration in gels by normalizing all the TRF profile data to the same total fluorescence value by calculating the sum of peak heights in each profile (13). The total fluorescence was standardized between samples by proportionally decreasing the height of each peak in the average profiles until the sum of peak heights (total fluorescence) for each profile equaled the lowest value represented among the samples in a comparison group. Adjustment of larger sample sizes resulted in the elimination of a small proportion of peaks from a profile, as some adjusted peak heights dropped below the noise threshold (peak height of 25). A low total fluorescence sample can remove from comparison much information from other profiles having higher total fluorescence values. For this reason, DNA concentrations must be kept similar across experimental treatments during gel electrophoresis if this standardization method is used.
TRF profiles were analyzed using S+ version 3.2 (MathSoft, Inc. Seattle, Wash.). TRF profiles were converted to binary data (presence or absence of a peak) for distance comparisons using the Jaccard distance measure. Dendrograms were generated from Jaccard distance matrices using an unweighted paired group means analysis (UPGMA). t tests were used to determine whether there were significant differences between each pair of environments in a comparison group. Principal component analysis (PCA) was conducted on some of the comparison groups to confirm the groupings observed in the UPGMA dendrograms.
Bacterial plating assays.
Soil dilutions were plated within 24 h of soil collection. A 10-g subsample of each soil sample was suspended in sterile water to a final volume of 50 ml. Ten-milliliter serial dilutions to 50,000-fold dilution were prepared. Aliquots (100 μl) of the dilutions were plated onto 1/10 strength Trypticase soy broth (TSB) agar (3 g of powdered TSB [Difco] per liter and 15 g of agar [Difco] per liter). Three replicate plates were used for each one of the two soil dilutions for each soil sample. The final sample dilution on plates was 5 × 105 to 5 × 106. Inoculated plates were incubated at 26°C for 3 days before the colonies were counted. Dilution plates with 100 to 300 colonies per plate were counted. Plate count data were analyzed using analysis of variance on raw data and ranked data (to normalize the count data). Variables with significant F values (P ≤ 0.05) were further analyzed using Tukey's Studentized t test mean separation procedure. Statistical analyses were conducted using SAS software (35).
RESULTS
Replication and standardization of TRF profiles.
Preliminary analysis of the TRF profiles used all the technical replicate data as separate profiles for calculation of Jaccard distance matrices and construction of UPGMA dendrograms (three technical replicates for each field sample). Visualization of the data in this manner provided an assessment of the variability between technical replicates. The three technical replicates of each field sample clustered more closely to each other than to any other sample, with a maximum distance of 0.20 (data not shown).
Two standardization methods were applied to the data. The TRF profiles contain information about the number, location, and height of the peaks, and the two methods for standardizing data before comparison use this information in different ways. For the data set in this study, where the peak numbers and DNA concentrations were similar for all the sample profiles, the primary effect of the different standardization procedures was to slightly change the distance measure values without changing the observed relationships between the samples in UPGMA dendrograms. The topologies of the dendrograms generated with the two standardization procedures were essentially the same, and the results and conclusions discussed below could be drawn from use of either standardization procedure. The figures and tables shown in this study are derived from fluorescence standardized data.
The number of TRFs in each field sample (determined after fluorescence standardization) was compared by analysis of variance. The number of TRFs was not significantly different (P = 0.05) across the different soil environments. With fragments from two enzyme digests analyzed together, the fluorescence-standardized TRF profiles averaged 132 (standard deviation, 12) and 37 (standard deviation, 7) peaks for the total bacterial community and the Acidobacterium division, respectively.
Two types of analyses were conducted using the Jaccard distances calculated from the TRF profiles. The UPGMA dendrograms provided a visual illustration of the differences observed between field replicates for an environment and between the communities residing in different environments. The results of t tests determined whether the distance observed between each pair of environments was significantly greater than the distance between replicates within an environment.
Comparisons of the soil bacterial communities from interspaces and the rhizospheres of plants.
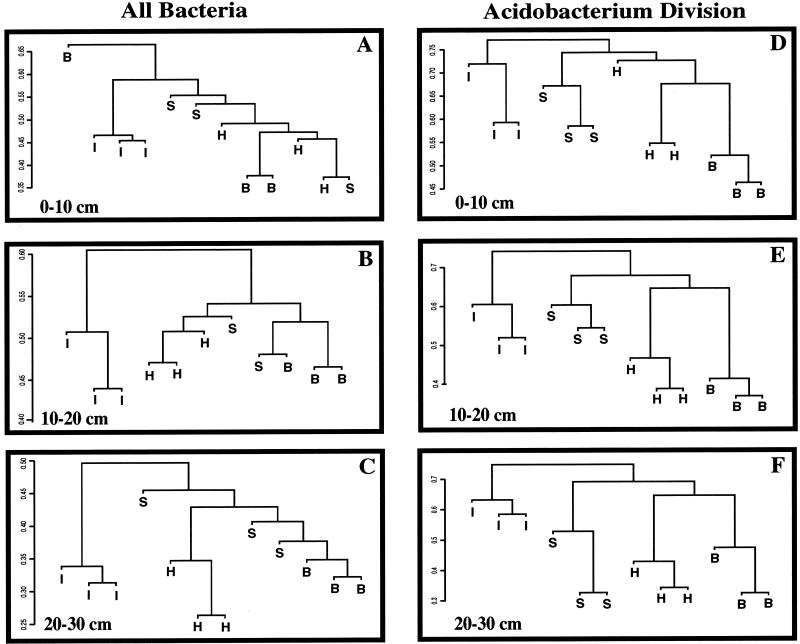
Figure 1 illustrates UPGMA dendrograms of the differences in soil bacterial communities from the interspaces and the rhizospheres of the three grass species. A dendrogram was generated to compare the plant and interspace communities at each soil depth. The Jaccard distances are illustrated on the y axis of each dendrogram. As expected, field replicate profiles of a given environment were somewhat variable (e.g., the three interspace samples). When the three field replicates representing an environment were more similar to each other than to the other samples in the comparison group, they appeared in the dendrograms as clusters with lower distance value than the between-environment comparisons. For example, for comparisons of the total bacterial community at the soil depth of 20 to 30 cm (Fig. 1C), the three interspace samples had a distance measure of 0.31 to 0.34 (on a scale where 0 represents identity and 1 represents no similarity). However, the distance between the interspace group and any of the plant groups was higher, at approximately 0.5.
FIG. 1.
Relationships between soil bacterial communities associated with interspaces (I) and the rhizospheres of Hilaria (H), Stipa (S), and Bromus (B) plants in an arid grassland, illustrated by UPGMA dendrograms generated from Jaccard distance matrices of TRF profiles. The y axis is the Jaccard distance measure. Panels A to C illustrate comparisons of the total bacterial community by PCR amplification of the 16S rRNA gene from soil DNA using primers conserved for all bacteria, followed by generation of TRF profiles. Panels D to F illustrate a similar analysis that focused on the Acidobacterium division. Each branch on a tree represents a field sample comprised of three pooled soil cores and a combination of three technical replicate TRF profiles that were averaged as described in the text. Because of significant differences between communities associated with different soil depths, comparisons between the plants are confined to a single depth. For the total bacterial community comparison, one of the Stipa samples at a depth of 10 to 20 cm was lost during processing and is missing from the figure.
The interspace samples clustered separately and were distinct from the plant rhizospheres at all three soil depths in TRF analysis of both the total bacterial community and the Acidobacterium division (Fig. 1A to C and D to F, respectively). t tests conducted between the interspaces and each plant species supported the observation from the UPGMA dendrograms. Each pair of t tests (comparing interspace and Bromus, interspace and Stipa, and interspace and Hilaria) was significantly different (P < 0.001, with P values ranging from 1 × 10−7 to 1 × 10−9), indicating that distinctly different bacterial communities were present in the interspaces and the plant rhizospheres. This was true for the analysis at each soil depth, using data from the total bacterial community or only the Acidobacterium division.
Soil organic matter was very low (0.25 to 0.30%), and measurements were typical of soils from this region where plants are sparse and water is scarce. The soil organic matter and cation concentrations measured from the between-plant interspaces and plant rhizospheres were similar, and the amounts of N, P, and K were slightly higher for the between-plant interspaces than for the plant rhizospheres (Table 1). The cultured bacterial counts from the interspaces and plants were not significantly different, but the amount of extracted DNA from the interspaces was lower than those from the plant rhizospheres (Table 2).
TABLE 1.
Organic matter, nutrient, and cation content for soils collected from Stipa and Bromus rhizospheres and from the interspaces
| Location | Organic matter (%) | Nutrient content (ppm)
|
Cation content (milliequivalents/liter)
|
||||
|---|---|---|---|---|---|---|---|
| N | P | K | Mg | Ca | Na | ||
| Interspace | 0.29 | 4.0 | 2.4 | 105 | 0.73 | 2.80 | 0.83 |
| Stipa | 0.25 | 2.4 | 1.9 | 93 | 0.64 | 2.74 | 0.87 |
| Bromus | 0.30 | 2.7 | 1.5 | 82 | 2.70 | 1.50 | 0.82 |
TABLE 2.
Plate counts of bacterial heterotrophs and the quantity of DNA extracted from soil in four environments at three soil depths
| Location | Soil depth (cm) | Bacterial count (105 CFU/g of soil)a | Amt of extracted DNA (μg/0.5 cm3 of soil) |
|---|---|---|---|
| Stipa | 0-10 | 73 (43) A | 2.5 |
| 10-20 | 19 (14) AB | 2.0 | |
| 20-30 | 13 (9) B | 1.5 | |
| Hilaria | 0-10 | 105 (25) A | 2.5 |
| 10-20 | 33 (10) AB | 1.5 | |
| 20-30 | 16 (14) B | 0.35 | |
| Bromus | 0-10 | 96 (60) A | 2.5 |
| 10-20 | 55 (25) AB | 1.5 | |
| 20-30 | 8 (7) B | 0.25 | |
| Interspace | 0-10 | 51 (12) A | 0.80 |
| 10-20 | 44 (39) AB | 0.35 | |
| 20-30 | 19 (26) B | 0.10 |
Bacterial heterotrophs were cultured on 1/10× TSB medium. Bacterial counts are averages of three plates per dilution per field sample × three field samples = nine observations. Standard deviations of the means are shown in parentheses. Bacterial counts followed by the same letter are not significantly different (P = 0.05) by Tukey's Studentized t test.
Comparisons of the soil bacterial communities from plant species.
The field replicates for a plant species did not always cluster together visually in the UPGMA dendrograms from TRF analysis of the total bacterial community. The exception was Hilaria, which clustered separately from the other two plants in the subsurface layers (Fig. 1B and C). Despite the observed field replicate variability, the t tests comparing the total bacterial community between each pair of plant species (Stipa and Bromus, Stipa and Hilaria, and Hilaria and Bromus) were significantly different (P < 0.001, with actual P values ranging from 1 × 10−7 to 1 × 10−9). For the Acidobacterium division comparisons, major differences in community composition are apparent from the UPGMA dendrograms. Each grass species rhizosphere community forms a distinct cluster at all three soil depths (Fig. 1D to F). Each set of three field samples for a plant species clustered most closely together and was distinct from the other plant species. The t tests comparing Acidobacterium division members between each pair of plant species were also significantly different (P < 0.001, with actual P values ranging from 1 × 10−7 to 1 × 10−9) at each depth. The numbers of cultured bacteria were not significantly different for the different plant species (Table 2), and the concentrations of DNA extracted from the three plant species were similar for samples from soil depths of 0 to 10 cm and 10 to 20 cm. For the soil from a depth of 20 to 30 cm, more DNA was extracted from Stipa (Table 2), which has a deeper root system than the other two plant species.
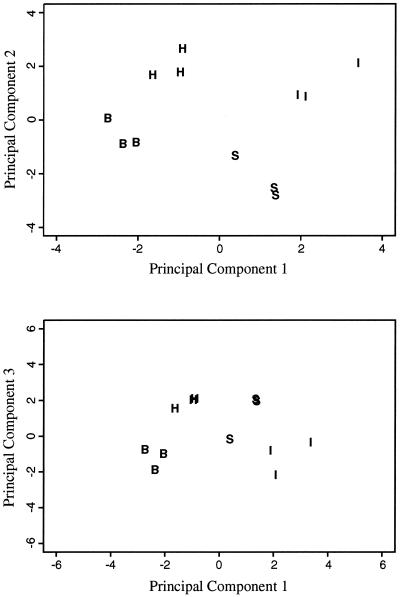
The clustering patterns obtained with UPGMA dendrograms generated from Jaccard distance matrices were compared with those obtained from PCA (Fig. 2 and data not shown). PCA and the Jaccard distance measure use the binary TRF data in different ways, and conducting the two different analyses improved our confidence in the observed UPGMA clusters. For example, the results of PCA for the Acidobacterium community at a soil depth of 20 to 30 cm (Fig. 2) illustrate the same clustering pattern as the UPGMA dendrogram (Fig. 1F).
FIG. 2.
Relationships between the Acidobacterium division bacterial communities by PCA. Acidobacterium division bacterial communities from the interspaces (I) and from the rhizospheres of Hilaria (H), Stipa (S), and Bromus (B) plants at a soil depth of 20 to 30 cm are shown. The proportion of variance for the first, second, and third components was 0.2441, 0.1913, and 0.1523, respectively. The first and second components explained 43.5% of the variability, and the first and third components explained 39.6% of the variability.
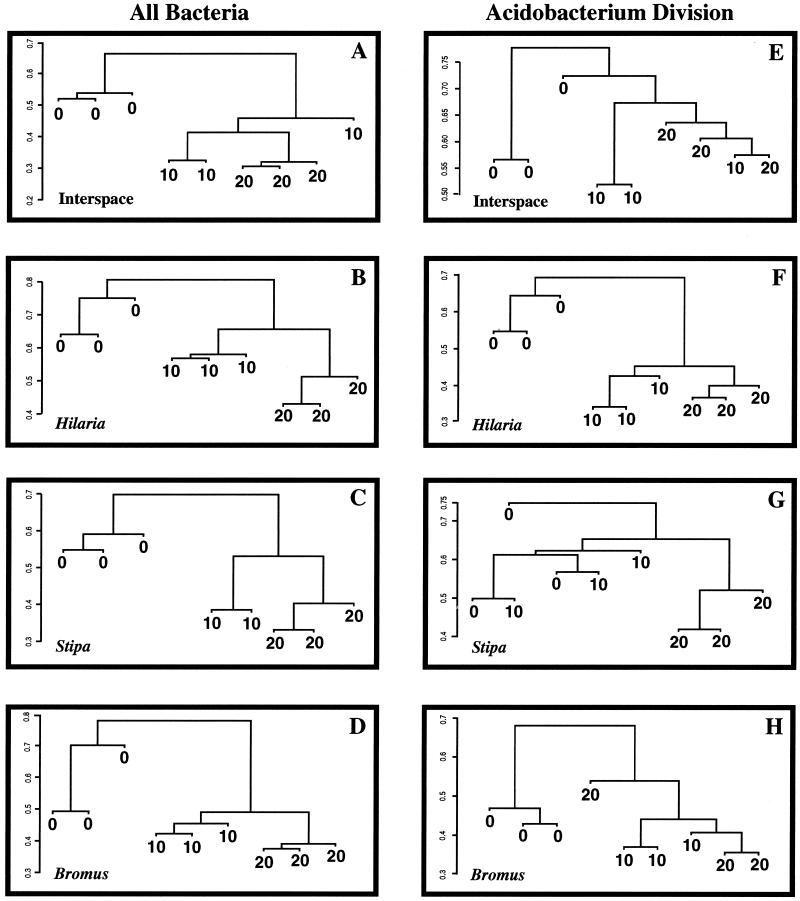
Soil bacterial community differences with depth.
Figure 3 illustrates the UPGMA dendrograms generated comparing the total bacterial community (Fig. 3A to D) and members of the Acidobacterium division (Fig. 3E to H) associated with the plants or interspaces at three soil depths. For all locations (plants and interspaces), the TRF profiles representing the surface environment (0 to 10 cm deep) clustered independently from the two subsurface environments in the total bacterial community analysis. Variability between field replicates was lower in the two subsurface environments than in the surface environment. The Jaccard distances between the surface and subsurface samples were large: 0.65 in the interspace (Fig. 3A) and 0.70 to 0.80 in the plant rhizosphere environments (Fig. 3B to D). Differences between the total bacterial communities in the soil layers at depths of 10 to 20 and 20 to 30 cm were clearly defined for all three plant species but not for the interspace samples. t tests of the values for different soil depths (0 to 10 versus 10 to 20 cm, 0 to 10 versus 20 to 30 cm, and 10 to 20 versus 20 to 30 cm) were significantly different (P < 0.001, with actual P values ranging from 1 × 10−7 to 1 × 10−9) for each depth comparison in each of the four plant and between-plant interspace environments.
FIG. 3.
Relationships between soil bacterial communities at three depths in four different environments in an arid grassland, illustrated by UPGMA dendrograms generated from Jaccard distance matrices of TRF profiles. The y axis is the Jaccard distance measure. Panels A to D illustrate comparisons of the total bacterial community by PCR amplification of 16S rRNA genes from soil DNA using primers conserved for all bacteria, followed by generation of TRF profiles. Panels E to H illustrate a similar analysis that focused on the Acidobacterium division. Each branch on a tree represents a field sample comprised of three pooled soil cores and a combination of three technical replicate TRF profiles that were averaged as described in the text. Because of significant differences between communities associated with the different grass species, comparisons between soil depths are confined to a single plant species or the interspace. In the figure, 0, 10, and 20 represent soil from a depth of 0 to 10, 10 to 20, and 20 to 30 cm, respectively. For the total bacterial community comparison, one of the Stipa samples at a depth of 10 to 20 cm was lost during processing and is missing from the figure.
The composition of Acidobacterium division members was also affected by soil depth (Fig. 3E to H). The differences were not resolved as well as the differences in the total bacterial community analysis, suggesting that different bacterial groups are affected differentially by soil depth. The surface community (soil depth of 0 to 10 cm) clustered separately from the two subsurface layers in the Hilaria and Bromus rhizospheres (Fig. 3F and H, respectively), and community differences were resolved at all three soil depths in Hilaria rhizospheres. In the interspace and Stipa rhizospheres (Fig. 3E and G, respectively), dendrogram results were more variable. As with the total community comparisons, the t tests of Acidobacterium division communities for all pairwise depth comparisons were significantly different (P < 0.001, with actual P values ranging from 1 × 10−7 to 1 × 10−9) for each depth comparison in each of the four plant and interspace environments.
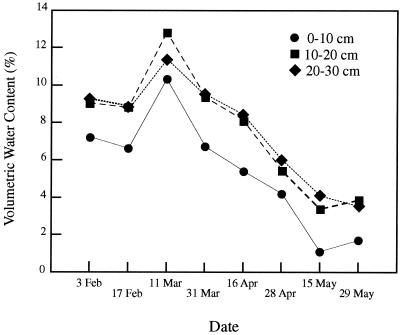
Soil chemistry also varied with depth. The concentration of nitrogen measured in the interspaces was higher in the soil layer at a depth of 0 to 10 cm (4.7 and 2.0 ppm of NH4+ and NO3−, respectively) than in the subsurface at a depth of 20 to 30 cm (1.3 and 1.5 ppm of NH4+ and NO3−, respectively). The volumetric water content in the soil layer at a depth of 0 to 10 cm was consistently lower than in the subsurface layers for 4 months prior to sampling and at the time samples were collected (Fig. 4). Consistent with precipitation trends in this area, soil moisture decreased over time from February to May.
FIG. 4.
Seasonal changes in volumetric water content in study site soils at three soil depths from February to May in 1998.
Bacterial plate counts were significantly different with soil depth (type I sum of squares on unranked and ranked data had P > F values of 0.0236 and 0.0001, respectively). Comparisons for the different soil depths using the unranked data and Tukey's Studentized t test (95% confidence interval) indicated that bacterial plate counts were significantly higher for soil from a depth of 0 to 10 cm than that from 20 to 30 cm, with the value for soil from 10 to 20 cm intermediate (Table 2). High plate counts in the surface soil are possibly due to the presence of gram-positive spore-forming bacteria that are commonly isolated on 1/10 × TSB (11). The concentration of DNA extracted from soil samples of equal volume decreased with depth in both rhizosphere and between-plant interspace locations (Table 2). The subsurface DNA concentration was highest in the deep-rooted Stipa rhizosphere and lowest in the between-plant interspaces.
DISCUSSION
Soil bacterial communities and the soil processes mediated by bacteria are critical for ecosytem functioning and productivity in arid lands. There is a need to integrate the soil bacterial community into our understanding of ecosystem interactions at a scale relevant to the whole-plant, between-plant, and landscape levels. However, successfully accomplishing this has been extremely difficult. The microscopic nature and the immense diversity of soil bacteria have so far precluded accurate or comprehensive surveys of soil bacterial species, and heterogeneity within the soil matrix has made it difficult to obtain meaningful ecological information from single-sample investigations. TRF analysis can provide a reliable, albeit low-resolution, fingerprint of complex bacterial communities. We conducted the present field study using TRF analysis to determine if there were differences in the soil bacterial communities associated with the rhizospheres of different plant species and the interspace areas in an arid grassland and if the soil bacterial communities in a location changed with soil depth. We found that generation of 16S rRNA gene TRF profiles, coupled with Jaccard distance measure or PCA, was a useful technique for comparing the broad-scale composition of complex soil bacterial communities. By employing field replication, we were able to identify consistent differences in soil bacterial communities across the natural heterogeneity in soils and to differentiate the soil bacterial communities in different locations in an arid grassland.
A TRF profile derived from the pool of 16S rRNA genes amplified from an environmental sample contains information on the number of different fragments, their length (in base pairs), and their relative abundance. Use of TRF peak number as a measure of species richness in bacterial communities has been attempted (12, 13) but has been problematic for two reasons. An individual TRF does not always represent an individual species or genus, and different restriction enzymes can suggest different levels of phylotype richness within a community or different trends between communities. Despite the limitations in identifying individual species or in describing species richness, the TRF method was useful for detecting compositional differences in complex soil communities at different soil locations. Community differences were detected in the total bacterial community as well as within a single bacterial division. Confining the analysis to a single division did not always improve the resolution of the TRF method. The Acidobacterium division results for interspace comparisons were less variable than the results for total community analysis, but the reverse was true for the depth comparisons. By comparing TRF profiles, sets of TRFs unique to a particular environment were identified (data not shown). These unique TRFs are useful for continued monitoring of community responses to changes in the soil environment. Sequence analysis of individual TRFs extracted from gels can identify the bacterial species comprising that TRF, determine whether the TRF represents one or multiple species, and identify species unique to a particular environment.
Our TRF results demonstrated that the bacterial communities from the soil rhizospheres were very different from communities from the interspaces and that reliable TRF community fingerprints that define the plant rhizosphere and interspace environments could be generated. In arid landscapes, plants are considered to be “islands of fertility” (43), with total soil nutrients, microbial biomass C and N, and bulk measurements of microbial activity generally higher in surface soils around grasses and shrubs than in the interspaces (2, 7, 8, 43). As plants invade the soil environment, they alter the local soil conditions. Invasion of Bromus into the landscape displaces soil crusts and results in changes in the chemical and nutritional status of the soil (4, 17). We have documented large differences in the soil bacterial communities from the uninvaded interspaces and Bromus-invaded soils, indicating that Bromus invasion alters the composition of the soil bacterial community. Continued monitoring of the interspace soil bacterial community as Bromus invades the grassland may help identify shifts in the community responsible for the observed changes in soil chemistry and nutrition that occur with Bromus invasion.
Our measurement of total soil biomass (extracted DNA) was lower in the interspace than in the plant rhizospheres and supports the idea of plants as “islands of fertility.” However, our measurements of soil organic matter content and abundance of cultured bacterial heterotrophs for the interspaces and plant rhizospheres were similar. In a study of an arid shrub-steppe community, Bolton et al. (7) also found cultured bacterial counts to be similar between the shrub and interspace soils. The interspaces in our study were colonized by cyanobacteria (cyanobacterial soil crusts), which improve soil nutritional conditions. Soil crusts are an important source of nitrogen (15) and substantial contributors to carbon input (28) in arid ecosystems. The input of nitrogen and carbon by soil crusts has been shown to benefit interspace soil bacteria (4, 17) and to impact the nutritional status of neighboring plants (3, 19). The numbers of TRFs (number of peaks) in profiles from the interspaces were similar in the cyanobacterium-colonized surface and the two subsurface soil samples and were similar to the number of TRFs in the plant rhizosphere profiles. The interspace and rhizosphere profiles differed only in the sizes of TRFs (presence and absence of different peaks) in the profiles. These results indicate that the interspace areas contain complex bacterial communities in the crusted surface and subsurface environments and that the interspaces contain many bacterial members that are distinct from those of the rhizosphere communities.
Differences in rhizosphere bacterial community composition were detected between plant species and were most pronounced with the Acidobacterium division comparisons. The three plant species compared in this study have different root growth habits that may alter local soil conditions and thus contribute to the observed bacterial community differences. Stipa is the most deeply rooted species of the three, with a fibrous root system that can extend to a depth of 1.5 m (33). Hilaria is generally more shallow rooted than Stipa. It reproduces vegetatively by forming rhizomes that extend across the soil surface with associated roots extending into the soil below. In contrast to the native bunchgrasses, Bromus is an annual winter grass that germinates and sustains root growth in cold winter temperatures when the bunchgrasses are dormant. The Bromus root system typically extends 15 to 30 cm into the soil (40), with 75% of the roots found in the upper 15 to 20 cm (9, 33). Our study has provided a baseline for further characterization of changes in rhizosphere bacterial composition as this grassland responds to changing plant cover and environmental conditions.
Bacterial abundance and community composition were affected by soil depth. In the low organic matter soils of this region, the most limiting resources are nitrogen and water, and gradients of these two resources at different depths could affect soil bacterial communities. Our measurements indicate that the soil layer at a depth of 0 to 10 cm from the interspaces is more nitrogen-rich and consistently drier than the subsurface layers. In the present study, total extractable DNA and cultured heterotroph abundance decreased with soil depth, suggesting a decrease in soil biomass with depth. This is consistent with results of other soil studies, where total microbial biomass (including bacteria, fungi, and other microscopic eukaryotes), as measured by biomass C and N (7, 18) and ATP and enzyme activities (7), was found to decrease with soil depth. Kloos et al. (24) observed that cultured denitrifiers and N-fixing bacteria in four soils in Germany decreased with depth, but this was not observed with heterotroph populations in arid shrub-steppe soils of eastern Washington (7).
Our TRF comparisons augment the above studies by showing that composition of the soil biomass changes with depth. This was true for the analysis of the total bacterial community and for the more focused analysis of the Acidobacterium division. Using phospholipid fatty acid patterns to examine total microbial community structure in soils of different depth, Fritze et al. (18) also found that the phospholipid fatty acid pattern changed with increasing soil depth, suggesting a shift in microbial community composition. The reasons for such large community shifts in soil at a depth of 0 to 30 cm are unknown. They probably include the observed changes in soil nitrogen and/or moisture and may also include plant root activity and exudates or other chemical and physical changes in the soil.
A diverse array of Acidobacterium division bacteria were found in soils of this arid grassland. The composition of Acidobacterium division members differed with soil depth and with the presence of plants or a cyanobacterial crust. The functional roles of these bacteria in soils are still unknown. Our study indicates that different members of this division occupy specific niches in the grassland soil, and continued monitoring of their dynamics in response to changing soil conditions may provide clues about their biology. Collecting TRFs for sequence analysis that are unique to different environments will help identify more precisely the abundant Acidobacterium division populations in different soil locations.
Acknowledgments
We thank the National Park Service staff at Needles District, Canyonlands National Park for their cooperation and support.
This work was funded in part by a grant to C.R.K. from the U.S. Department of Energy, OBER Program for Ecosystem Research. M.E.M. was supported through a Star graduate education fellowship awarded by the U.S. Environmental Protection Agency.
REFERENCES
- 1.Barns, S. M., S. L. Takala, and C. R. Kuske. 1999. Wide distribution and diversity of members of the Acidobacterium bacterial kingdom in the environment. Appl. Environ. Microbiol. 65:1731-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, R. C., and J. O. Klemmedson. 1978. Shrub-induced spatial patterns of dry matter, nitrogen, and organic carbon. Soil Sci. Soc. Am. J. 42:804-809. [Google Scholar]
- 3.Belnap, J., and K. T. Harper. 1995. Influence of cryptobiotic soil crusts on elemental content of tissue of two desert seed plants. Arid Soil Res. Rehabil. 9:107-115. [Google Scholar]
- 4.Belnap, J., and S. Phillips. 2001. Soil biota in an ungrazed semi-arid grassland: response to an exotic annual grass invasion. Ecol. Appl. 11:1261-1275. [Google Scholar]
- 5.Belnap, J., K. T. Harper, and S. D. Warren. 1994. Surface disturbance of cryptobiotic soil crusts: nitrogenase activity, chlorophyll content, and chlorophyll degradation. Arid Soil Res. Rehabil. 8:1-8. [Google Scholar]
- 6.Billings, D. B. 1990. Bromus tectorum, a biotic cause of ecosystem impoverishment in the Great Basin, p. 301-322. In G. M. Woodwell (ed.), The earth in transition: patterns and processes of biotic impoverishment. Cambridge University Press, New York, N.Y.
- 7.Bolton, H., Jr., J. L. Smith, and S. O. Link. 1993. Soil microbial biomass and activity of a disturbed and undisturbed shrub-steppe ecosystem. Soil Biol. Biochem. 25:545-552. [Google Scholar]
- 8.Charley, J. L., and N. E. West. 1977. Micro-patterns of nitrogen mineralization activity in soils of some shrub-dominated semi-desert ecosystems of Utah. Soil Biol. Biochem. 9:357-365. [Google Scholar]
- 9.Cline, J. F., D. W. Uresk, and W. H. Rickard. 1977. Comparison of soil water used by a sagebrush-bunchgrass and a cheatgrass community. J. Range Manag. 30:199-201. [Google Scholar]
- 10.D'Antonio, C. M., and P. M. Vitousek. 1992. Biological invasions by exotic grasses, the grass/fire cycle, and global change. Annu. Rev. Ecol. Syst. 23:63-87. [Google Scholar]
- 11.Dunbar, J., S. Takala, S. M. Barns, J. A. Davis, and C. R. Kuske. 1999. Levels of bacterial community diversity in four arid soils compared by cultivation and 16S rRNA gene cloning. Appl. Environ. Microbiol. 65:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2000. Assessment of microbial diversity in four southwestern United States soils by 16S rRNA gene terminal restriction fragment analysis. Appl. Environ. Microbiol. 66:2943-2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edwards, U., T. Rogall, H. Blöcker, M. Emde, and E. C. Böttger. 1989. Isolation and direct complete determination of entire genes. Nucleic Acids Res. 17:7843-7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Evans, R. D., and J. R. Ehleringer. 1993. A break in the nitrogen cycle in arid lands? Evidence from δ15N of soils. Oecologia 94:314-317. [DOI] [PubMed] [Google Scholar]
- 16.Evans, R. D., and J. R. Johansen. 1999. Microbiotic crusts and ecosystem processes. Crit. Rev. Plant Sci. 18:183-225. [Google Scholar]
- 17.Evans, R. D., R. Rimer, L. Sperry, and J. Belnap. Exotic plant invasion alters nitrogen dynamics in an arid grassland. Ecol. Appl., in press. [DOI] [PubMed]
- 18.Fritze, H., J. Pietikäinen, and T. Pennanen. 2000. Distribution of microbial biomass and phospholipid fatty acids in podzol profiles under coniferous forest. Eur. J. Soil Sci. 51:565-573. [Google Scholar]
- 19.Harper, K. T., T. Kimball, and J. Belnap. 2001. The influence of biological soil crusts on mineral uptake by associated vascular plants. J. Arid Environ. 47:347-357. [Google Scholar]
- 20.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaynes, R. A., and K. T. Harper. 1978. Patterns of natural re-vegetation in arid southeastern Utah. J. Range Manag. 31:407-411. [Google Scholar]
- 22.Johansen, J. R. 1993. Cryptogamic crusts of semiarid and arid lands of North America. J. Phycol. 29:140-147. [Google Scholar]
- 23.Kitts, C. L. 2001. Terminal restriction fragment patterns: a tool for comparing microbial communities and assessing community dynamics. Curr. Issues Intest. Microbiol. 2:17-25. [PubMed] [Google Scholar]
- 24.Kloos, K., U. M. Hüsgen, and H. Bothe. 1998. DNA-probing for genes coding for denitrification, N2-fixation and nitrification in bacteria isolated from different soils. Z. Naturforsch. Sect. C 53:69-81. [DOI] [PubMed] [Google Scholar]
- 25.Knapp, P. A. 1996. Cheatgrass (Bromus tectorum L.) dominance in the Great Basin desert. Global Environ. Change 6:37-52. [Google Scholar]
- 26.Kuske, C. R., S. M. Barns, and J. D. Busch. 1997. Diverse uncultivated bacterial groups from soils of the arid southwestern United States that are present in many geographic regions. Appl. Environ. Microbiol. 63:3614-3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuske, C. R., K. L. Banton, D. L. Adorada, P. C. Stark, K. K. Hill, and P. J. Jackson. 1998. Small-scale DNA sample preparation method for field PCR detection of microbial cells and spores in soil. Appl. Environ. Microbiol. 64:2463-2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lange, O. L. 2001. Photosynthesis of soil-crust biota as dependent on environmental factors, p. 217-240. In J. Belnap and O. L. Lange (ed.), Biological soil crusts: structure, function and management. Springer-Verlag, Berlin, Germany.
- 29.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 30.Lynch, J. M. (ed.) 1990. The rhizosphere. John Wiley and Sons, New York, N.Y.
- 31.Mack, R. N. 1981. Invasion of Bromus tectorum L. into western North America: an ecological chronicle. Agroecosystems 7:145-165. [Google Scholar]
- 32.Marsh, T. L. 1999. Terminal restriction fragment length polymorphism (T-RFLP): an emerging method for characterizing diversity among homologous populations of amplification products. Curr. Opin. Microbiol. 2:323-327. [DOI] [PubMed] [Google Scholar]
- 33.Melagoza, G., and R. S. Nowak. 1991. Competition between cheatgrass and two native species after fire: implications from observations and measurements of root distribution. J. Range Manag. 44:27-33. [Google Scholar]
- 34.Miller, M. E. 2000. Effects of resource manipulations and soil characteristics on Bromus tectorum L. and Stipa hymenoides R.& S. in calcareous soils of Canyonlands National Park, Utah. Ph.D. dissertation. University of Colorado, Boulder.
- 35.SAS Institute, Inc. 1989. SAS/STAT users' guide, version 6, fourth edition, volume 2. Cary, N.C.
- 36.Scala, D. J., and L. J. Kerkoff. 2000. Horizontal heterogeneity of denitrifying bacterial communities in marine sediments by terminal restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 66:1980-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skogley, E. O. 1992. The universal bioavailability environment/soil test: UNIVEST. Commun. Soil Sci. Plant Anal. 23:2225-2246. [Google Scholar]
- 38.Skogley, E. O., and A. Dobermann. 1996. Synthetic ion-exchange resins: soil and environmental studies. J. Environ. Qual. 25:13-24. [Google Scholar]
- 39.Steppe, T. F., J. B. Olson, H. W. Paerl, R. W. Litaker, and J Belnap. 1996. Consortial N2 fixation: a strategy for meeting nitrogen requirements of marine and terrestrial cyanobacterial mats. FEMS Microb. Ecol. 21:149-156. [Google Scholar]
- 40.Svejcar, T. J. 1990. Root length, leaf area, and biomass of crested wheatgrass and cheatgrass seedlings. J. Range Manag. 43:446-448. [Google Scholar]
- 41.Ticknor, L. O., A.-B. Kolstø, K. K. Hill, P. Keim, M. T. Laker, M. Tonks, and P. J. Jackson. 2001. Fluorescent amplified fragment length polymorphism analysis of Norwegian Bacillus cereus and Bacillus thuringiensis soil isolates. Appl. Environ. Microbiol. 67:4863-4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.USDA Soil Conservation Service. 1991. Soil survey of Canyonlands area, Utah: parts of Grand and San Juan counties. USDA Soil Conservation Service, Government Printing Office, Washington, D.C.
- 43.West, N. E., and J. A. Young. 2000. Intermountain valleys and lower mountain slopes, p. 255-284. In M.G. Barbour and W. D. Billings (ed.), North American terrestrial vegetation, 2nd ed. Cambridge University Press, Cam-bridge, United Kingdom.
- 44.Wilson, K. H., R. B. Blitchington, and R. C. Green. 1990. Amplification of bacterial 16S ribosomal DNA with polymerase chain reaction. J. Clin. Microbiol. 28:1942-1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang, J. E., E. O. Skogley, S. J. Georgitis, B. E. Schaff, and A. H. Ferguson. 1991. Phytoavailability soil test: development and verification of theory. Soil Sci. Soc. Am. J. 55:1358-1365. [Google Scholar]