Abstract
In spite of the importance of many members of the genus Burkholderia in the soil microbial community, no direct method to assess the diversity of this genus has been developed so far. The aim of this work was the development of soil DNA-based PCR-denaturing gradient gel electrophoresis (DGGE), a powerful tool for studying the diversity of microbial communities, for detection and analysis of the Burkholderia diversity in soil samples. Primers specific for the genus Burkholderia were developed based on the 16S rRNA gene sequence and were evaluated in PCRs performed with genomic DNAs from Burkholderia and non-Burkholderia species as the templates. The primer system used exhibited good specificity and sensitivity for the majority of established species of the genus Burkholderia. DGGE analyses of the PCR products obtained showed that there were sufficient differences in migration behavior to distinguish the majority of the 14 Burkholderia species tested. Sequence analysis of amplicons generated with soil DNA exclusively revealed sequences affiliated with sequences of Burkholderia species, demonstrating that the PCR-DGGE method is suitable for studying the diversity of this genus in natural settings. A PCR-DGGE analysis of the Burkholderia communities in two grassland plots revealed differences in diversity mainly between bulk and rhizosphere soil samples; the communities in the latter samples produced more complex patterns.
The genus Burkholderia is an important component of the soil microbial community (18). For instance, Burkholderia cepacia was first described as the causative agent of onion soft rot (11), but several strains of this species are not phytopathogenic and play an important role in promoting plant health (5). Moreover, many species belonging to the genus Burkholderia have the ability to produce compounds with antimicrobial activity (13, 20, 28, 30) and thus can be used as biocontrol agents with activity against phytopathogens. In addition, other Burkholderia strains have been shown to be plant-growth-promoting rhizobacteria (42), and introduction of Burkholderia species in crops such as maize and sorghum has resulted in increases in both root and shoot dry weights (4, 14). The mechanisms involved in plant growth promotion may range from production of phytohormones to fixation of atmospheric nitrogen, as shown for Burkholderia vietnamiensis (42). Estrada-De Los Santos et al. (21) recently showed that nitrogen fixation is a common property in the genus Burkholderia, after they isolated new diazotrophic Burkholderia species which were phylogenetically unrelated to B. vietnamiensis from coffee and maize plants. Furthermore, nonculturable bacteria belonging to the genus Burkholderia have been found as endosymbionts of arbuscular mycorrhizal fungi (6), and genes involved in nitrogen fixation have been shown to be active at least during the germination of spores (33). The endosymbionts were detected mainly in members of the family Gigasporaceae and were present as homogeneous populations throughout the fungal life cycle (7). In addition to all these features, the great nutritional versatility of the genus Burkholderia, reflected in its ability to use a wide range of organic compounds as carbon sources (24), certainly contributes to its capacity to successfully compete for root exudates and thus to efficiently colonize habitats such as the plant root. This nutritional versatility has also led to the use of Burkholderia strains for biodegradation of environmental pollutants (22).
Concomitant with the use of members of the genus Burkholderia, there is increasing concern about the risk of using this group of bacteria in processes such as biological control and bioremediation (12) since some species are important pathogens in cystic fibrosis patients (25, 43).
The list of species belonging to the genus Burkholderia has changed several times since 1992, when Yabuuchi et al. (47) proposed that seven former Pseudomonas species belonging to so-called rRNA group II should be grouped in this new genus, based on the results of a polyphasic taxonomic study. Now, the genus Burkholderia comprises 21 species (1, 8, 43, 46, 48). Moreover, several strains previously identified as B. cepacia were grouped in the so-called B. cepacia complex, which comprises at least six genomic species or genomovars (16).
The microbial community in soil is inherently complex, and assessments performed with such a complex population do not always reveal its specific components. Moreover, cultivation-based methods are limited because they do not assess the nonculturable fraction of the soil microbiota (44). Hence, an analysis of distinct phylogenetic groups of bacteria on the basis of soil DNA is required, because such an analysis reduces the complexity and thereby facilitates assessment of the subgroups that contribute to the total diversity (35). This can be achieved by denaturing gradient gel electrophoresis (DGGE) of PCR-amplified 16S ribosomal DNA (rDNA) fragments, a technique that has been widely used to assess the diversity of various phylogenetic groups (34).
Burkholderia spp. have been identified by techniques such as DNA-DNA hybridization, sodium dodecyl sulfate-polyacrylamide gel electrophoresis, amplified fragment length polymorphism fingerprinting, and PCR performed with primers with different degrees of specificity (3, 17, 43). In addition, assessment of Burkholderia species in environmental samples has been based mainly on analyses of the B. cepacia complex in which restriction fragment length polymorphism analyses of the recA gene or 16S rDNA have been used (5, 18, 23). However, none of these methods, including the PCR-based approaches, can be used to directly evaluate the diversity of the genus Burkholderia in natural settings.
The main goal of this work was to develop a method, based on PCR-DGGE, that allows direct analysis of the diversity of Burkholderia species in environmental samples. To achieve this goal, primers specific for the genus Burkholderia were developed based on the 16S rRNA gene. The PCR system was first evaluated for specificity and sensitivity by using DNAs isolated from Burkholderia and non-Burkholderia species. After optimization of the method, the PCR-DGGE system specific for Burkholderia was used to assess the diversity of this genus in soil samples.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study and their growth characteristics are listed in Table 1. All species were stored at −80°C in 20% glycerol.
TABLE 1.
Strains used in this study
| Group | Taxon | Strain(s) | Growth conditionsa |
|---|---|---|---|
| β-Proteobacteria | Burkholderia andropogonis | ATCC 19311, LMG 6872, ATCC 2361 | TB-T, 27°C |
| Burkholderia caribensis | LMG18531, WD3 | TB-T, 27°C | |
| Burkholderia caryophylli | NCPPB353, ATCC 25418 | TB-T, 27°C | |
| Burkholderia cepacia | IPO1718, NCPPB945, NCPPB946, ATCC 25416, LMG16656, LMG18941, P2b | TB-T, 27°C | |
| Burkholderia gladioli | ATCC 3664 | TB-T, 27°C | |
| Burkholderia glathei | ATCC 29195, WD1 | TSB, 37°Cc | |
| Burkholderia glumae | NCPPB3708, ATCC 33617 | TB-T, 27°C | |
| Burkholderia graminis | WD2 | TB-T, 27°C | |
| Burkholderia multivorans | LMG13010 | TB-T, 27°C | |
| Burkholderia phenazinium | LMG2247 | TB-T, 27°C | |
| Burkholderia plantarii | NCPPB3590, ATCC 43733 | TB-T, 27°C | |
| Burkholderia pyrrocinia | ATCC 15958 | TSB, 37°C | |
| Burkholderia stabilis | LMG14294 | TB-T, 27°C | |
| Burkholderia vietnamiensis | LMG10929 | TB-T, 27°C | |
| Alcaligenes faecalis | A1501d | 10% TSB, 27°C | |
| Alcaligenes sp. | Isolated | 10% TSB, 27°C | |
| Delftia acidovorans | Q3-4-6-9d | 10% TSB, 27°C | |
| Ralstonia eutropha | 815d | LB, 27°C | |
| Ralstonia solanacearum | IPO1609d | LB, 27°C | |
| Variovorax paradoxus | Q2-5-27-9d | 10% TSB, 27°C | |
| α-Proteobacteria | Agrobacterium radiobacter | Isolated | LB, 27°C |
| Rizobium leguminosarum bv. trifolii | ANV794d | 10% TSB, 27°C | |
| Rizobium meliloti | L530d | LB, 27°C | |
| Sphingomonas chlorophenolica | ATCC 33790 | TSA, 27°Ce | |
| Xanthobacter autotrophicus | GJ 10d | NB, 27°Cf | |
| γ-Proteobacteria | Acinetobacter calcoaceticus | BD413jd | LB, 27°C |
| Enterobacter agglomerans | Isolated | LB, 27°C | |
| Enterobacter cloacae | BE1d | LB, 27°C | |
| Pseudomonas aeruginosa | PAO25d | LB, 27°C | |
| Pseudomonas chlororaphis | PC8d | LB, 27°C | |
| Pseudomonas cichorii | PC170d | LB, 27°C | |
| Pseudomonas corrugata | PD704d | LB, 27°C | |
| Pseudomonas fluorescens | R2fd | LB, 27°C | |
| Pseudomonas glycinea | Pg1d | LB, 27°C | |
| Pseudomonas putida | UWC1d | LB, 27°C | |
| Pseudomonas stutzeri | JM303d | LB, 27°C | |
| Pseudomonas syringae | Isolated | LB, 27°C | |
| Stenotrophomonas maltophilia | Pd1484d | 10% TSB, 27°C | |
| Actinomycetes | Mycobacterium chlorophenolicum | PCP-1d | DSM, 27°C |
| Streptomyces griseus | ISP5236d | TSBy, 27°Cg | |
| Gram-positive bacteria | Bacillus cereus | FoTc-30d | LB, 27°C |
| Bacillus subtilis | 168 TrpC2d | LB, 27°C | |
| Listeria innocua | ALM105d | LBg, 27°Ch | |
| Paenibacillus azotofixans | ATCC 35681 | TBN, 27°C |
Strain P2 was obtained from the culture collection of Cluster MIBU, Plant Research International, Wageningen, The Netherlands.
TSB, Trypticase soy broth containing (per liter) 17 g of pancreatic digested casein peptone, 3 g of papaic digested soybean meal, 5 g of NaCl, 2.5 g of K2HPO4, and 2.5 g of dextrose (pH 7.3).
Strain obtained from the culture collection of Cluster MIBU, Plant Research International, Wageningen, The Netherlands.
TSA, tryptone soya agar containing (per liter) 15 g of tryptone, 5 g of soya peptone, 5 g of NaCl, and 15 g of agar (pH 7.3).
NB, nutrient broth containing (per liter) 3 g of Bacto Beef Extract (Difco) and 5 g of Bacto Peptone (Difco) (pH 7.3).
TSBy, Trypticase soy broth supplemented with 5% yeast extract.
LBg, Luria-Bertani medium supplemented with 5 g of glucose per liter.
Soil samples.
Samples from grassland bulk and rhizosphere soil were collected in a field (Wildekamp) located in Wageningen, The Netherlands. This site has been under permanent grassland for approximately 50 years. The soil used is a loamy sand soil (3% clay, 10% silt, 87% sand) with about 2% organic matter and a pH of 4.2. Samples were taken with a soil core sampler (diameter, 3 cm) from the surface (depth, 0 to 10 cm) in two replicate plots (plots 47 and 31) in the same area. One composite sample was prepared for each plot by combining 100 such samples. Bulk soil was obtained from each composite grassland sample by removing loosely adhering soil from the plant material and mixing it thoroughly. Rhizosphere samples were prepared from the remaining root material with tightly adhering soil by removing the soil from the roots.
DNA extraction.
Genomic DNAs were extracted from all bacterial strains (Table 1) by the method described by Duineveld et al. (19). DNA was extracted from bulk and rhizosphere soil samples by using the MO BIO UltraClean soil DNA isolation kit (BIOzymTC, Landgraaf, The Netherlands) according to the protocol described by the supplier, except that the cells were disrupted by bead beating for 60 s with a cell homogenizer (Braun, Melsungen, Germany). The bead-beating step was included to ensure maximal cell lysis without severe shearing of the DNA (45).
Primer design.
16S rDNA sequences belonging to members of the genus Burkholderia were retrieved from GenBank (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/) and were aligned using Clustal_X (41). After alignment, the sequences were manually searched for homologous regions specific for this genus. The regions selected were analyzed further by BLAST (2) to search for homologous nucleotide sequences in the GenBank database. This procedure was repeated until the desired specificity for the genus Burkholderia was obtained. After the optimal sequences for the forward primer (Burk3; 5′CTGCGAAAGCCGGAT3′) and the reverse primer (BurkR; 5′TGCCATACTCTAGCYYGC3′) were determined, a GC clamp (5′CGCCCGGGGCGCGCCCCGGGCGGGGCGGGGGCACGGGGGG 3′) (27) was attached to the 5′ end of the forward primer so that it could be used in a DGGE system.
PCR.
Amplification of 16S rDNA from genomic DNA was performed in 50-μl reaction mixtures containing 1 μl of DNA (5 to 10 ng), 200 μmol of each deoxyribonucleoside triphosphate per liter, 400 μmol of each primer per liter, 1× TaqPlus Precision buffer (Stratagene, Leusden, The Netherlands), and 2 U of TaqPlus Precision polymerase mixture (Stratagene). Amplification from soil or rhizosphere DNA extracts using a direct PCR was performed in 50-μl reaction mixtures containing 5 to 10 ng of target DNA, 10 mmol of Tris-HCl (pH 8.3) per liter, 10 mmol of KCl per liter, 3.75 mmol of MgCl2 per liter, 200 μmol of each deoxyribonucleoside triphosphate per liter, 400 nmol of each primer (GC-clamped Burk3 and BurkR) per liter, 1% (vol/vol) formamide, 0.25 μg of T4 gene 32 protein (Boehringer, Mannheim, Germany), and 5 U of AmpliTaq DNA polymerase (Stoffel fragment; Perkin-Elmer, Nieuwerkerk, The Netherlands). Amplification was performed with a PTC-100 thermal cycler (MJ Research, Inc., Tilburg, The Netherlands). Before the start of the reaction, the temperature was maintained at 95°C for 4 min. To enhance the specificity of the reaction, a touchdown PCR was carried out as follows. The annealing temperature was initially 62°C, and it was decreased by 1°C every fifth cycle until it was 60°C, after which 25 additional cycles were carried out at 58°C. Denaturation was performed at 94°C for 1 min, primer annealing was performed at the temperatures described above for 90 s, and primer extension occurred at 72°C for 2 min. After the thermal cycle, there was a final extension step consisting of 72°C for 10 min, followed by cooling to 10°C. The nested PCR procedure consisted of performing a first PCR with primer Burk3 in combination with universal eubacterial primer R1378 (27), using the PCR conditions described by Rosado et al. (36). The products from the first PCR were diluted 1:1,000 and used as the template in the second PCR, which was performed with primers Burk3 (GC clamped) and BurkR, as described above for genomic DNA. The PCR products, expected to be approximately 500 bp long, were analyzed by electrophoresis in a 1.5% (wt/vol) agarose gel in 0.5× TBE buffer (38). When necessary, products were stored at −20°C before they were used for DGGE analysis.
DGGE.
The DGGE analysis was performed by using the phorU2 system (Ingeny, Leiden, The Netherlands) and the method described by Rosado et al. (36), except that 50 to 60% denaturant gradients were used and the gels were electrophoresed at a constant voltage of 100 V for 15 h. After electrophoresis, the gels were stained with SYBR Gold I nucleic acid gel stain (Molecular Probes Europe, Leiden, The Netherlands) and with a silver staining kit (Bio-Rad, Veenendaal, The Netherlands). The Molecular Analyst software (version 1.61; Bio-Rad) was used to analyze the DGGE profiles. The tolerance with respect to band positions was set at 0.8%. Cluster analysis was done with the Molecular Analyst software, using the unweighted pair group method with mathematical averages. Correlations were calculated using the Dice coefficient of similarity (95% probability). A relatedness tree was produced with the algorithm of the Molecular Analyst software.
PCR-DGGE system sensitivity.
To evaluate the sensitivity of the PCR-DGGE system with soil DNA, a mixture of Burkholderia strains (Burkholderia andropogonis ATCC 19311, Burkholderia caribensis LMG18531, B. cepacia LMG18941, and Burkholderia multivorans LMG13010) was added to 50-g portions of nonsterile Wildekamp soil at three cell densities (5 × 103, 5 × 104, and 5 × 105 cells of each strain per g of soil). In control pots, sterile water was added to the soil. All treatments were done in duplicate. After overnight incubation at room temperature, soil DNA was extracted as described above. The sensitivity of the PCR-DGGE method was evaluated by using both direct and nested PCRs and was analyzed further by DGGE.
Soil clones and sequence analyses.
PCR products generated with DNA extracted from bulk soil and grass rhizosphere samples were purified with a High Pure PCR product purification kit (Boehringer). The products were then cloned into the pGEM-T easy vector by using Escherichia coli strain JM109 for transformation according to the procedure recommended by the manufacturer (Promega Benelux, Leiden, The Netherlands). Clones were randomly selected and grown, and after plasmid extraction with the Wizard Plus SV miniprep DNA purification system (Promega Benelux) they were used as templates in PCRs to produce products for controls in agarose gels. Soil clones producing PCR fragments of the appropriate size were then subjected to sequencing with an ABI Prism automatic sequencer (Greenomics, Plant Research International, Wageningen, The Netherlands). Sequence identities were determined by BLAST analyses (2).
Sequence alignment.
Sequences recovered from the GenBank/EMBL database or generated in this study were aligned by using Clustal_X (41). Phylogenetic trees were constructed by the neighbor-joining method (37) based on distance estimates calculated by the method of Jukes and Cantor (29). This analysis was performed with the TREECON program, version 1.3b (Yves van de Peer, Department of Biochemistry, University of Antwerp, Antwerp, Belgium).
Nucleotide sequence accession numbers.
The sequences generated in this study have been deposited in the GenBank database under accession numbers AF407341 to AF407358.
RESULTS AND DISCUSSION
Primer design.
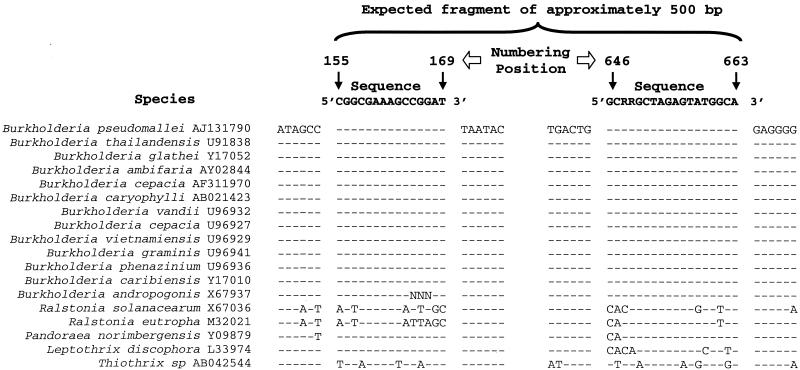
A comparison of 19 Burkholderia 16S rDNA sequences and 19 non-Burkholderia 16S rDNA sequences obtained from GenBank revealed one region that was potentially specific for all of the Burkholderia sequences analyzed (Fig. 1). A 15-mer forward primer was selected based on this region and was analyzed to determine its specificity for the genus Burkholderia by using all 16S rDNA sequences deposited in the GenBank database, estimated to represent more than 10,000 different sequences, and BLAST. The results showed that 51% of the 97 hits obtained belonged to members of the genus Burkholderia, 38% belonged to unculturable clones or as-yet-unidentified bacteria, and 11% belonged to members of other genera, such as Pandoraea (6%), Ralstonia (1%), Thiothrix (3%), and Lautropia (1%). Subsequently, all 16S rDNA sequences of strains classified as Burkholderia were recovered from the database, and 92% of those containing the primer region (178 sequences) showed complete homology with the primer sequence; the remainder differed by insertions or deletions at the 3′ end of the primer.
FIG. 1.
Alignment of 16S rDNA sequences from Burkholderia and non-Burkholderia species, corresponding to the region amplified by the Burkholderia-specific primers. The numbers correspond to E. coli 16S rDNA sequence numbers (10). Dashes indicate nucleotides that were identical to the nucleotides in the sequences at the top, which correspond to the 16S rDNA regions homologous to the Burkholderia-specific primers in the same DNA strand. R = A or G.
The 16S rDNA sequences of several of the non-Burkholderia species which produced hits in the BLAST assay were included in additional alignments to search for a region that could be used as a reverse primer. This analysis revealed a consensus region at positions 646 to 663 (E. coli numbering [10]) found only in members of the genus Burkholderia despite some variation in the third and fourth bases at the 3′ end (T-to-C conversions) (Fig. 1). A BLAST search was performed with this putative reverse primer sequence, including all C-T variations observed in Burkholderia spp. at the third and fourth nucleotides (CC, CT, and TT; positions 648 and 649). The BLAST report revealed that sequences containing the nucleotides C and T at positions 648 and 649 were widespread in Burkholderia species, occurring in 65 (45%) of the 145 Burkholderia 16S rDNA sequences which were available in the database and contained that region. Sequences with the nucleotide motifs CC and TT were less common, occurring in 43 (30%) and 23 (16%) Burkholderia 16S rDNA sequences, respectively. Only 9% of the remaining Burkholderia 16S rDNA sequences exhibited low levels of homology to the 3′ end of the reverse primer sequence. Moreover, the BLAST search also identified 28 sequences from nonculturable or unidentified bacteria which might represent Burkholderia sequences. A few sequences belonging to other genera were also detected. However, since those sequences were not detected in the BLAST output obtained with the forward primer, the specificity of the primer system was not affected.
In order to evaluate the positions of the Burkholderia species whose sequences displayed low levels of homology with one or both primers, a cluster analysis of complete 16S rDNA sequences from a range of Burkholderia species was performed (data not shown). The tree obtained showed that the low-homology sequences clustered in distinct groups, quite apart from the other Burkholderia species. One cluster comprised sequences from one recently described species, Burkholderia kururiensis (48), from two as-yet-undescribed closely affiliated nitrogen-fixing species, tentatively designated Burkholderia tropicalis and Burkholderia brasiliensis, and from three strains identified as Burkholderia sp. (accession numbers AF262932, AF074712, and AF074711). Another separate branch with low-homology Burkholderia sequences encompassed two putative Burkholderia spp. (X92188 and AJ011509) together with Pandoraea norimbergensis (Y09879), which has recently been removed from the genus Burkholderia (15). Finally, other low-homology sequences from strains identified as B. cepacia (AF244133) and Burkholderia sp. (AB011287, AY0055032, U76088, and AY0055039) were also separated from the main Burkholderia cluster.
Thus, the analysis of all sequences from the database reported to belong to the genus Burkholderia showed that only a minority of the sequences (15 of 145) exhibited low levels of homology with either of the primers developed in this study. However, phylogenetic analysis showed that six of these sequences might actually belong to members of genera other than Burkholderia.
Finally, the forward (Burk3) and reverse (BurkR) primers were checked for possible secondary structures that could prevent annealing of the primers to the target region during the PCR. Due to the formation of a strong hairpin structure in the forward primer, the second base at the 5′ end, a guanidine, was replaced by a thymidine (Fig. 1). Although this change reduced the identity of the forward primer sequence with the Burkholderia 16S rDNA sequences, it did not affect the specificity of the primers.
Sensitivity and specificity of the PCR-DGGE system for Burkholderia spp.
The specificity of the PCR-DGGE system was tested by using pure-culture DNAs from 14 Burkholderia species and 30 non-Burkholderia species as templates (Table 1). Products of the appropriate size (i.e., 500 bp) were detected with all strains of the Burkholderia species tested but not with any of the non-Burkholderia species. This indicated that the primer pair used exhibited 100% specificity for species of the genus Burkholderia.
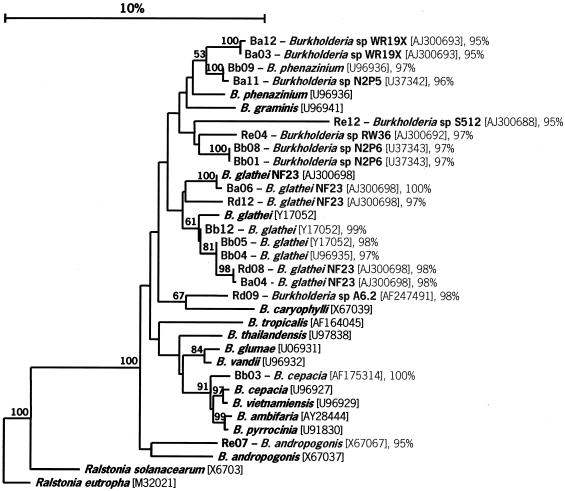
The specificity of the primers was also confirmed by performing sequence analyses of randomly chosen soil-derived clones (see Fig. 3). All 18 clones sequenced were identified as clones affiliated with Burkholderia species, and the levels of similarity were greater than 95% and often greater than 97% (it has been suggested that 97% similarity is a level that can be used to define species).
FIG. 3.
Phylogenetic tree showing the relationship between some Burkholderia species and soil-derived clones. The tree was constructed based on the fragment amplified by the Burkholderia-specific primers at positions 155 to 663 of the 16S rDNA (E. coli numbering [10]), using the neighbor-joining method (37). A bootstrap analysis was performed with 100 repetitions, and only values greater than 50 are shown. The GenBank accession number for each strain is enclosed in brackets. The bacterial sequence most closely related to each clone and the level of identity are shown after the clone designation. Soil clones Ba03, Ba12, Bb01, Bb04, and Bb05 also exhibited similarity to unculturable eubacterium WD2116 (accession number AJ292648) (96, 96, 98, 98, and 98% identity, respectively). Soil clone Bb08 exhibited 98% identity with unculturable eubacterium WD2120 (accession number AJ292661); soil clone Rd12 exhibited 99% identity with unculturable eubacterium WD263 (accession number AJ262641); and soil clone Re07 exhibited 96% similarity with unculturable eubacterium WD211 (accession number AJ292651). The letters in the clone designations indicate the following: B, bulk soil; R, rhizosphere soil; a and d, plot 47; b and e, plot 31.
The sensitivity of the PCR-DGGE method was evaluated with DNA extracted from a mixture of four Burkholderia species after incorporation of the cells into soil. The concentrations of the inoculated cells were 5 × 103, 5 × 104, and 5 × 105 cells g of soil−1. The detection limit of the direct PCR-DGGE system in soil was high (5 × 105 cells g of soil−1), and in order to increase the sensitivity, a nested PCR procedure, in which the clamped primer was used only in the second PCR, was performed. In this case, the detection limit was 5 × 103 cells per g of soil (data not shown). The nested PCR procedure increased the sensitivity of the method but did not interfere with the specificity, since the DGGE patterns obtained with the two methods (nested PCR and direct PCR) were equivalent (data not shown). The increase in sensitivity due to the use of a nested PCR procedure was expected, especially when the target organism was present in an environment containing compounds that might inhibit PCR, such as plant-derived compounds (32). Although the soil used in this study was not an organic soil, its organic matter content was high enough to affect the PCR when the direct approach was used. Due to the presence of potential inhibitors, the nested approach is more convenient for sensitive detection of Burkholderia communities in soil samples.
PCR-DGGE analyses.
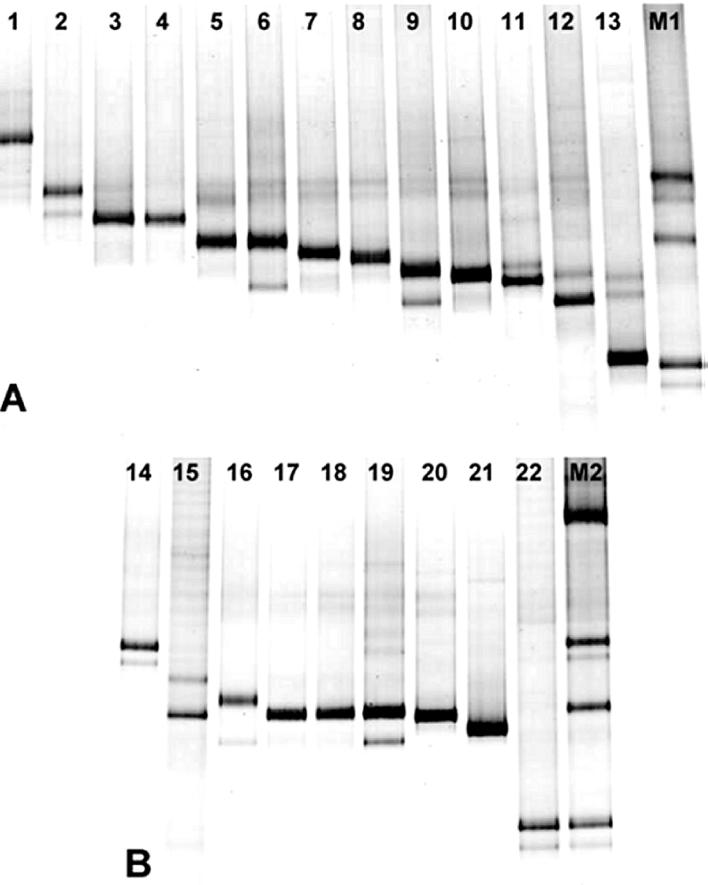
PCR-DGGE analyses of genomic DNAs of various Burkholderia strains showed that there were sufficient differences in the migration of the amplicons to discriminate between the majority of the Burkholderia strains listed in Table 1 (Fig. 2). Products obtained from different strains of the same Burkholderia species displayed the same electrophoretic mobility, except for two strains of B. caribensis (Fig. 2A) and several strains of B. cepacia (Fig. 2B). On the other hand, the region amplified by the specific primers failed to distinguish Burkholderia plantarii from Burkholderia gladioli, as well as B. cepacia genomovar I and B. vietnamiensis, due to their similar electrophoretic mobilities. Sequence alignments showed that the species that could not be differentiated by DGGE exhibited very high levels of similarity in the 16S rDNA region amplified by the primers (99.2 to 99.4%). However, sequence analysis of DGGE bands can be used to differentiate between species, and sequencing in combination with DGGE is now routinely used in several laboratories (34).
FIG. 2.
DGGE patterns of 16S rDNA fragments of Burkholderia species (A) and members of the B. cepacia complex (B) generated by PCR with Burkholderia-specific primers (positions 155 to 663 [E. coli numbering] [10]) in a 50 to 60% denaturing gradient. Lane 1, B. glathei WD1; lanes 2 and 14, B. multivorans LMG13010; lane 3, B. plantarii NCPPB3590; lane 4, B. gladioli ATCC 33664; lane 5, B. pyrrocinia ATCC 15958; lanes 6 and 16, B. stabilis LMG14294; lanes 7 and 18, B. vietnamiensis LMG10929; lane 8, B. phenazinium LMG2247; lane 9, B. caribensis WD3; lane 10, B. glumae NCPPB3708; lane 11, B. graminis WD2; lane 12, B. caribensis LMG18531; lane 13, B. caryophylli NCPPB353; lane 15, B. cepacia LMG16656 (genomovar III); lane 17, B. cepacia ATCC 25416 (genomovar I); lane 19, B. cepacia NCPPB945; lane 20, B. cepacia P2; lane 21, B. cepacia IPO1718; lane 22, B. cepacia LMG18941 (genomovar VI); lane M1, Burkholderia marker containing (from top to bottom) B. multivorans LMG13010, B. cepacia ATCC 25416, and B. cepacia LMG18941; lane M2, Burkholderia marker containing (from top to bottom) B. andropogonis LMG6872, B. multivorans LMG13010, B. cepacia ATCC 25416, and B. cepacia LMG18941.
To evaluate if the patterns obtained for each strain were reproducible in a complex community, DNAs of four strains (B. andropogonis LMG6872, B. multivorans LMG13010, and B. cepacia ATCC 25416 and LMG18941) were mixed in a 1:1:1:1 ratio, and the mixture was used as a template in a PCR. The DGGE profiles obtained were in line with the profiles obtained for each strain separately (data not shown). In addition, the intensities of the bands corresponding to each strain were similar, showing that there was no preferential amplification. Some Burkholderia species produced DGGE patterns comprising more than one band (Fig. 2), which could be explained either by the use of a degenerate reverse primer or by the presence of different 16S rDNA operons in one cell. To assess whether the use of a degenerate primer was the cause of the multiple bands, genomic DNAs from all Burkholderia species (Table 1) were used as the templates in PCRs performed with each of the three possible reverse primers separately. Each strain tested produced strong PCR products with only one of the three reverse primers. DGGE analyses of these PCR products revealed that the patterns obtained with nondegenerate primers (only one sequence) were similar to those obtained with the degenerate primers (mixture of three sequences) (data not shown). The similarity of the DGGE patterns obtained with the degenerate and nondegenerate primers suggested that the multiple bands could not be explained by the use of a combination of three sequences as a reverse primer. Another plausible explanation for the multiple bands is the fact that bacterial species have multiple rRNA genes, which might exhibit microheterogeneity. According to Klappenbach et al. (31), the number of rRNA operons per bacterial genome can vary from 1 to 15. This probably reflects ecological strategies of bacteria, such as the rate at which some bacteria respond to nutritional changes (upshift) in the environment. The B. cepacia genome was estimated to contain a maximum of six rRNA operons (rRNA Operon Copy Number Database [http://rrndb.cme.msu.edu/rrndb/servlet/controller]), but this estimate was based on a limited number of strains. Indeed, the multiple bands detected in some species with nondegenerate primers indicate that these species have multiple 16S rDNA operons with different sequences in the fragment amplified by PCR. Based on this hypothesis, the number of bands obtained by PCR-DGGE may well be higher than the number of actual species present in a Burkholderia community. The fact that an organism might be represented by more than one band and the fact that one band might correspond to more than one organism suggest that the number of bands in DGGE profiles does not provide an accurate estimate of richness. Therefore, diversity indices obtained by analysis of DGGE gels must be evaluated carefully. However, the DGGE profiles can certainly be used to detect shifts in the Burkholderia communities due to different environmental conditions and/or over time.
Analysis of soil bacterial populations.
Analysis of the sequences of 18 randomly picked clones obtained from grassland-derived DNA revealed that all of these sequences exhibited high levels of similarity to sequences typical of species of the genus Burkholderia (Fig. 3). These results confirmed that the primer set used is probably specific for the genus Burkholderia. The most abundant species to which similarity was found among the soil clones was Burkholderia glathei, which was detected as the closest hit for seven different clones. Similarity to Burkholderia phenazinium and similarity to B. andropogonis were also detected, albeit in only one clone each. Although the remaining soil clones could not be identified to the species level, their relationship to Burkholderia species could be confirmed by phylogenetic analyses. A phylogenetic tree based on the 16S rDNA region amplified by the primers showed that three clones were closely related to B. phenazinium, one clone was closely related to Burkholderia caryophylli, and one clone was closely related to species belonging to the B. cepacia complex (Fig. 3). Four soil clones formed a separate cluster closely related to the cluster formed by B. phenazinium and Burkholderia graminis. Two clones belonging to the latter cluster showed a high level of similarity to Burkholderia sp. isolate N2P6, a strain that was found to be closely related to Burkholderia fungorum and Burkholderia caledonica, two recently described species (15a). Interestingly, almost one-half of the clones were included in this branch of the phylogenetic tree, which contains organisms known for their ability to produce antimicrobial compounds, such as B. phenazinium, and for their ability to degrade xenobiotic compounds, such as Burkholderia sp. strain N2P6 (15a) (Fig. 3).
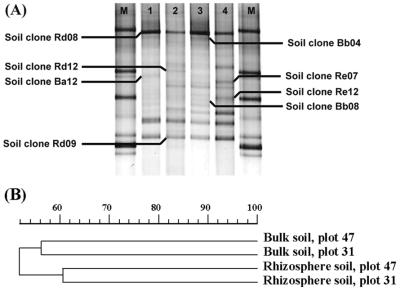
The DGGE profiles of the total Burkholderia populations in bulk and rhizosphere soil samples from the grassland were complex, comprising between 13 and 20 bands for each sample (Fig. 4A). The analysis of the DGGE profiles generated a dendrogram which showed clear grouping of the samples in two clusters, one composed of the two bulk soil samples and the other composed of the two rhizosphere soil samples (Fig. 4B). Therefore, this analysis demonstrated that grass roots had a clear influence on the structure of the Burkholderia populations. As some strong bands were detected in all samples, the main differences among samples were identified by analyzing the weaker bands. A comparison of the DGGE profiles obtained directly with soil DNA and the DGGE profiles obtained with the soil clones allowed presumptive identification of some of the bands. Thus, two strong bands present in all samples were identified as bands produced by organisms related to B. glathei (clones Rd08 and Bb04) and by organisms related to Burkholderia sp. strain A6.2 (clone Rd09), a strain closely related to B. caryophylli. In addition, several bands that were present in only one plot were also identified. Thus, two bands detected only in the rhizosphere soil of plot 31 were identified as bands produced by organisms related to B. andropogonis (clone Re07) and Burkholderia sp. strain S512 (clone Re12) (Fig. 4A).
FIG. 4.
Comparison of DGGE patterns for bulk and rhizosphere soil communities in a grassland field. Samples were taken in two different plots at the same location. (A) DGGE patterns for bulk soil from plot 47 (lane 1), rhizosphere soil from plot 47 (lane 2), bulk soil from plot 31 (lane 3), and rhizosphere soil from plot 31 (lane 4). Lane M contained Burkholderia markers (from top to bottom, B. andropogonis LMG6872, B. multivorans LMG13010, B. cepacia ATCC 25416, and B. cepacia LMG18941). (B) Clustering by the unweighted pair group method with mathematical averages, showing the levels of similarity of the microbial communities obtained by using the Burkholderia-specific PCR-DGGE system.
DGGE analyses of the PCR products obtained from both pure-culture and soil DNAs revealed that this technique was useful for evaluating the diversity of Burkholderia in soil samples. This is an advantage compared to the methods used previously, which relied on evaluation of specific groups within the genus Burkholderia, such as the B. cepacia group (5, 23). PCR-DGGE proved to be a powerful tool for detecting the dominant members of the Burkholderia community since it combined the sensitivity and specificity of the genus-specific PCR with direct screening of the dominant sequences, visualized on the basis of sequence divergence, via DGGE. Using this system, an effect of the grass rhizosphere on the selection of specific groups of Burkholderia species could be observed. Since this effect occurs because of the presence of compounds released by the roots, changes in the composition of these compounds are likely to induce changes in the rhizosphere populations. In fact, different crops can induce shifts in diversity by selecting different bacterial communities in their rhizospheres (40). Therefore, agricultural practices can induce changes in microbial diversity, and these changes presumably lead to changes in the ecological roles of Burkholderia spp. The PCR-DGGE system described here is now being used to study the effect of crop rotation on the diversity of Burkholderia populations, particularly measures that result in an increase in the presumably beneficial (plant-growth-promoting or antagonistic) Burkholderia species. Thus, PCR-DGGE targeting specific groups of microorganisms should be a useful monitoring tool for predicting the effects of agricultural practices on microbial communities in soil.
Acknowledgments
We thank P. Vandamme for providing Burkholderia strains ATCC 2361, LMG18531, ATCC 25418, ATCC 25416, LMG16656, LMG18941, ATCC 33664, ATCC 29195, ATCC 33617, LMG13010, LMG2247, ATCC 43733, ATCC 15958, LMG14294, and LMG10929 and E. Top for providing strains WD1, WD2, and WD3. We also thank J. A. van Veen for his comments on the manuscript and Rogier Doornbos for his assistance with the Molecular Analyst fingerprinting software (Bio-Rad).
This work was supported by CNPq/Brazil and by DWK, NL.
REFERENCES
- 1.Achouak, W., R. Christen, M. Barakat, M. H. Martel, and T. Heulin. 1999. Burkholderia caribensis sp. nov., an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique. Int. J. Syst. Bacteriol. 49:787-794. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. H. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bauernfeind, A., I. Schneider, R. Jungwirth, and C. Roller. 1999. Discrimination of Burkholderia multivorans and Burkholderia vietnamiensis from Burkholderia cepacia genomovars I, III, and IV by PCR. J. Clin. Microbiol. 37:1335-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bevivino, A., C. Dalmastri, S. Tabacchioni, and L. Chiarini. 2000. Efficacy of Burkholderia cepacia MCI 7 in disease suppression and growth promotion of maize. Biol. Fertil. Soils 31:225-231. [Google Scholar]
- 5.Bevivino, A., S. Sarrocco, C. Dalmastri, S. Tabacchioni, C. Cantale, and L. Chiarini. 1998. Characterization of a free-living maize-rhizosphere population of Burkholderia cepacia: effect of seed treatment on disease suppression and growth promotion of maize. FEMS Microbiol. Ecol. 27:225-237. [Google Scholar]
- 6.Bianciotto, V., C. Bandi, D. Minerdi, M. Sironi, H. V. Tichy, and P. Bonfante. 1996. An obligately endosymbiotic mycorrhizal fungus itself harbors obligately intracellular bacteria. Appl. Environ. Microbiol. 62:3005-3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bianciotto, V., E. Lumini, L. Lanfranco, D. Minerdi, P. Bonfante, and S. Perotto. 2000. Detection and identification of bacterial endosymbionts in arbuscular mycorrhizal fungi belonging to the family Gigasporaceae. Appl. Environ. Microbiol. 66:4503-4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brett, P. J., D. DeShazer, and D. E. Woods. 1998. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei-like species. Int. J. Syst. Bacteriol. 48:317-320. [DOI] [PubMed] [Google Scholar]
- 9.Briglia, M., R. I. Eggen, J. D. van Elsas, and W. M. De Vos. 1994. Phylogenetic evidence for transfer of pentachlorophenicol-mineralizing Rhodococcus chlorophenolicus PCP-I(T) to the genus Mycobacterium. Int. J. Syst. Bacteriol. 44:494-498. [DOI] [PubMed] [Google Scholar]
- 10.Brosius, J., T. J. Dull, D. D. Sleeter, and H. F. Noller. 1981. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J. Mol. Biol. 148:107-127. [DOI] [PubMed] [Google Scholar]
- 11.Burkholder, W. H. 1950. Sour skin, a bacterial rot of onion bulbs. Phytopathology 40:115-117. [Google Scholar]
- 12.Butler, S. 1995. Burkholderia cepacia and cystic fibrosis: do natural environments present a potential hazard. J. Clin. Microbiol. 33:1001-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cartwright, D. K., W. S. Chilton, and D. M. Benson. 1995. Pyrrolnitrin and phenazine production by Pseudomonas cepacia, strain 5.5B, a biocontrol agent of Rhizoctonia solani. Appl. Microbiol. Biotechnol. 43:211-216. [Google Scholar]
- 14.Chiarini, L., A. Bevivino, S. Tabacchioni, and C. Dalmastri. 1998. Inoculation of Burkholderia cepacia, Pseudomonas fluorescens and Enterobacter sp. on Sorghum bicolor: root colonization and plant growth promotion of dual strain inocula. Soil Biol. Biochem. 30:81-87. [Google Scholar]
- 15.Coenye, T., E. Falsen, B. Hoste, M. Ohlen, J. Goris, J. R. W. Govan, M. Gillis, and P. Vandamme. 2000. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int. J. Syst. Evol. Microbiol. 50:887-899. [DOI] [PubMed] [Google Scholar]
- 15a.Coenye, T., S. Laevens, A. Willems, M. Ohlen, W. Hannant, J. R. W. Govan, M. Gillis, E. Falsen, and P. Vandamme. 2001. Burkholderia fungorum sp. nov. and Burkholderia caledonica sp. nov., two new species isolated from the environment, animals and human clinical samples. Int. J. Syst. Evol. Microbiol. 51:1099-1107. [DOI] [PubMed] [Google Scholar]
- 16.Coenye, T., J. J. LiPuma, D. Henry, B. Hoste, K. Vandemeulebroecke, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia cepacia genomovar VI, a new member of the Burkholderia cepacia complex isolated from cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 51:271-279. [DOI] [PubMed] [Google Scholar]
- 17.Coenye, T., L. M. Schouls, J. R. W. Govan, K. Kersters, and P. Vandamme. 1999. Identification of Burkholderia species and genomovars from cystic fibrosis patients by AFLP fingerprinting. Int. J. Syst. Bacteriol. 49:1657-1666. [DOI] [PubMed] [Google Scholar]
- 18.Dalmastri, C., L. Chiarini, C. Cantale, A. Bevivino, and S. Tabacchioni. 1999. Soil type and maize cultivar affect the genetic diversity of maize root-associated Burkholderia cepacia populations. Microb. Ecol. 38:273-284. [DOI] [PubMed] [Google Scholar]
- 19.Duineveld, B. M., A. S. Rosado, J. D. van Elsas, and J. A. Van Veen. 1998. Analysis of the dynamics of bacterial communities in the rhizosphere of the chrysanthemum via denaturing gradient gel electrophoresis and substrate utilization patterns. Appl. Environ. Microbiol. 64:4950-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El Banna, N., and G. Winkelmann. 1998. Pyrrolnitrin from Burkholderia cepacia: antibiotic activity against fungi and novel activities against streptomycetes. J. Appl. Microbiol. 85:69-78. [DOI] [PubMed] [Google Scholar]
- 21.Estrada-De Los Santos, P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fain, M. G., and J. D. Haddock. 2001. Phenotypic and phylogenetic characterization of Burkholderia (Pseudomonas) sp. strain LB400. Curr. Microbiol. 42:269-275. [DOI] [PubMed] [Google Scholar]
- 23.Fiore, A., S. Laevens, A. Bevivino, C. Dalmastri, S. Tabacchioni, P. Vandamme, and L. Chiarini. 2001. Burkholderia cepacia complex: distribution of genomovars among isolates from the maize rhizosphere in Italy. Environ. Microbiol. 3:137-143. [DOI] [PubMed] [Google Scholar]
- 24.Gillis, M., V. Tran Van, R. Bardin, M. Goor, P. Hebbar, A. Willems, P. Segers, K. Kerstens, T. Heulin, and M. P. Fernadez. 1995. Polyphasic taxonomy in the genus Burkholderia leading to an amended description of the genus and proposition of Burkholderia vietnamiensis sp. nov. for N2-fixing isolates from rice in Vietnam. Int. J. Syst. Bacteriol. 45:274-289. [Google Scholar]
- 25.Govan, J.-R. W., J. E. Hughes, and P. Vandamme. 1996. Burkholderia cepacia: medical, taxonomic and ecological issues. J. Med. Microbiol. 45:395-407. [DOI] [PubMed] [Google Scholar]
- 26.Hagedorn, C., W. D. Gould, T. R. Bardinelli, and D. R. Gustavson. 1987. A selective medium for enumeration and recovery of Pseudomonas cepacia biotypes from soil. Appl. Environ. Microbiol. 53:2265-2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. M. H. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16 rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, F. P., and J. M. Young. 1998. Biocidal activity in plant pathogenic Acidovorax, Burkholderia, Herbaspirillum, Ralstonia and Xanthomonas spp. J. Appl. Microbiol. 84:263-271. [DOI] [PubMed] [Google Scholar]
- 29.Jukes, T. H., and C. R. Cantor. 1969. Evolution of protein molecules, p. 21-132. In H. N. Munro (ed.), Mammalian protein metabolism. Academic Press, New York, N.Y.
- 30.Kang, Y. W., R. Carlson, W. Tharpe, and M. A. Schell. 1998. Characterization of genes involved in biosynthesis of a novel antibiotic from Burkholderia cepacia BC11 and their role in biological control of Rhizoctonia solani. Appl. Environ. Microbiol. 64:3939-3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klappenbach, J. A., J. M. Dunbar, and T. M. Schmidt. 2000. rRNA operon copy number reflects ecological strategies of bacteria. Appl. Environ. Microbiol. 66:1328-1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, I. M., I. M. Bartoszyk, D. E. Gundersen, B. Mogen, and R. E. Davis. 1997. Nested PCR for ultrasensitive detection of the potato ring rot bacterium, Clavibacter michiganensis subsp. sepedonicus. Appl. Environ. Microbiol. 63:2625-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minerdi, D., R. Fani, R. Gallo, A. Boarino, and P. Bonfante. 2001. Nitrogen fixation genes in an endosymbiotic Burkholderia strain. Appl. Environ. Microbiol. 67:725-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 35.Øvreås, L., and V. Torsvik. 1998. Microbial diversity and community structure in two different agricultural soil communities. Microb. Ecol. 36:303-315. [DOI] [PubMed] [Google Scholar]
- 36.Rosado, A. S., G. F. Duarte, L. Seldin, and J. D. van Elsas. 1998. Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl. Environ. Microbiol. 64:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 39.Seldin, L., J. D. van Elsas, and E. G. C. Penido. 1984. Bacillus azotofixans sp. nov., a nitrogen-fixing species from Brazilian soils and grass roots. Int. J. Syst. Bacteriol. 34:451-456. [Google Scholar]
- 40.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tran Van, V., O. Berge, K. S. Ngo, J. Balandreau, and T. Heulin. 2000. Repeated beneficial effects of rice inoculation with a strain of Burkholderia vietnamiensis on early and late yield components in low fertility sulphate acid soils of Vietnam. Plant Soil 218:273-284. [Google Scholar]
- 43.Vandamme, P., B. Holmes, M. Vancanneyt, T. Coenye, B. Hoste, R. Coopman, H. Revets, S. Lauwers, M. Gillis, K. Kersters, and J.-R. W. Govan. 1997. Occurrence of multiple genomovars of Burkholderia cepacia in cystic fibrosis patients and proposal of Burkholderia multivorans sp. nov. Int. J. Syst. Bacteriol. 47:1188-1200. [DOI] [PubMed] [Google Scholar]
- 44.van Elsas, J. D., G. F. Duarte, A. S. Rosado, and K. Smalla. 1998. Microbiological and molecular biological methods for monitoring microbial inoculants and their effects in the soil environment. J. Microbiol. Methods 32:133-154. [Google Scholar]
- 45.van Elsas, J. D., V. Mantynen, and A. C. Wolters. 1997. Soil DNA extraction and assessment of the fate of Mycobacterium chlorophenolicum strain PCP-1 in different soils by 16S ribosomal RNA gene sequence based most-probable-number PCR and immunofluorescence. Biol. Fertil. Soils 24:188-195. [Google Scholar]
- 46.Viallard, V., I. Poirier, B. Cournoyer, J. Haurat, S. Wiebkin, K. K. Ophel, and J. Balandreau. 1998. Burkholderia graminis sp. nov., a rhizospheric Burkholderia species, and reassessment of [Pseudomonas] phenazinium, [Pseudomonas] pyrrocinia and [Pseudomonas] glathei as Burkholderia. Int. J. Syst. Bacteriol. 48:549-563. [DOI] [PubMed] [Google Scholar]
- 47.Yabuuchi, E., Y. Kosako, H. Oyaizu, I. Yano, H. Hotta, Y. Hashimoto, T. Ezaki, and M. Arakawa. 1992. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 36:1251-1275. [DOI] [PubMed] [Google Scholar]
- 48.Zhang, H., S. Hanada, T. Shigematsu, K. Shibuya, Y. Kamagata, T. Kanagawa, and R. Kurane. 2000. Burkholderia kururiensis sp. nov., a trichloroethylene (TCE)-degrading bacterium isolated from an aquifer polluted with TCE. Int. J. Syst. Evol. Microbiol. 50:743-749. [DOI] [PubMed] [Google Scholar]