Abstract
The genetic heterogeneity of nitrite reductase gene (nirK and nirS) fragments from denitrifying prokaryotes in forested upland and marsh soil was investigated using molecular methods. nirK gene fragments could be amplified from both soils, whereas nirS gene fragments could be amplified only from the marsh soil. PCR products were cloned and screened by restriction fragment length polymorphism (RFLP), and representative fragments were sequenced. The diversity of nirK clones was lower than the diversity of nirS clones. Among the 54 distinct nirK RFLP patterns identified in the two soils, only one pattern was found in both soils and in each soil two dominant groups comprised >35% of all clones. No dominance and few redundant patterns were seen among the nirS clones. Phylogenetic analysis of deduced amino acids grouped the nirK sequences into five major clusters, with one cluster encompassing most marsh clones and all upland clones. Only a few of the nirK clone sequences branched with those of known denitrifying bacteria. The nirS clones formed two major clusters with several subclusters, but all nirS clones showed less than 80% identity to nirS sequences from known denitrifying bacteria. Overall, the data indicated that the denitrifying communities in the two soils have many members and that the soils have a high richness of different nir genes, especially of the nirS gene, most of which have not yet been found in cultivated denitrifiers.
Biological denitrification is ubiquitous in soils and involves a stepwise reduction of nitrogen oxides associated with electron transport phosphorylation and producing the gases nitric oxide (NO), nitrous oxide (N2O), and in most situations, primarily molecular nitrogen (N2). Denitrification is of major importance to the cycling of nitrogen in soils and causes the loss of nitrogen from agricultural soils (4). Furthermore, any nitrous oxide and nitric oxide released could contribute to global warming (19) and to the destruction of stratospheric ozone (12). The capacity for denitrification is found among a wide variety of taxonomic groups mainly within the bacteria. Nearly 130 species of bacteria and archaea belonging to more than 50 genera can denitrify (33). Denitrifiers are found in all major physiological groups except for the Enterobacteriaceae, obligate anaerobes, and endospore-forming gram-positive bacteria other than Bacillus spp. (28).
The reduction of nitrite to nitric oxide by nitrite reductase is the first step that distinguishes denitrifiers from nitrate-respiring bacteria, which do not reduce nitrite to gas. Two structurally different nitrite reductases are found among denitrifiers, although never in the same cell (34): one contains copper (Cu-Nir) encoded by the nirK gene and one contains heme c and heme d1 (cd1-Nir) encoded by the nirS gene. To our knowledge, no functionally significant differences between the two nitrite reductases have been reported.
Different denitrifiers (1, 28) and different denitrifying communities in soils (10, 18) show differences in, e.g., oxygen threshold, carbon requirement, and kinetic parameters, which may influence competition with aerobic heterotrophs and emission rates of nitrous oxide. Hence, knowledge of the underlying composition and diversity of denitrifier communities may help us better understand, and ultimately manage, nitrogen cycling in soils.
Classical cultivation-based techniques are insufficient for studying the diversity of naturally occurring prokaryotic communities since the majority of prokaryotes is believed to be unculturable by traditional techniques (3). Approaches based on 16S rDNA gene sequences avoid the limitations of culturability and have been used to detect and analyze a larger portion of the prokaryotic community in soil samples (14, 22, 25). But, the high phylogenetic diversity among denitrifiers excludes a 16S rDNA-based approach and instead suggests the use of genes involved in the denitrification process, e.g., nir genes (7, 8, 17) or the nosZ gene coding for nitrous oxide reductase (23, 24).
The primers specific for the nirK and nirS genes were developed based on sequences available from a limited number of species for both genes, and only sequences from proteobacteria were considered (7). It is not known to what extent the primers will amplify nir fragments from other denitrifying bacteria, e.g., gram-positive species. The nirK sequence of an archaean denitrifier has recently been sequenced, but the two nirK primers do not show any homology to this sequence. All known archaean denitrifiers are halophilic, so great care should be taken if amplifiying nirK gene fragments from environments with a high salinity.
In the present report, we used a PCR method with subsequent cloning and sequencing to characterize the heterogeneity of nirK and nirS gene fragments in soil samples from two forested sites with contrasting water regime.
MATERIALS AND METHODS
Soil sampling.
Soil samples were collected on four dates in September 1999 (and at dates in October to November 1999 and April to May 2000 for additional experimentation) from 0 to 5 cm below the litter layer from sites near Turkey Marsh at the W. K. Kellogg Biological Station, Hickory Corners, Mich. (42°24′N, 85°22′W). The average annual temperature (1988 to 1999) is 8.9°C, and annual precipitation (1988 to 1999) is 860 mm year−1 (http://lter.kbs.msu.edu/Weather/Data/LTER/AnnualPrecip.html). One site was a small (<5 ha) mixed deciduous marsh, and the other was a mixed deciduous forest ca. 80 m up slope from the marsh site. The marsh soil is a Sebewa soil (mesic Typic Argiaquoll; loamy at 0- to 30-cm soil depth) (H. P. Collins, G. P. Robertson, M. J. Rosek, S. H. Gage, and J. R. Crum, personal communication) which is poorly drained, and the water table is typically at or near the soil surface from September to March and the site is flooded from March to May (5). The upland soil is an Oshtemo soil (mesic Typic Hapludalf; sandy loam at 0- to 25-cm soil depth) (H. P. Collins, G. P. Robertson, M. J. Rosek, S. H. Gage, and J. R. Crum, personal communication), which is well drained. At 0 to 5 cm below the litter layer, the pH was 7.8 in the marsh soil, and in the upland soil, the pH was 5.5, while carbon content estimated from weight loss by ignition was 8.2% in the marsh soil and 1.6% in the upland soil. Soil samples were sieved (2 mm) before isolation of DNA, which was initiated within 4 h of soil sampling.
Extraction of DNA.
DNA was isolated from 0.25 g of soil sample consisting of several aggregates picked randomly from the batch of sieved soil. DNA was isolated using UltraClean Soil DNA Isolation Kit (Mo Bio Laboratories, Solana Beach, Calif.) as specified by the manufacturer. DNA from the four September sampling dates was pooled and purified using Wizard DNA Clean-Up System (Promega, Madison, Wis.).
PCR amplification of nir fragments.
Fragments of the nirK and nirS genes were amplified using primer pairs nirK1F-nirK5R for nirK and nirS1F-nirS6R for nirS (7). PCR was done in a model 9600 thermal cycler (Perkin-Elmer, Norwalk, Conn.) using Ready To Go PCR Beads (Amersham Pharmacia, Piscataway, N.J.) and 35 pmol of each primer for amplification of DNA isolated from soil samples. Amplification of DNA from clones or plasmids was performed in a total volume of 50 μl of PCR buffer (500 mM KCl, 25 mM MgCl2, 200 mM Tris-HCl [pH 8.4], 0.1% Triton X-100), 200 μM concentrations of each deoxyribonucleoside triphosphate, 1 U of Taq polymerase (Gibco BRL, Rockville, Md.), 20 μg of bovine serum albumin (Boehringer Mannheim), and 25 pmol of each primer. The following PCR conditions were used for amplification. A denaturation step of 6 min at 95°C was followed by a touchdown PCR of 10 cycles with denaturation at 95°C for 30 s, primer annealing at 56°C to 51.5°C for 40 s, and extension at 72°C for 30 s, and then by 25 cycles with annealing at 54°C. The amplification ended with 7 min at 72°C. Annealing temperatures during PCR amplification of clones or plasmids were 66 to 61.5°C during touchdown and 64°C during the remaining cycles (touchdown PCR was followed by 20 cycles).
Cloning of nirK and nirS PCR products from soil samples.
PCR products were purified by cutting out the band of expected size from a low-melt agarose gel. The PCR products were purified from the gel using the Wizard PCR Preps DNA Purification Systems (Promega). Eluted products were cloned using the Original TA Cloning Kit (Invitrogen, Carlsbad, Calif.), and 55 to 95 clones from each cloning experiment were randomly selected for further analysis.
RFLP screening of nirK and nirS clones.
A small amount of cell material from a clone picked up with a toothpick was resuspended in 35 μl of pre-prepared PCR mixture, and the inserts were amplified as described above. Aliquots were run on an agarose gel for verification of products of the expected size. Clones which showed no product or a product of unexpected size were not analyzed further. PCR products from clones were screened by restriction fragment length polymorphism (RFLP) after digestion by 1 U of restriction enzyme for 6 h at 37°C. The products from nirK clones were digested in two separate reactions with the restriction enzymes HaeIII (New England BioLabs, Beverly, Mass.) and MspI (Gibco BRL, Rockville, Md.), while nirS clones were digested with HhaI (New England BioLabs) and MspI. The digested products were separated by gel electrophoresis in 3.5% Metaphor agarose (FMC Bioproducts, Rockland, Maine) in 1× Tris-borate-EDTA buffer at 4°C for 3.5 h at 7 V cm−1. The RFLP patterns were compared by eye.
Calculation of diversity indices.
To evaluate richness and evenness, diversity statistics were calculated by using the RFLP data as representations of different phylotypes. We are aware that any given RFLP pattern may represent sequences from multiple phylogenetic groups and may therefore not represent a true phylotype in the traditional sense. We use the term phylotype to indicate groups for richness calculations. Phylotype richness (S) was calculated as the total number of distinct RFLP patterns in a soil. The Shannon-Weiver diversity index (20) was calculated as follows: H = −Σ(pi)(log2pi), where p is the proportion of a distinct RFLP pattern relative to the sum of all distinct patterns. Evenness (20) was calculated from the Shannon-Weiver diversity index: E = H/Hmax, where Hmax = log2(S).
Sequencing of nir products and phylogenetic analysis.
Clones representing each distinct RFLP pattern were selected for sequencing. In addition, nirK products from a selection of clones with identical RFLP patterns were sequenced. Clones were grown for 14 to 16 h, and nir-containing plasmids were isolated using Wizard Plus SV Minipreps DNA Purification System (Promega). DNA sequences were determined by direct sequencing with an ABI 377 DNA sequencer (Applied Biosystems, Foster City, Calif.) using dye primer sequencing with the primers M13 reverse or T7, which bind to sites next to the nir insert in the plasmids. The amplified nirS product consisted of ca. 890 bp, but only approximately 515 bases were sequenced. Before sequencing the nirS fragments, we identified the orientation of the nirS insert in the plasmid by subjecting each plasmid to two PCR amplifications with the plasmid-specific primer M13 reverse and either nirS1F or nirS6R. With knowledge on the orientation of the fragment, we sequenced ca. 515 bases of the fragment from the downstream end using either M13 reverse or T7 primer. Thus, the sequenced region corresponds to position 1139 to 1653 in the nirS gene of Pseudomonas stutzeri ZoBell (accession no. X56813).
Nucleotide sequences and deduced amino acid sequences were aligned with nir sequences from marine sediment samples (see reference 8; only marine clones that were sequenced in the same region of the nir gene as the soil clones were included in the analysis) and from the National Center for Biotechnology Information (NCBI) database using the CLUSTAL W program from the BioEdit program package (16). The putative nirK sequence from Nitrosomonas europaea was obtained from the preliminary whole-genome sequence (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html). Identity matrices were constructed by using PAUP* version 4.0 beta version (27), and dendrograms were constructed with a neighbor-joining algorithm using TREECON version 1.3b (31). In order to avoid problems during calculation of the dendrograms, a single sequence was chosen to represent all sequences in each cluster consisting of >98% identical sequences.
RESULTS
Amplification of nir gene fragments.
High-molecular-mass DNA (7 to 10 kb) was isolated and purified from soil samples from both study sites. nirK gene fragments were successfully amplified from both soils, and nirS gene fragments were successfully amplified from the marsh soil. Despite an extensive effort involving soil samples from nine different sampling dates, additional purification of isolated DNA, nested PCR, and changes in PCR conditions (like annealing temperature and template concentration), we were not able to amplify nirS gene fragments from the upland soil. In addition, we spiked DNA isolated and purified from the upland soil with pCR 2.1 plasmids (Invitrogen) containing nirS fragments from the marsh soil. Spiking DNA isolated from 0.1 g of upland soil with 200 copies of the plasmid produced a band of the expected size (890 bp) visible on an agarose gel, indicating that fewer than 2,000 copies of nirS genes were present in 1 g of upland soil.
RFLP analysis of clones.
A total of 143 nirK clones (50 from the upland soil and 93 from the marsh soil) and 64 nirS clones (from the marsh soil) were screened by RFLP. For nirK clones from the upland soil, the RFLP analysis showed two dominant groups of clones comprising 42% of the clones. In the marsh soil, two dominant nirK groups comprised 35% of all nirK clones. Among the 54 distinct nirK RFLP patterns identified in the two soils, only one pattern was found in both soils. In contrast to nirK clones, no dominant nirS group was found among the 45 distinct RFLP patterns from the marsh soil.
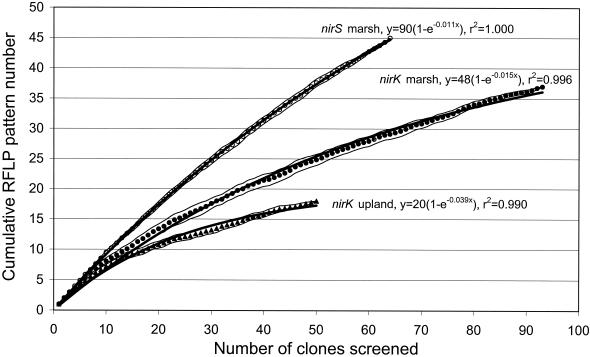
To determine how well the sampling captured the total diversity of RFLP patterns, the cumulative number of distinct RFLP patterns was plotted as a function of the number of clones screened (Fig. 1). This technique is analogous to generating a rarefaction curve to estimate species richness from species abundance data (30). We repeated the rarefaction analysis 26 times by assigning a random number to each clone (using the SLUMP function in Microsoft Excel 97) and sorting the clones according to the random numbers. Populations are assumed to be well-sampled if continuous effort does not produce new distinct RFLP patterns (that is, the cumulative RFLP pattern curve reaches an asymptotic value). To estimate the asymptotic value, we fitted the average data from the 26 replicate rarefaction curves (using SPSS Advanced Statistics 6.1; SPSS Inc., Chicago, Ill.) to a simple exponential model: y = a(1 − e−bx), where x = number of clones screened, y = cumulative number of RFLP patterns, and a and b are constants. a is equivalent to the total number of distinct nirK or nirS RFLP patterns in the two soils, while b is equivalent to a first-order constant. a was estimated to be 48 ± 0.71 (estimate ± one asymptotic standard error) for nirK marsh clones, 20 ± 0.41 for nirK upland clones, and 90 ± 0.60 for the nirS marsh clones. These values indicate that our 17 nirK upland clones and 37 nirK marsh clones with different RFLP patterns cover most of the nirK diversity in the two soils. It should be noted that the sequences from 9 to 11% of the distinct RFLP patterns did not indicate any relationship to known nir sequences, and these sequences may not have originated from denitrifying bacteria. Thus, the number of distinct nir RFLP patterns may be lower than indicated.
FIG. 1.
Rarefaction curves indicating diversity of denitrifying bacteria as revealed by RFLP analysis of cloned nitrite reductase gene fragments (nirK and nirS) from upland and marsh soil samples. nirK gene fragments were digested with the restriction enzymes MspI and HaeIII, and nirS gene fragments were digested with MspI and HhaI. Data points represent average values from 26 replicate rarefaction curves, and thin lines are 95% confidence limits of the average values. The thick lines represent fit to an exponential model: y = a × (1 − e−bx), where x = number of clones screened, y = cumulative number of RFLP patterns, and a and b are constants.
Sequence analysis of clones.
Partial nirK gene fragments (ca. 515 bases) from 21 upland soil clones and 41 marsh soil clones were sequenced. We sequenced both strands on 17 nirK clones and one strand in the remaining nirK clones. The only dissimilarities between complementary sequences were at bases recorded as ambiguous by the sequence analysis software. The sequenced fragments represented all distinct RFLP patterns except one and also included several clones with a similar RFLP pattern. Comparison with the NCBI database by using BLAST search revealed that 57 (92%) of the 62 sequences showed homology to known nirK sequences. Also, the deduced amino acid sequence within the amplified region was conserved in all sequenced nirK clones of the copper ligands His-145, Met-150, and His-306 (Achromobacter cycloclastes [accession no. Z48635] numbering) and His-255, which is not a copper ligand in the crystal form of the enzyme but is close to the active site of the enzyme (2), indicating that the sequences could produce functional enzymes. A matching sequence for the remaining five sequences could not be confirmed by BLAST search. Levels of nucleotide identity from pairwise comparisons of the confirmed nirK clones in the upland soil ranged from 74.0 to 99.7% and from 70.0 to 100% for the marsh soil. The average level of identity for all confirmed nirK sequences with different RFLP patterns was 90.0% ± 0.90% (average ± standard error; n = 1,522 pairwise comparisons), which was significantly lower (t test, P < 0.001) than the average level of identity of 97.1% ± 0.01% (n = 16) for sequences with pairwise identical RFLP patterns. Levels of identity of deduced amino acids ranged from 74.7 to 100% for the upland soil and from 61.1 to 100% for the marsh soil.
We sequenced 46 nirS gene fragments (ca. 515 bases) from clones representing all distinct RFLP patterns except one and included three clones with a similar RFLP pattern. Three clones were sequenced twice, while the remaining clones were sequenced once. Comparison with the NCBI database by using BLAST search revealed that 41 (89%) of the 46 sequences showed similarity to known nirS sequences. The deduced amino acid sequence within the sequenced region showed a conservation in all sequenced nirS clones of His-388 (Pseudomonas aeruginosa [accession no. X16425] numbering), which is believed to participate in the protonation of the nitrite oxygen atom (15). A matching sequence for the remaining five sequences could not be confirmed by BLAST search. Levels of nucleotide identity of the confirmed nirS clones ranged from 61.7% to 100%, with an average level of identity for the sequences with different RFLP patterns of 72.4% ± 0.3% (average ± standard error; n = 741), which was significantly lower (t test, P < 0.001) than the average level of identity of 100% ± 0.0% (n = 3) for sequences with identical RFLP patterns.
The aniA gene from Neisseria gonorrhoeae encodes a putative nitrite reductase with homologies to copper-containing nitrite reductase from denitrifying bacteria (21) and was used as an outgroup for phylogenetic distance analysis of the nirK sequences. Regions of insertion or deletion were omitted from the analysis because of uncertain alignment. The dendrogram of deduced amino acids showed five major clusters of nirK sequences (Fig. 2, I to V). No clone showed more than 90% identity to nirK sequences from known denitrifying bacteria, and the distant relatedness indicates that denitrifiers with novel nirK genes inhabit the upland soil.
FIG. 2.
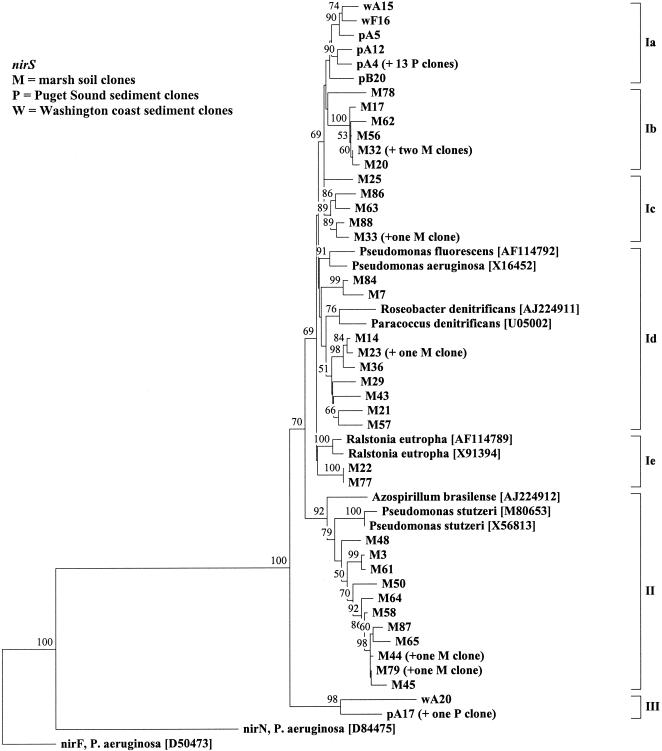
Neighbor-joining analysis of partial nirK gene products (171 amino acids) cloned from DNA isolated from upland (U) and marsh (M) soil samples or from marine sediment samples from the Washington coast (w) (see reference 8). The tree shows the topology of branching. Accession numbers in brackets. The sequence from Nitrosomonas europaea was obtained from the preliminary whole-genome sequence (http://www.jgi.doe.gov/tempweb/JGI_microbial/html/index.html). aniA from Neisseria gonorrhoeae (accession no. M97926) was used as outgroup. A single sequence was chosen to represent all sequences in clusters consisting of >98% identical sequences, and in each case, the origin and number of the other sequences in the cluster are indicated in parentheses following the sequence name. Values indicate the percentage of 100 replicate trees supporting the branching order; values below 50 are omitted.
nirF and nirN proteins have recognizable sequence relatedness to nirS (34), and nirF and nirS may have arisen from gene duplication (6). We used nirF (accession no. D50473) and nirN (accession no. D84475) sequences from P. aeruginosa as outgroups for phylogenetic distance analysis of the nirS sequences. Regions of insertion or deletion were omitted from the analysis because of uncertain alignment. The dendrogram of deduced amino acids showed three major clusters of nirS sequences (Fig. 3, I to III). No soil clones showed more than 80% identity to nirS sequences from any isolate, indicating that denitrifiers with novel nirS genes exist in the marsh soil.
FIG. 3.
Neighbor-joining analysis of partial nirS gene products (167 amino acids) cloned from DNA isolated from marsh soil samples (M followed by number) or from marine sediment samples (p and w followed by number; p from Puget Sound and w from Washington coast [8]). The tree shows the topology of branching. Accession numbers are in brackets. nirF (accession no. D50473) and nirN (accession no. D84475) from Pseudomonas aeruginosa were used as outgroups. A single sequence was chosen to represent all sequences in clusters consisting of >98% identical sequences and in each case the origin and number of the other sequences in the cluster are indicated in parentheses following the sequence name. Values indicate the percentage of 100 replicate trees supporting the branching order; values below 50 are omitted.
DISCUSSION
It was not possible to amplify a 890-bp nirS gene fragment from upland soil samples. In contrast, Braker et al. (8) amplified the nirS gene fragment from all five marine sediment samples explored and were able to amplify the nirK fragment only from the Pacific coast sediments off Washington state and not from Puget Sound, Wash. It is not possible to explain the nondetectable level of nirS in the upland soil in an ecological context, as virtually nothing is known about the environmental preferences of nirK- and nirS-containing denitrifiers. But, oxygen threshold, carbon requirement, and kinetic parameters vary between different denitrifiers (28), and between different denitrifying soil communities (10, 18), and the Cu-Nir and cd1-Nir enzymes may also have unknown differences related to, e.g., pH, kinetic parameters, or oxygen tolerance. The apparent predominance of either nirK or nirS in soils and sediments may reflect differences in denitrifier enzyme properties in soils where this process is strongly selected, or it may be due to traits of the organism, which are not connected to denitrification. The two soils in this study are contrasting with respect to several parameters, e.g., oxygen status, organic carbon content, and pH, which may have profound influence on the composition of the denitrifying communities. pH is a strong negative selector for denitrifiers and the low pH in the upland soil may contribute to the low nirK phylotype richness, the low nirK sequence diversity, and the very low nirS occurrence.
The RFLP analysis indicated a low diversity of nirK compared to nirS clones (Fig. 1 and Table 1), which was also indicated by the high average sequence identity of the nirK clones relative to the average sequence identity of known nirK denitrifying bacteria. The range of nucleotide identity among nirK clones from marine sediment samples was 78.6 to 100% (8) and was similar to the range found in the present study (Table 1).
TABLE 1.
Summary of data from RFLP and nucleotide analyses of nirK and nirS gene fragments from denitrifier communities in upland soil and marsh soil
| Gene | Source | RFLP analysis results
|
Nucleotide sequence analysis results
|
||||
|---|---|---|---|---|---|---|---|
| Sa | Estimated no. of distinct patternsc | Hd | H/Hmaxe | Ave sequence identity (%)f | Sequences with <90% identity to any published sequence (%) | ||
| nirK | Upland | 18 (50)b | 20 ± 0.41 | 4.40 | 0.91 | 93.5 ± 0.9 | 100 |
| Marsh | 37 (93) | 48 ± 0.71 | 3.55 | 0.88 | 88.2 ± 1.1 | 100 | |
| Isolatesg | 74.7 ± 0.7 | ||||||
| nirS | Marsh | 45 (64) | 90 ± 0.60 | 5.27 | 0.97 | 72.4 ± 0.3 | 100 |
| Isolatesg | 70.8 ± 1.0 | ||||||
Phylotype richness, S, was calculated as the total number of distinct RFLP patterns in a soil.
Numbers in parentheses are the total numbers of clones screened by RFLP.
Estimated from rarefaction curve; see text for details. Data are mean ± standard error.
Shannon-Weiver diversity index (20) was calculated as follows: H = −Σ(pi) (log2pi), where p is the proportion of a distinct RFLP pattern relative to the sum of all distinct patterns.
Evenness (20) was calculated from the Shannon-Weiver diversity index: E = H/Hmax where Hmax= log2(S).
Average sequence identity (±1 standard error of the mean) from pairwise comparisons of gene fragments.
Sequences from isolates (nirK from Nitrosomonas europaea was excluded from the analysis; see text) recorded in the NCBI database.
The heme cd1-containing Nir (nirS) occurred in a majority of numerically dominant isolates and aerobic soil enrichments (11). For nirK, it is not known if the gene has an inherently low diversity in natural habitats or if the restricted diversity of nirK sequences is due to parameters related to the studied soils (this study) and sediments (8). However, primer preference for more conserved nir genes and possible bias during PCR (26) may partly explain the low nirK diversity. Thus, the nirK1F and nirK5R primers used in this study and by Braker et al. (8) do not show any homology to the recently published nirK sequence from the archaean denitrifier Haloarcula marismortui (accession no. AJ278286). On the other hand, all known archaean denitrifiers are halophilic (33) and may not have any ecological significance in the two soils. Also, the nirK1F primer binds to a region of the nirK gene that has an insert of three bases in two sequences from known denitrifying bacteria, Bradyrhizobium japonicum (accession no. AJ002516) and Rhodobacter sphaeroides (accession no. U62291). The six other published sequences which encompass this region have no insert. The insert is located in the 5′ end of the primer and may not have a dramatic effect on the specificity of the primer, and we were able to PCR amplify the gene fragment from Rhodobacter sphaeroides using nirK1F under the PCR conditions used in this study.
The average pairwise sequence identity of nirK clones (88.2 and 93.5%; Table 1) was higher (t test, P < 0.001) compared to the identity of known nirK-containing bacteria (74.7%; Table 1) (in some cases the NCBI database had several nirK sequences from the same strain or from the same species with sequence identity of >99%; in those cases we avoided redundant sequences and included only one of the sequences in the analysis). Nitrosomonas europaea was excluded from this analysis as it remains unresolved whether the nirK sequence annotated in the Nitrosomomas europaea genome encodes the protein previously described as a nitrite reductase (9). It should be noted that some nirK sequences from known denitrifying bacteria show a comparatively high (>88%) sequence identity; this is true for four similar sequences from Achromobacter xylosoxidans (accession no. AF051831), Paracoccus denitrificans Pd1222 (accession no. AF114788), Alcaligenes sp. strain (accession no. AJ224903), and Alcaligenes xylosoxidans (accession no. AJ224904) and two similar sequences from Bradyrhizobium japonicum (accession no. AJ002516) and Blastobacter denitrificans (accession no. AJ224906).
In contrast to the nirK clones, the nirS clones showed an average pairwise sequence identity that was similar (t test, P > 0.05) to the average identity of known nirS-containing isolates (Table 1). In cases where the NCBI database had two or more nirS sequences from the same denitrifying strain or species with a sequence identity of >99%, only one sequence was included in the analysis. Six groups of nirS sequences from known denitrifying bacteria have >83% identity. One group consists of four similar sequences (Pseudomonas stutzeri [accession no. X56813], Pseudomonas stutzeri [accession no. M80653], Alcaligenes faecalis [accession no. AJ224913], and an unidentified strain [accession no. AJ224910]), another group consists of two sequences from Paracoccus denitrificans (accession no. U05002 and U75413), a third group consists of two sequences from Ralstonia eutropha (accession no. X91394 and AF114789), and another group consists of Achromobacter xylosoxidans and Alcaligenes xylosoxidans, which are synonymous (32); so, these four groups with a high level of nucleotide identity generally consisted of a single species. The exceptions are the Alcaligenes faecalis-in-Pseudomonas stutzeri group, a group with the very closely related Bradyrhizobium japonicum and Blastobacter denitrificans, and the Paracoccus denitrificans Pd1222-in-Achromobacter xylosoxidans group. The inclusion of Paracoccus denitrificans Pd1222 in the Achromobacter xylosoxidans group is surprising, as Paracoccus denitrificans PD1222 has been shown to have the nirS gene (13, 29). It should be noted that Hallin and Lindgren (17) found PCR products from Paracoccus denitrificans PD1222 when amplifying with a set of primers specific to nirK and not with a set of primers specific to nirS. Paracoccus denitrificans PD1222 is the first denitrifying strain with evidence of having both the nirK and the nirS gene, but the occurrence of nirK in Paracoccus denitrificans has yet to be confirmed by biochemical studies. However, at present, nirK or nirS has been sequenced from only a limited number of denitrifying prokaryotes, and it would be premature to use nirK or nirS sequences from clones to distinguish different species of denitrifiers.
The nirK tree revealed three important patterns (Fig. 2). First, the majority of clones did not branch with any known denitrifying bacteria, indicating that the two soils have unique denitrifiers not known among cultivated denitrifiers. Second, the five clones that branched with the known denitrifying bacteria were from the marsh soil. It is not known whether this reflects a more intense isolation effort in habitats similar to the marsh soil than in habitats similar to the upland soil or a predominance of easily isolated denitrifiers in the marsh soil compared to those of the upland soil. In contrast to the nirK clones, the majority of nirS clones were found in subclusters within the major clusters formed by the known denitrifying bacteria (Fig. 3). Third, the tree topology indicated that all the nirK sequences in the upland soil are closely related to sequences found in the marsh soil, suggesting that the majority of the diversity of nir genes in the upland soil is also found in the marsh soil. In contrast, the nirK clones from marine sediment samples formed a distinct cluster (Fig. 2, cluster III) and phylogenetic analysis of marine sediment samples grouped nirS clones into subclusters that corresponded to the habitats from which the clones were obtained. Also, nirS clones from sediment (Fig. 3, clusters Ia and III) and soil (Fig. 3, clusters Ib to Ie and II) formed distinct clusters, albeit at fairly low bootstrap values (Fig. 3 and reference 8).
The upland soil showed a less diverse denitrifying community than the marsh soil. This is supported by the rarefaction curves (Fig. 1) and the nirK tree (Fig. 2). Also, the upland soil had a low abundance of nirS genes. It may not be surprising that the marsh soil has the higher denitrifier diversity since this is a soil with a high organic matter content and fluctuating water level, which are likely to lead to fluctuating oxygen concentrations. This apparently good denitrifier habitat may allow denitrifiers to flourish and fill niches that are likely to be filled with aerobic bacteria in predominantly aerobic soils.
RFLP analysis proved to be a good, but not perfect, tool for screening cloned nir gene fragments. nir sequences with a similar RFLP pattern are likely to have DNA sequences with a very high level of identity. However, RFLP data from clones from complex prokaryotic communities should be analyzed with caution. The RFLP analysis indicated widely different nirK communities in the two soils, with only one RFLP pattern common to both soils. The RFLP analysis alone may have led to the conclusion that widely different denitrifying communities inhabit the two soils while the sequence analysis showed that the two communities were overlapping (Fig. 2).
Acknowledgments
We thank Stephen C. Nold for his generous help and Stephen K. Hamilton for his advice when selecting the study sites.
This work was supported by Danish Natural Science Research Council grant 9800420, NSF grant BIR 9120006, and the Biotechnological Investigations-Oceans Margin Program, U.S. Department of Energy grant DE-FG02-98ER62535.
REFERENCES
- 1.Abou-Seada, M. N. I., and J. C. G. Ottow. 1988. Einfluss chemischer Bodeneigenschaften auf Ausmass and Zusammensetzung der Denitrifikationsverluste drei verschiedener Bakterien. Z. Pflanzenernähr. Bodenk. 151:109-115. [Google Scholar]
- 2.Adman, E. T., J. W. Godden, and S. Turley. 1995. The structure of copper-nitrite reductase from Achromobacter cycloclastes at five pH values, with NO2−bound and with type II copper depleted. J. Biol. Chem. 270:27458-27474. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K.-H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aulakh, M. S., J. W. Doran, and A. R. Mosier. 1992. Soil denitrification—significance, measurement, and effects of management. Adv. Soil Sci. 18:1-57. [Google Scholar]
- 5.Austin, F. R., and G. R. Konwinski. 1979. Soil survey of Kalamazoo County, Michigan. USDA-Soil Conservation Service and Michigan Agricultural Experiment Station, East Lansing, Mich.
- 6.Baker, S. C., N. F. W. Saunders, A. C. Willis, S. J. Ferguson, J. Hajdu, and V. Fülöp. 1997. Cytochrome cd1 structure; unusual haem environments in a nitrite reductase and analysis of factors contributing to β-propeller folds. J. Mol. Biol. 269:440-455. [DOI] [PubMed] [Google Scholar]
- 7.Braker, G., A. Fesefeldt, and K.-P. Witzel. 1998. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 64:3769-3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braker, G., J. Zhou, L. Wu, A. H. Devol, and J. M. Tiedje. 2000. Nitrite reductase genes (nirK and nirS) as functional markers to investigate diversity of denitrifying bacteria in Pacific northwest marine sediment communities. Appl. Environ. Microbiol. 66:2096-2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casciotti, K. L., and B. B. Ward. 2001. Dissimilatory nitrite reductase genes from autotrophic ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 67:2213-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cavigelli, M., and G. P. Robertson. 2000. The functional significance of denitrifier community composition in a terrestrial ecosystem. Ecology 81:1402-1414. [Google Scholar]
- 11.Coyne, M. S., A. Arunakumari, A. Averill, and J. M. Tiedje. 1989. Immunological identification and distribution of dissimilatory heme cd1 and nonheme copper nitrite reductases in denitrifying bacteria. Appl. Environ. Microbiol. 55:2924-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crutzen, P. 1970. The influence of nitrogen oxides on the atmospheric ozone content. Q. J. R. Meteorol. Soc. 96:320-325. [Google Scholar]
- 13.de Boer, A. P., W. N. Reijnders, J. G. Kuenen, A. H. Stouthamer, and R. J. van Spanning. 1994. Isolation, sequencing and mutational analysis of a gene cluster involved in nitrite reduction in Paracoccus denitrificans. Antonie Leeuwenhoek 66:111-127. [DOI] [PubMed] [Google Scholar]
- 14.Felske, A., and A. D. L. Akkermans. 1998. Spatial homogeneity of abundant bacterial 16S rRNA molecules in grassland soils. Microb. Ecol. 36:31-36. [DOI] [PubMed] [Google Scholar]
- 15.Fülöp, V., J. W. B. Moir, S. J. Ferguson, and J. Hajdu. 1995. The anatomy of a bifunctional enzyme: structural basis for reduction of oxygen to water and synthesis of nitric oxide by cytochrome cd1. Cell 81:369-377. [DOI] [PubMed] [Google Scholar]
- 16.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 17.Hallin, S., and P.-E. Lindgren. 1999. PCR detection of genes encoding nitrite reductase in denitrifying bacteria. Appl. Environ. Microbiol. 65:1652-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holtan-Hartwig, L., P. Dörsch, and L. R. Bakken. 2000. Comparison of denitrifying communities in organic soils: kinetics of NO3− and N2O reduction. Soil Biol. Biochem. 32:833-843. [Google Scholar]
- 19.Houghton, J. T., B. A. Callander, and S. K. Varney. 1992. Climate change 1992: the supplementary report to the IPCC scientific assessment. Cambridge University Press, New York, N.Y.
- 20.Margalef, R. 1958. Information theory in ecology. Gen. Syst. 3:36-71. [Google Scholar]
- 21.Mellies, J., J. Jose, and T. F. Meyer. 1997. The Neisseria gonorrhoeae gene aniA encodes an inducible nitrite reductase. Mol. Gen. Genet. 256:525-532. [DOI] [PubMed] [Google Scholar]
- 22.Nüsslein, K., and J. M. Tiedje. 1998. Characterization of the dominant and rare members of a young Hawaiian soil bacterial community with small-subunit ribosomal DNA amplified from DNA fractionated on the basis of its guanine and cytosine composition. Appl. Environ. Microbiol. 64:1283-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scala, D. J., and L. J. Kerkhof. 1998. Nitrous oxide reductase (nosZ) gene-specific PCR primers for detection of denitrifiers and three nosZ genes from marine sediments. FEMS Microbiol. Lett. 162:61-68. [DOI] [PubMed] [Google Scholar]
- 24.Scala, D. J., and L. J. Kerkhof. 1999. Diversity of nitrous oxide reductase (nosZ) genes in continental shelf sediments. Appl. Environ. Microbiol. 65:1681-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:624-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 1998. PAUP*. Phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 28.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, New York, N.Y.
- 29.Timkovich, R., R. Dhesi, K. J. Martinkus, M. K. Robinson, and T. M. Rea. 1982. Isolation of Paracoccus denitrificans cytochrome cd1: comparative kinetics with other nitrite reductases. Arch. Biochim. Biophys. 215:47-58. [DOI] [PubMed] [Google Scholar]
- 30.Tipper, J. C. 1979. Rarefaction and rarefiction—the use and abuse of a method in paleoecology. Paleobiology 5:423-434. [Google Scholar]
- 31.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Appl. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 32.Yabuuchi, E., Y. Kawamura, Y. Kosako, and T. Ezaki. 1998. Emendation of genus Achromobacter and Achromobacter xylosoxidans (Yabuuchi and Yano) and proposal of Achromobacter ruhlandii (Packer and Vishniac) comb. nov., Achromobacter piechaudii (Kiredjian et al.) comb. nov., and Achromobacter xylosoxidans subsp. denitrificans (Ruger and Tan) comb. nov. Microbiol. Immunol. 42:429-438. [DOI] [PubMed] [Google Scholar]
- 33.Zumft, W. G. 1992. The denitrifying prokaryotes, p. 554-582. In A. Balows (ed.), The prokaryotes: a handbook of the biology of bacteria: ecophysiology, isolation, identification, applications. Springer-Verlag, New York, N.Y.
- 34.Zumft, W. G. 1997. Cell biology and molecular basis of denitrification. Microbiol. Mol. Biol. Rev. 61:533-616. [DOI] [PMC free article] [PubMed] [Google Scholar]