Abstract
A genetically engineered Escherichia coli cell expressing both organophosphorus hydrolase (OPH) and a cellulose-binding domain (CBD) on the cell surface was constructed, enabling the simultaneous hydrolysis of organophosphate nerve agents and immobilization via specific adsorption to cellulose. OPH was displayed on the cell surface by use of the truncated ice nucleation protein (INPNC) fusion system, while the CBD was surface anchored by the Lpp-OmpA fusion system. Production of both INPNC-OPH and Lpp-OmpA-CBD fusion proteins was verified by immunoblotting, and the surface localization of OPH and the CBD was confirmed by immunofluorescence microscopy. Whole-cell immobilization with the surface-anchored CBD was very specific, forming essentially a monolayer of cells on different supports, as shown by electron micrographs. Optimal levels of OPH activity and binding affinity to cellulose supports were achieved by investigating expression under different induction levels. Immobilized cells degraded paraoxon rapidly at an initial rate of 0.65 mM/min/g of cells (dry weight) and retained almost 100% efficiency over a period of 45 days. Owing to its superior degradation capacity and affinity to cellulose, this immobilized-cell system should be an attractive alternative for large-scale detoxification of organophosphate nerve agents.
The family of organophosphorus compounds is used extensively for agricultural and domestic pest control and as chemical warfare agents. These compounds function by inhibiting the acetylcholinesterase of many living organisms, including human beings (5). High-level exposure to organophosphates results in acetylcholine accumulation, which interferes with muscular responses and leads to serious damage of vital organs and eventually death. Due to the environmental concern associated with the accumulation of these compounds in food products and water supplies, there is a great need to develop safe, convenient, and economically feasible methods for their detoxification.
Organophosphorus hydrolase (OPH) isolated from soil microorganisms has been shown to detoxify organophosphates effectively (15, 20). However, practical applications of large-scale enzymatic degradation have always been limited by the cost and stability of OPH. As a cost-effective alternative, whole cells (either growing or nongrowing) instead of enzymes can be immobilized on the support (such as in an immobilized-cell bioreactor). However, the transport of pesticides across the cell membrane presents a major problem, since the outer membrane acts as a permeability barrier and prevents the pesticides from interacting with the OPH residing within the cells (8). This bottleneck, however, could be eliminated if OPHs were displayed on the cell surface.
Recently, active OPH has been successfully expressed on the cell surface of Escherichia coli by using either the Lpp-OmpA fusion system (18) or the truncated ice nucleation protein (INPNC) anchor (21). Cultures with surface-expressed OPH degraded parathion and paraoxon very effectively without the transport limitation observed in cells expressing OPH intracellularly. It has been demonstrated that immobilization of these live biocatalysts on solid supports by physical adsorption provides an attractive and economical means for pesticide detoxification (16). However, gradual cell detachment from the support reduced the effectiveness of the immobilized-cell system for long-term operation. A significant improvement, both in terms of economics and technology, could be achieved by improving the affinity of the OPH-expressing cells towards the support surface.
Endocellulases and exocellulases isolated from a variety of organisms consist of a cellulose-binding domain (CBD), which binds to cellulose and increases the rate of hydrolysis (3, 4, 17). The discrete CBD is usually separated from the catalytic domain by a Pro-Thr linker (3, 7, 24) and has been shown to retain binding function independent of the catalytic domain. Because of its high affinity towards cellulose, the CBD has been exploited as an affinity tag for the purification and immobilization of heterologous proteins (1, 2, 10, 11, 13, 19, 22). Specific adhesion of whole cells to cellulosic materials with high affinity has also been demonstrated by anchoring the CBD from Cellulomonas fimi exoglucocase (CBDcex) onto the surface of E. coli (6). Since recombinant cells with surface-expressed CBDs have been shown to bind tightly and rapidly to cellulose fibers, the CBD could be exploited to enable very strong attachment of the organophosphate-degrading cells to cellulose supports for long-term usage. However, the coexpression of two functional moieties on the cell surface of any bacterial cell has never been demonstrated. In this paper, we report for the first time the genetic coimmobilization of the CBD and OPH on the surface of E. coli by use of two different surface anchors. The resulting recombinant strains could be immobilized tightly on cellulose supports and could sustain long-term degradation of organophosphates.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
E. coli strain XL1-Blue {recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 lac (F′ proAB lacIqZΔM15 Tn10 [Tetr])} was used in this study. Plasmid pUCBD (23), a pUC18 derivative, was used for expression of Lpp-OmpA-CBD on the cell surface. Plasmid pPNCO33 was used to express ice nucleation protein (INP)-OPH on the cell surface. The ina-opd fragment was PCR amplified from pINCOP (21) and subcloned into EcoRI/HindIII-digested pVLT33 (14), a medium-copy-number vector, to give pPNCO33. Cells carrying pVLT33 and pK184 were used as the controls for the OPH activity measurement and cellulose binding assay, respectively. Plasmids pET38b and pJK33 were used for the intracellular expression of the CBD and OPH, respectively.
E. coli cells carrying plasmids were cultivated in buffered Luria broth medium (10 g of Difco tryptone per liter, 5 g of Difco yeast extract per liter, 10 g of NaCl per liter, 1 g of KH2PO4 per liter, 3 g of K2HPO4 per liter), pH 7.0, and supplemented with either ampicillin (100 μg/ml) or kanamycin (20 μg/ml). Except for cells bearing pUCBD alone, induction of Lpp-OmpA-CBD or INPNC-OPH was achieved by adding isopropyl-β-d-thiogalactopyranoside (IPTG) at an optical density at 600 nm (OD600) of 0.5. For cells expressing OPH, 1 mM CoCl2 was added 24 h after induction. Cells were harvested 48 h after induction.
Cell fractionation.
Cell fractionation experiments were conducted as follows. Cells were harvested by centrifugation, resuspended in 150 mM citrate-phosphate buffer (pH 8), and lysed by passage through a French press operated at 20,000 lb/in2. The lysates were centrifuged for 5 min at 1,500 × g to remove any unbroken cells and then ultracentrifuged for 45 min at 115,000 × g. The membrane pellets were resuspended in the same citrate-phosphate buffer.
Immunoblotting.
Two hundred microliters of cells (OD600 = 1) was combined with 40 μl of disruption buffer (62.5 mM Tris-HCl [pH 6.8], 10% glycerol, 2% [wt/vol] sodium dodecyl sulfate, 5% 2-mercaptoethanol, 0.05% [wt/vol] bromophenol blue) and boiled for 10 min. Samples were then electrophoresed through sodium dodecyl sulfate-12.5% (wt/vol) polyacrylamide gels prior to immunoblotting analysis. Immunoblotting was performed with an Immun-Blot GAR-AP kit (Bio-Rad, Hercules, Calif.). Antiserum against the CBDcex, OPH, or INPNC was used as the first antibody. Antiserum to the CBDcex was purchased from Novagen, and antiserum to OPH was prepared by Robert Sergeant Polyclonal Antibody Production Services (San Diego, Calif.). Antiserum against INPNC was a gift from J.-G. Pan (9). Prestained low-range protein markers (Bio-Rad) were utilized for the estimation of protein molecular weights.
Immunofluorescence microscopy.
Cells were harvested, washed and resuspended in phosphate-buffered saline (PBS) supplemented with 3% bovine serum albumin containing either rabbit anti-CBDcex antiserum (Novagen) at a dilution of 1:1,000 or rabbit anti-OPH antiserum at a dilution of 1:1,000 (PBS buffer), and incubated at 4°C for 4 h. After five washes with PBS buffer, the cell-antibody complex was incubated overnight at 4°C with goat anti-rabbit immunoglobulin G (IgG) conjugated with fluorescein isothiocyanate (FITC) at a dilution of 1:3,000. Prior to microscopy, cells were washed five times with PBS buffer, and then a photograph was taken with a fluorescence microscope (Nikon).
Whole-cell immobilization.
Cells were harvested and resuspended in 150 mM carbonate-bicarbonate buffer (30% [vol/vol] 150 mM sodium carbonate, 70% [vol/vol] 150 mM sodium bicarbonate), pH 9.5, to an OD600 of 2. For each immobilization experiment, a piece (16 by 5 cm) of nonwoven cellulose fiber (Bemliese #601; Asahi Chemical Industry Co., Ltd., Japan) (a gift from S.-I. Suye) was wrapped on a plastic frame (4.7 cm in diameter by 6 cm high) and immersed in 150 ml of cell suspension at an OD600 of 0.2 in a 250-ml glass beaker. The beaker was put on a stirring plate, and the cell suspension was mixed at 200 rpm for 24 h at 25°C. After removal of the support, the amount of cells in the suspension was determined before and after the incubation to determine the cell amount bound on the support. The fiber was then carefully removed and washed in 150 ml of buffer for 24 h, and the washing buffer was collected and concentrated to determine the amount of cells being washed out. As a comparison, cells were also immobilized on cellulose beads (Biomaterials Ltd., Fukui, Japan) under similar incubation conditions.
The protein content measurement method was coupled to cell dry weight measurement to determine the cell amounts in suspensions at very low cell densities. Cell suspension was collected and concentrated to a final volume of 50 μl. After the addition of 425 μl of 0.1 N NaOH, the sample was heated at 80°C for 10 min to break down the cells and release the proteins. Four hundred twenty-five microliters of 0.1 N HCl was added to neutralize the solution, and 100 μl of 0.5 M citric phosphate buffer (pH 8.0) was added to give a total volume of 1 ml. Eight hundred microliters of this solution was mixed with 200 μl of dye reagent concentrate (Bio-Rad), and after 5 min of incubation the OD595 was read and converted to protein content (in milligrams per milliliter) according to the Bradford method (3a) with bovine serum albumin as the standard.
For cell dry weight determination, cell suspensions were centrifuged at 16,000 × g in preweighed Eppendorf tubes and the buffer was decanted. The pellet was dried overnight to a constant weight at 105°C in a THELCO oven (model 130DM). The cell dry weight was obtained by subtracting the weight of the tube from the total weight of the tube and the dry pellet. The protein content of this cell pellet was determined by the protein content measurement method described above. A calibration for the conversion of cell dry weight to protein content was conducted for each cell line and binding experiment.
OPH assay.
Cells were centrifuged and resuspended in 150 mM carbonate-bicarbonate buffer, pH 9.5. Activity assays were conducted in 1.5-ml disposable methacrylate cuvettes (Fisher). For each assay, 900 μl of cells (OD600 = 0.5) was combined with 100 μl of 20 mM paraoxon (Sigma) dissolved in methanol. Cells with no OPH activity were used as the control. The change in absorbance (at 412 nm; E410 = 16,500 M−1 cm−1 for p-nitrophenol) was measured with a Beckman DU-60 spectrophotometer for 2 min at 37°C. Whole-cell activities were expressed as units (micromoles of paraoxon hydrolyzed per minute) per milligram of protein.
Scanning electron microscopy.
The cellulose material with immobilized E. coli cells was washed once with 150 mM carbonate-bicarbonate buffer (pH 9.5) and then soaked overnight in the same buffer containing 2% (wt/vol) glutaraldehyde to fix the cells. The material was washed five times with 150 mM carbonate-bicarbonate buffer (pH 9.5), followed by secondary fixation with 1% (wt/vol) osmium tetroxide for 1 h at 4°C and an additional washing with carbonate-bicarbonate buffer. The fixed sample was dehydrated by successive treatments in 50, 70, 80, 90, and 95% (vol/vol) ethanol solutions for 10 min and two treatments in 100% ethanol for 60 min to ensure complete dehydration of the sample. The ethanol was removed with a Balzers critical-point dryer (model CPD0202). The material was then cut and placed on a scanning electron microscope sample holder and coated with gold by using an Emscope sputter coater (model ES500). The gold-coated samples were viewed with a Philips electron microscope (model XL30-FEG).
Degradation of paraoxon by immobilized cells in repeated-batch operations.
The plastic frame with the immobilized cells was placed in a 250-ml beaker containing 150 ml of 150 mM carbonate-bicarbonate buffer (pH 9.5) supplemented with 0.05 mM CoCl2, 5% methanol, and 0.25 mM paraoxon. The reaction mixture was stirred at 200 rpm and kept at 25°C. The rate of hydrolysis was determined by measuring the formation of p-nitrophenol at OD412. After the degradation was completed, the support was removed and washed repeatedly in the same buffer until no yellow color was visible. The apparatus was kept at 25°C under constant mixing until the next experiment.
RESULTS AND DISCUSSION
Regulated coexpression of the CBD and OPH on the cell surface.
The initial goal of our study was to coexpress the CBD and OPH on the surface of E. coli. Since the CBD and OPH have been separately displayed on the cell surface with the Lpp-OmpA anchor system, it may seem logical to use the same anchor for the coexpression of the CBD and OPH on the surface. This initial strategy proved to be unsuccessful because the resulting cell lines coexpressing Lpp-OmpA-OPH and Lpp-OmpA-CBD were extremely unstable, with massive plasmid deletions and rearrangements. This is probably due to competition for the translocation pathway by the same Lpp-OpmA anchors. As a result, two different surface anchors were employed to target OPH and the CBD to the cell surface in order to minimize direct competition for the same translocation machinery. An IPTG-inducible, low-copy-number plasmid, pPNCO33, which has been shown to express INPNC-OPH on the surface of E. coli (21), was used for genetic immobilization of OPH. A pUC18-based compatible vector, pUCBD (23), was used to target Lpp-OmpA-CBD to the cell surface. Introduction of plasmids was carried out stepwise, and the resulting strain was named XL1-Blue/pUCBD/pPNCO33. Expression of both INPNC-OPH and Lpp-OmpA-CBD was induced by the addition of 1 mM IPTG.
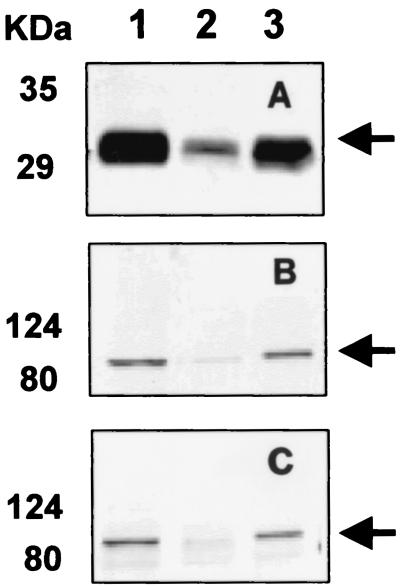
Production of the Lpp-OmpA-CBD fusion protein was verified by immunoblotting with CBDcex antiserum. A band corresponding to Lpp-OmpA-CBD at 30 kDa was detected (Fig. 1A). The localization of Lpp-OmpA-CBD in the membrane fraction was also demonstrated by immunoblotting (Fig. 1A, lane 3). Expression of the INPNC-OPH fusions in the membrane fraction was probed by immunoblotting, and the expected 82-kDa band was detected with both OPH (Fig. 1B, lane 3) and INPNC antisera (Fig. 1C, lane 3). Overnight cultures of XL1-Blue/pUCBD/pPNCO33 also exhibited significant OPH activity in the membrane fraction (0.227 U/mg of protein). These results suggest that both the CBD and OPH were efficiently synthesized and that the coexpression of the CBD did not inhibit the expression of functional OPH on the cell surface.
FIG. 1.
Western blot analysis of protein fractions from XL1-Blue/pUCBD/pPNCO33 with anti-CBD serum (A), anti-OPH serum (B), and anti-INPNC serum (C). Anti-CBD serum, anti-OPH serum, and anti-INP serum were used at dilutions of 1:1,000, 1:1,000, and 1:3,000, respectively. Lanes 1, 2, and, 3 indicate total protein, soluble fraction, and membrane fraction, respectively. The desired fusion proteins are marked with arrows.
Immunofluorescence microscopy.
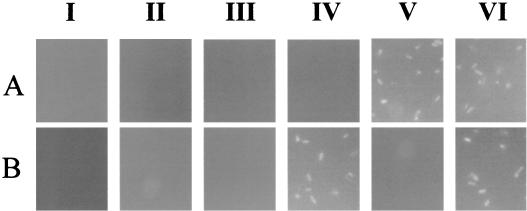
To confirm the presence of the CBD and OPH on the cell surface, immunofluorescence labeling of cells was performed by probing with either CBD or OPH antisera, followed by incubation with FITC-conjugated goat anti-rabbit IgG as the second antibody. The results are shown in Fig. 2. While XL1-Blue cells expressing either the CBD (pET-38b) or OPH (pJK33) intracellularly were not labeled at all with either antibody, XL1-Blue/pUCBD/pPNCO33 cells were brightly fluorescent labeled with both antisera. As a comparison, XL1-Blue/pUCBD cells were labeled only with CBD antisera and XL1-Blue/pPNCO33 cells were labeled only with OPH antisera, confirming that only XL1-Blue/pUCBD/pPNCO33 cells codisplayed the CBD and OPH on the cell surface.
FIG. 2.
Immunofluorescence micrographs of E. coli cells harboring different plasmids.E. coli XL1-Blue (I), XL1-Blue/pET38b (II), XL1-Blue/pJK33 (III), XL1-Blue/pUCBD (IV), XL1-Blue/pPNCO33 (V), and XL1-Blue/pUCBD/pPNCO33 (VI) were probed with either anti-OPH serum (A) or anti-CBDcex serum (B) and fluorescently stained with goat anti-rabbit IgG-FITC conjugate.
Whole-cell immobilization on cellulose supports.
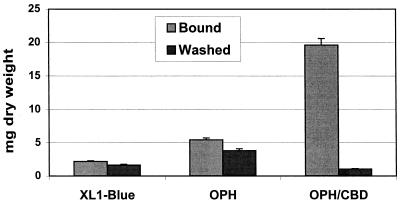
To confirm the functionality of the surface-exposed CBD, whole-cell immobilization on a nonwoven cellulosic fiber was investigated. Cell suspensions were incubated with a 16- by 5-cm cellulose fiber mat at room temperature for 24 h. After the support was carefully removed from the cell suspension, the amount of cells bound to the support was measured. Cells with both the CBD and OPH on the surface bound extensively to the support (Fig. 3). Significantly lower levels of binding were observed with the control XL1-Blue cells and cells with only OPH anchored on the surface. The stability of the immobilized cells was determined by subjecting the supports to overnight washing in buffer. While less than 10% of the immobilized XL1-Blue/pUCBD/pPNCO33 cells were removed after 24 h, over 80% of the bound cells were removed for the other cell lines with no CBDs on the surface (Fig. 3). This result confirms that surface-exposed CBDs endowed the XL1-Blue cells with the ability to bind tightly to cellulose supports.
FIG. 3.
Comparison of binding capacities of XL1-Blue cells to cellulose fiber. The amount of cells immobilized or washed out is expressed in milligrams of cells (dry weight) per support. XL1-Blue/pPNCO33 and XL1-Blue/pUCBD/pPNCO33 cells are designated OPH and OPH/CBD, respectively. E. coli strain XL1-Blue was used as a control, with no protein anchored on the cell surface. Data are mean values ± standard deviations from four experiments.
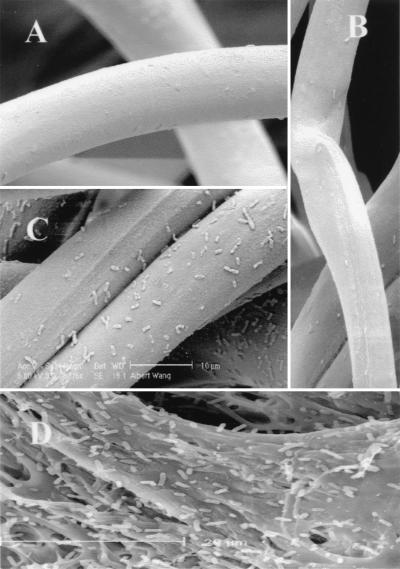
The washed supports were also observed with a scanning electron microscope. XL1-Blue cells with surface-exposed CBDs formed a monolayer on the supports (Fig. 4C), while cells without CBDs on the surface showed virtually no binding (Fig. 4A and B). In addition to the cellulose fibers, whole-cell immobilization was also observed with cellulose beads (Biomaterials Ltd.) with a significantly higher efficiency of immobilization (Fig. 4D). This difference may be attributed to the different modifications of the cellulose surfaces, which affect the surface sites available for immobilization.
FIG. 4.
Scanning electron micrographs of cellulose fiber showing immobilized XL1-Blue/pK184 (A), XL1-Blue/pPNCO33 (B), and XL1-Blue/pUCBD/pPNCO33 (C) cells after 24 h of washing. (D) Immobilization of XL1-Blue/pUCBD/pPNCO33 on cellulose beads.
Optimal display of the CBD and OPH on the cell surface.
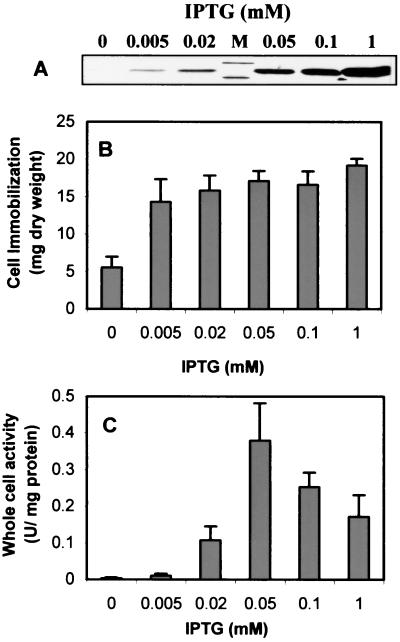
Since OPH and the CBD targeted toward the cell surface are competing for the same surface sites, it is essential to investigate the optimal level of the surface-exposed CBD for immobilization without sacrificing OPH activity in order to achieve both tight immobilization and high-level degradation of organophosphate pesticides. The levels of Lpp-OpmA-CBD and INPNC-OPH expressed on the surface were varied by adding different amounts of IPTG. As shown in Fig. 5A, expression of Lpp-OmpA-CBD was detected when IPTG induction was done at a concentration as low as 0.005 mM and significant cell immobilization was observed. Although the level of Lpp-OmpA-CBD increased dramatically with induction at higher IPTG concentrations, only a slight increase in cell immobilization was achieved (Fig. 5B). This result is in line with the earlier observation that even a very low level of surface-exposed CBD is sufficient to provide maximum cell immobilization efficiency (23). In comparison, OPH activity was virtually undetected at 0.005 mM IPTG but increased significantly with higher IPTG concentrations (Fig. 5C). Whole-cell activity reached a maximum (0.37 U/mg of protein) at an IPTG concentration of 0.05 mM, an activity level fivefold higher than that observed with cells expressing only OPH on the surface with the INPNC anchor (18). Further induction resulted in a gradual decline in the activity. These experiments suggest that induction with 0.05 mM IPTG provides the highest whole-cell OPH activity and a very high efficiency of cell immobilization. This condition was adopted for preparing immobilized cells for repeated-batch degradation experiments.
FIG. 5.
(A) Expression levels of Lpp-OmpA-CBD at increasing concentrations of IPTG. Total proteins were probed with anti-CBD serum as described in the legend to Fig. 1. (B) Binding capacities of XL1-Blue/pUCBD/pPNCO33 cells under different levels of induction. The amount of cells immobilized is expressed in milligrams of cells (dry weight) per support. (C) Whole-cell activities of XL1-Blue/pUCBD/pPNCO33 under different levels of induction. Data are mean values ± standard deviations from four experiments.
Detoxification of paraoxon by immobilized cells in repeated-batch operations.
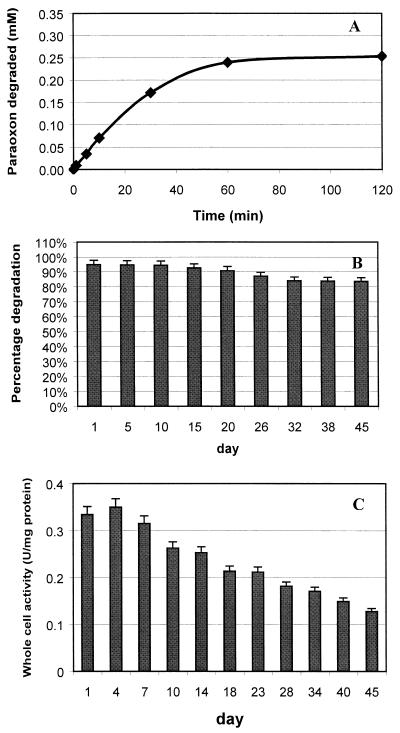
The ability of the immobilized XL1-Blue/pUCBD/pPNCO33 cells to rapidly degrade paraoxon was investigated. Cells were induced under optimal conditions, and immobilization was carried out as described above. Repeated-batch degradation experiments were performed by placing the immobilized support in a 250-ml beaker containing 150 ml of 150 mM carbonate-bicarbonate buffer (pH 9.5) supplemented with 0.05 mM CoCl2, 5% methanol, and 0.25 mM paraoxon. The reaction mixture was stirred at 200 rpm and kept at 25°C. The time course of paraoxon degradation on day 1 of the repeated-batch experiments is shown in Fig. 6A. Immobilized cells hydrolyzed paraoxon at an initial rate of 0.65 mM/min/g of cells (dry weight), and the added paraoxon was completely degraded within 70 min. This hydrolysis rate is fourfold higher than the rate of 0.16 mM/min/g of cells (dry weight) observed with the previously described immobilized-cell system involving physical adsorption onto a polypropylene fabric (16). This increase in specific activity is mainly attributed to the fivefold-higher specific OPH activity achieved with the double-surface expression system. The rate of degradation is also very favorable when compared to the rate of OPH immobilization into polyurethane foams. The rate of 7.0 μM/min for 0.24 g of cellulose fabric is comparable to the rate of 6.25 μM/min obtained with 0.22 g of polyurethane foam-entrapped OPH (12).
FIG. 6.
Degradation of paraoxon by immobilized XL1-Blue/pUCBD/pPNCO33 cells in repeated-batch operations. Degradation experiments were conducted at 25°C. (A) Degradation of 0.25 mM paraoxon by immobilized cells on day 1. (B) Percentage of paraoxon degraded after 1 h by immobilized cells. (C) Whole-cell activity for cell suspensions in phosphate-citrate buffer. Data are mean values ± standard deviations from four experiments.
Immobilization of cells also facilitated their recycle and reuse. Figure 6B depicts the recyclability and reusability of the immobilized cells with both the CBD and OPH on the cell surface. Although cells were no longer viable after 1 week, a decline in paraoxon hydrolysis efficiency of less than 10% was detected over a 45-day period. More importantly, no cell detachment was detected in the reaction or washing buffer, indicating that cell immobilization via the action of the surface-exposed CBD was very stable over this period. This is a significant improvement over immobilization by physical adsorption, where more than 10% of the immobilized cells were washed away within the first 5 days (16).
Immobilization also enhanced the stability of the biocatalysts; whole-cell activity of the resuspended cultures declined by 60% over the same 45-day period (Fig. 6C). This enhanced stability and the ease of repeated usage reinforce the need for immobilization. In this case, whole-cell biocatalysts with OPH and CBDs on the surface can be regarded as immobilized-enzyme particles and could be employed in a fashion similar to that of conventional immobilized OPH without tedious purification or loss of activity.
Conclusions.
We have demonstrated the construction of a genetically engineered E. coli strain that coexpresses the CBD and OPH on the cell surface. The end result is a cell line endowed with the abilities to rapidly degrade organophosphorus compounds and to bind tightly to cellulose materials. This is a superior system in terms of both biodegradation capacity and immobilization stability. The specific degradation rate for this CBD-based immobilized system is more than 10-fold higher than that of the previously described immobilized-cell system (16) based on physical adsorption onto a polypropylene fabric. More importantly, the CBD-based immobilization system is stable (no cell detachment) over a 45-day period, with a decline in the degradation rate of less than 10%. In comparison, the same decline in degradation rate was observed for immobilization based on physical adsorption because more than 10% of the immobilized cells were washed away within the first 5 days. Therefore, the CBD-based immobilization system is far superior in terms of the specific degradation rate and long-term stability.
More importantly, we have demonstrated for the first time the successful coexpression of two functional moieties on the surface of a bacterial cell, which enables it to carry out concomitantly two independent functions. This strategy should pave the way for the future use of engineered cells with multiple functional moieties displayed on the cell surface.
Acknowledgments
This work was supported by the UC Biotechnology Research and Education Program, the National Science Foundation (BES9731513), and the U.S. Environmental Protection Agency (R827227). A.W. was partially supported by a UCR Ph.D. dissertation grant.
We thank S.-I. Suye of Fukui University, Fukui, Japan, for providing the cellulose fiber and Stephen McDaniel in the UCR Central Facility for Advanced Microscopy and Microanalysis for technical support.
REFERENCES
- 1.Ahn, D. H., H. Kim, and M. Y. Pack. 1997. Immobilization of β-glucosidase using the cellulose-binding domain of Bacillus subtilis endo-β-1,4-glucanase. Biotechnol. Lett. 19:483-486. [Google Scholar]
- 2.Assouline, Z., H. Shen, D. G. Kilburn, and R. A. J. Warren. 1993. Production and properties of a factor X-cellulose-binding domain fusion protein. Protein Eng. 6:787-792. [DOI] [PubMed] [Google Scholar]
- 3.Black, G. W., J. E. Rixon, J. H. Clarke, G. P. Hazlewood, L. M. A. Ferreira, D. N. Bolam, and H. J. Gilbert. 1997. Cellulose binding domains and linker sequences potentiate the activity of hemicellulases against complex substrates. J. Biotechnol. 57:59-69. [DOI] [PubMed] [Google Scholar]
- 3a.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Carrard, G., A. Koivula, H. Söderlund, and P. Béguin. 2000. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. USA 97:10342-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Donarski, W. J., D. P. Dumas, D. P. Heitmeyer, V. E. Lewis, and F. M. Raushel. 1989. Structure-activity relationships in the hydrolysis of substrates by the phosphotriesterase from Pseudomonas diminuta. Biochemistry 28:4650-4655. [DOI] [PubMed] [Google Scholar]
- 6.Francisco, J. A., C. Stathopoulos, R. A. J. Warren, D. G. Kilburn, and G. Georgiou. 1993. Specific adhesion and hydrolysis of cellulose by intact Escherichia coli expressing surface anchored cellulase or cellulose binding domains. Bio/Technology 11:491-495. [DOI] [PubMed] [Google Scholar]
- 7.Greenwood, J. M., E. Ong, N. R. Gilkes, R. A. J. Warren, R. C. Miller, Jr., and D. G. Kilburn. 1992. Cellulose-binding domains: potential for purification of complex proteins. Protein Eng. 5:361-365. [DOI] [PubMed] [Google Scholar]
- 8.Hung, S.-C., and J. C. Liao. 1996. Effects of ultraviolet light irradiation in biotreatment of organophosphates. Appl. Biochem. Biotechnol. 56:37-47. [DOI] [PubMed] [Google Scholar]
- 9.Kim, E.-J., and S.-K. Yoo. 1999. Cell surface display of hepatitis B virus surface antigen by using Pseudomonas syringae ice nucleation protein. Lett. Appl. Microbiol. 29:292-297. [DOI] [PubMed] [Google Scholar]
- 10.Kyaw, C. M., R. C. Araújo, F. F. de Valência, M. S. S. Felipe, and S. Astolfi-Filho. 1994. Construction of a bifunctional protein with IgG- and cellulose-binding activities. Bioresour. Technol. 50:31-35. [Google Scholar]
- 11.Le, K. D., N. R. Bilkes, D. G. Kilburn, R. C. Miller, Jr., J. N. Saddler, and R. A. J. Warren. 1994. A streptavidin-cellulose-binding domain fusion protein that binds biotinylated proteins to cellulose. Enzyme Microb. Technol. 16:496-500. [DOI] [PubMed] [Google Scholar]
- 12.LeJeune, K. E., and A. J. Russell. 1996. Covalent binding of a nerve agent hydrolyzing enzyme within polyurethane foams. Biotechnol. Bioeng. 51:450-457. [DOI] [PubMed] [Google Scholar]
- 13.Linder, M., T. Nevanen, L. Söderholm, O. Bengs, and T. T. Teeri. 1998. Improved immobilization of fusion proteins via cellulose-binding domains. Biotechnol. Bioeng. 60:642-647. [DOI] [PubMed] [Google Scholar]
- 14.Lorenzo, V., L. Eltis, B. Kessler, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Pirp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 15.Mulbry, W. W., and J. S. Karns. 1989. Parathion hydrolase specified by the Flavobacterium opd gene: relationship between the gene and protein. J. Bacteriol. 171:6740-6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulchandani, A., I. Kaneva, and W. Chen. 1999. Detoxification of organophosphate nerve agents by immobilized Escherichia coli with surface-expressed organophosphorus hydrolase. Biotechnol. Bioeng. 63:216-223. [DOI] [PubMed] [Google Scholar]
- 17.Ong, E., N. R. Gilkes, R. A. J. Warren, R. C. Miller, and D. G. Kilburn. 1989. Enzyme immobilization using the cellulose-binding domain of a Cellulomonas fimi exoglucanase. Bio/Technology 7:604-607. [Google Scholar]
- 18.Richins, R. D., I. Kaneva, A. Mulchandani, and W. Chen. 1997. Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nat. Biotechnol. 15:984-987. [DOI] [PubMed] [Google Scholar]
- 19.Richins, R. D., A. Mulchandani, and W. Chen. 2000. Expression, immobilization, and enzymatic characterization of cellulose-binding domain-organophosphorus hydrolase fusion proteins. Biotechnol. Bioeng. 69:591-596. [DOI] [PubMed] [Google Scholar]
- 20.Serdar, C. M., and D. T. Gibson. 1985. Enzymatic hydrolysis of organophosphates: cloning and expression of a parathion hydrolase gene from Pseudomonas diminuta. Bio/Technology 3:567-571. [Google Scholar]
- 21.Shimazu, M., A. Mulchandani, and W. Chen. 2001. Cell surface display of organophosphorus hydrolase using ice nucleation protein. Biotechnol. Prog. 17:76-80. [DOI] [PubMed] [Google Scholar]
- 22.Shpigel, E., A. Goldlust, G. Efroni, A. Avraham, A. Eshel, M. Dekel, and O. Shoseyov. 1999. Immobilization of recombinant heparinase I fused to cellulose-binding domain. Biotechnol. Bioeng. 65:17-23. [DOI] [PubMed] [Google Scholar]
- 23.Wang, A. A., A. Mulchandani, and W. Chen. 2001. Whole-cell immobilization with cell surface-exposed cellulose-binding domain. Biotechnol. Prog. 17:407-411. [DOI] [PubMed] [Google Scholar]
- 24.Wierzba, A., U. Reichl, R. F. B. Turner, R. A. J. Warren, and D. G. Kilburn. 1995. Production and properties of a bifunctional fusion protein that mediates attachment of Vero cells to cellulosic matrices. Biotechnol. Bioeng. 47:147-154. [DOI] [PubMed] [Google Scholar]