Abstract
Bitterness is a flavor defect in Cheddar cheese that limits consumer acceptance, and specificity of the Lactococcus lactis extracellular proteinase (lactocepin) is widely believed to be a key factor in the development of bitter cheese. To better define the contribution of this enzyme to bitterness, we investigated peptide accumulation and bitterness in 50% reduced-fat Cheddar cheese manufactured with single isogenic strains of Lactococcus lactis as the only starter. Four isogens were developed for the study; one was lactocepin negative, and the others produced a lactocepin with group a, e, or h specificity. Analysis of cheese aqueous extracts by reversed-phase high-pressure liquid chromatography confirmed that accumulation of αS1-casein (f 1-23)-derived peptides f 1-9, f 1-13, f 1-16, and f 1-17 in cheese was directly influenced by lactocepin specificity. Trained sensory panelists demonstrated that Cheddar cheese made with isogenic starters that produced group a, e, or h lactocepin was significantly more bitter than cheese made with a proteinase-negative isogen and that propensity for bitterness was highest in cells that produced group h lactocepin. These results confirm the role of starter proteinase in bitterness and suggest that the propensity of some industrial strains for production of the bitter flavor defect in cheese could be altered by proteinase gene exchange or gene replacement.
Proteolysis and its secondary reactions play a major role in the maturation of Cheddar and many other bacterium-ripened cheese varieties (16). Proteolysis in Cheddar cheese is a complex process that involves endogenous milk enzymes, coagulant, and microbial proteinases and peptidases. Hydrolysis of intact casein (CN) is catalyzed almost exclusively by the added coagulant and endogenous milk enzymes, while proteinases and peptidases from Lactococcus lactis starter bacteria and adventitious (nonstarter) lactic acid bacteria are responsible for the production of water-soluble peptides and free amino acids (17). The contribution of individual enzymes in the cheese matrix to this process will also be influenced by specificity, relative activity, stability in the cheese matrix, and in the case of intracellular enzymes, access to appropriate substrates.
In many bacterium-ripened cheeses, the L. lactis cell envelope-associated proteinase (lactocepin, EC 3.4.21.96) is the most important microbial enzyme for the conversion of large-molecular-weight (water-insoluble) peptides produced by coagulant or plasmin into the small water-soluble peptides needed for flavor development (10, 17, 35). Lactocepin is a 180- to 190-kDa membrane-anchored enzyme that belongs to the subtilisin family of serine proteases. Although lactocepins exhibit more than 98% amino acid sequence identity, purified enzymes may be differentiated by their relative affinity and specificity for individual CNs (23). Genetic studies showed that most differences in lactocepin specificity could be traced to amino acid substitutions in the enzyme substrate-binding regions, and this property is now used as a classification system for lactocepin specificity (11, 23).
While substrate cleavage sites on αS1-, β-, and κ-CN have been identified for several purified lactocepins (23), the specificity of purified enzyme differs from the native (cell-bound) form (12) and may also be influenced by pH, salt content, and water activity of cheese (14, 15, 33, 34). Thus, even though lactocepin is widely believed to have an integral role in cheese proteolysis and flavor development (11, 35), the influence of lactocepin specificity on cheese quality remains unclear.
One aspect of the relationship between lactocepin specificity and cheese flavor that has attracted considerable research interest involves bitter flavor development. Bitterness is a serious quality problem in reduced- and full-fat Cheddar cheese (31, 42), and L. lactis starter bacteria play an important role in both the production and degradation of bitter peptides (27, 28). Bitterness develops when small to medium-sized hydrophobic peptides produced by the coagulant and some starter bacteria accumulate to levels that exceed desirable taste thresholds, whereas starter autolysis releases intracellular peptidases that can hydrolyze many of these peptides (27, 28). However, the degree of starter autolysis and the individual activity of peptidases varies widely among lactococci (23), and previous work has demonstrated that some lactocepin-derived peptides still accumulated in cheese made with a strongly autolytic starter (4). Thus, it is the hypothesis of our group that the most effective strategy to control bitterness in cheese is to develop a starter system that combines a low propensity for the production of bitter peptides with a high level of debittering peptidase activity. To test this hypothesis, however, we need to better understand the relationship between lactocepin specificity and bitterness. Although lactocepin specificity has been implicated in the production of bitter peptides (4, 25, 38), previous efforts to define this relationship have been hampered by strain-to-strain variability in the propensity for autolysis and intracellular peptidase activity. In an effort to overcome this limitation, we investigated peptide accumulation and bitter flavor development in 50% reduced-fat Cheddar cheeses manufactured with isogenic, single-strain L. lactis starters that lacked the major autolysin, AcmA (7), and which produced group a, e, or h or no lactocepin.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Lactococcus lactis strains and plasmids used in the study are listed in Table 1. Stock cultures were maintained at −80°C, and working cultures were prepared from frozen stocks by two transfers in M17 broth (37) at 30°C.
TABLE 1.
Bacteria and plasmids used in this studya
| Bacterium or plasmid | Relevant characteristics (phenotype) | Source or reference |
|---|---|---|
| L. lactis | ||
| LM0230 | Plasmid-cured derivative of L. lactis C2 | 9 |
| S1 | WT strain with group e cell envelope proteinase | 4, 11 |
| SK11 | WT strain with group a cell envelope proteinase | 39, 11 |
| S3 | WT strain with group h cell envelope proteinase | 4 |
| MBS3 | L. lactisLM0230 transformed with pMBS3 | This study |
| MG1363 acmAΔ1 | Plasmid-cured derivative of L. lactis 712 and isogenic host (AcmA− Opp+ Lac− LCP−) | 6 |
| PH | L. lactis MG1363 acmAΔ1 transformed with pPN-1 (AcmA− Opp+ Lac+ LCP−) | This study |
| PHa | L. lactis PH transformed with pNZ521 (AcmA− Opp+ Lac+ LCP+) | This study |
| PHe | L. lactis PH transformed with pGKV552 (AcmA− Opp+ Lac+ LCP+) | This study |
| PHh | L. lactis PH transformed with pMBS3 (AcmA− Opp+ Lac+ LCP+) | This study |
| Plasmids | ||
| pPN-1 | Lac+ LCP− plasmid isolated from L. lactis C2 | 8 |
| pNZ521 | L. lactis SK11 prtP/prtM genes cloned into pNZ122 (Cmr) | 40 |
| pGKV552 | L. lactis Wg2 prtP/prtM genes cloned into pGKV2 (Cmr Emr) | 22 |
| pMBS3 | L. lactis S3 prtP/prtM genes cloned into pGK12 (Cmr Emr) | This study |
Abbreviations: AcmA−, lacks major autolysin; LCP+, produces cell envelope proteinase; Lac+, able to ferment lactose; Opp+, has ATP-dependent oligopeptide transport system; Cmr, chloramphenicol resistance; Emr, erythromycin resistance; WT, wild type.
Cloning the L. lactis S3 group h lactocepin.
The L. lactis S3 prtP/prtM locus encoding a group h lactocepin and its maturation enzyme (4, 20, 40) was isolated by PCR. Oligonucleotide primers with SacI linkers (5′-CCGAGCTCAACGCAACGCA TGGACAGGC-3′ and 5′-CCGAGCTCATCAACTCTACTTGACGAAGAGCC-3′) were designed from conserved sequences that flank the prtM/prtP locus in L. lactis strains Wg2, SK11, and NCDO763 (19, 20, 39). Template DNA was isolated from L. lactis S3 as described previously (29), and a long-chain PCR was prepared as directed by the kit supplier (GeneAmp XL; PE Applied Biosystems, Foster City, Calif.). Amplification of the 7.5-kbp prtP/prtM region was performed in a Perkin-Elmer DNA Thermal Cycler, model 480, with a hot start at 94°C for 1 min followed by 16 cycles of 94°C for 30 s and 68°C for 10 min. This sequence was immediately followed by 12 cycles under similar conditions, except that 15 s per cycle was incrementally added to the 10-min elongation step at 68°C. The reaction was finished with a 10-min incubation at 72°C and then chilled to 4°C. An amplicon of the expected size was detected by horizontal gel electrophoresis in 1% agarose and collected with a Bio-Rad (Hercules, Calif.) Prep-a-Gene kit. Purified amplicon was digested with SacI (New England Biolabs, Beverly, Mass.), ligated into SacI-digested cloning vector pGK12 (21), and then transformed into L. lactis LM0230 (9). This and all other L. lactis transformations were performed by the electroporation procedure of Holo and Nes (18) using a Bio-Rad gene pulser (Bio-Rad Laboratories, Richmond, Calif.) set to the following parameters: 1.8-kV/cm field strength; 200-Ω resistance; 25-μF capacitance.
After electroporation, cells were incubated at 30°C for 48 h on M17-glucose agar that contained 0.5 M sucrose and 5 μg of chloramphenicol (CHL) (Sigma-Aldrich, St. Louis, Mo.) per ml. Chloramphenicol-resistant (Cmr) CFU were collected and tested for lactocepin production by overnight incubation at 30°C on citrated milk agar (5) that contained 0.5% glucose and 5 μg of CHL per ml. Plasmid DNA was isolated from lactocepin-positive (LCP+) isolates by the method of Anderson and McKay (2), and DNA sequence analysis of substrate-binding regions was performed as described previously (4). Finally, the specificity of cloned L. lactis S3 lactocepin (native cell-bound form) toward αS1-CN (f 1-23) in 25 mM Tris-NaH2PO4-Na acetate buffer (pH 5.2) with 4% NaCl was analyzed by reverse-phase high-pressure liquid chromatography (HPLC) as described previously (4). After these steps, a representative S3 prtP/prtM clone confirmed to have group h lactocepin substrate binding regions and specificity toward αS1-CN (f 1-23) at pH 5.2 in the presence of 4% NaCl (4) was selected and designated pMBS3 (Table 1).
Construction of L. lactis lactocepin isogens.
Cheese starter strains of L. lactis have a fast milk coagulation phenotype (FMC+) that requires lactose-fermenting ability, lactocepin, and an ATP-dependent oligopeptide transport system (43). L. lactis strain MG1363ΔacmA (6) has a chromosomally encoded oligopeptide transport system but cannot ferment lactose or produce lactocepin. To construct a series of isogenic strains that differed only in proteinase specificity, L. lactis MG1363 acmAΔ1 was first transformed with the naturally occurring L. lactis plasmid pPN-1, which encodes lactose utilization (8). Lactose-positive transformants were collected after 48 h of incubation at 30°C on lactose indicator agar (30) that contained 0.5 M sorbitol, and the presence of pPN-1 in cell lysates was confirmed by agarose gel electrophoresis. A representative isolate, designated L. lactis PH, was then separately transformed with plasmids pGKV552, pNZ521, and pMBS3 (Table 1). Transformants were selected by incubation on M17-lactose agar that contained 0.5 M sucrose and 5 μg of CHL per ml, and plasmid uptake was confirmed by agarose gel electrophoresis. The ability of individual clones to coagulate 11% reconstituted skim milk within 18 h at 21°C was confirmed, and then lactocepin specificity of representative isolates was determined by HPLC after incubation of whole cells with αS1-CN (f 1-23) in 25 mM Tris-NaH2PO4-Na acetate buffer (pH 5.2) with 4% NaCl as described previously (4).
Cheddar cheese manufacture and compositional analysis.
Vats of 50% reduced-fat Cheddar cheese were manufactured in duplicate at the University of Wisconsin-Madison from 250-kg lots of milk with 1.3% fat as described previously (4). L. lactis PH, PHa, PHe, and PHh were grown separately at 30°C for 12 to 14 h in skim milk that had been steamed for 45 min. Duplicate vats were inoculated with 2 to 3% (wt/wt) single-strain PHa, PHe, or PHh, or 3 to 4% strain PH to obtain a uniform rate of acid production in each cheese. Fat, moisture, salt content, and pH of cheese were determined on day 1 as described previously (41), and then the 9-kg cheese blocks were vacuum-packaged and stored at 7°C for ripening.
Cheese samples (approximately 200 g) were collected once per month, and 11-g portions were homogenized in 100 ml of 2% citrate for enumeration of starter and nonstarter CFU. Starter counts were collected by the pour plate method with Elliker's agar (Difco, Becton Dickinson, Sparks, Md.) that contained 5 μg of CHL per ml (except L. lactis PH, which was enumerated without antibiotic). Nonstarter lactobacilli were enumerated using Rogosa SL agar (Difco). Both types of agar plates were incubated anaerobically for 2 to 3 days at 30°C before colony enumeration.
Cheese proteolysis.
Production and accumulation of water-soluble peptides in experimental cheeses was monitored by reverse-phase HPLC in a Beckman gradient HPLC system equipped with a 125 dual pump, a 168 diode array detector, and a personal computer-based data system controller (Beckman System Gold, version 8.1). Sample preparation, HPLC columns, elutants, and separation parameters were as described previously (4). Peptide detection was performed at 214 nm, and peaks were identified by coelution with purified αS1-CN (f 1-9), αS1-CN (f 1-13), αS1-CN (f 1-14), αS1-CN (f 1-16), αS1-CN (f 1-17), and αS1-CN (f 1-23) peptide standards.
Peptide quantification in experimental cheeses.
Levels of β-CN (f 193-209) and αS1-CN (f 1-9) in experimental cheeses were quantified by HPLC after determination of the extraction efficiency for each peptide from a spiked model cheese. Cheese for the model system was manufactured with the lactocepin-negative (LCP−) isogen L. lactis PH as described above, and then 9-kg blocks were cooled for 3 to 4 days at 7°C, cut into 0.5-kg samples, vacuum packaged, and frozen at −80°C until needed. Purified β-CN (f 193-209) and αS1-CN (f 1-9) were synthesized on a Rainin Symphony (Protein Technologies, Inc., Woburn, Mass.) instrument and then collected by preparative HPLC and lyophilized. Synthetic peptides were dissolved at 10 to 50 mg per ml in sterile, double-deionized water and stored at −20°C. Model cheese was spiked by addition of 2 ml of a standard peptide solution (range, 25 to 200 mg per ml) and 40 μl of single-strength annatto (DSM Foods, Inc., Millville, Utah) to 50 g of grated L. lactis PH cheese in an 8- by 15-cm Teflon-coated pan. Control cheese was prepared with 2 ml of water without peptides. The mixtures were stirred well with a glass rod, and the pan was covered and heated at medium setting on a hot plate until the cheese had melted completely (approximately 10 min). The melted cheese blends were then stirred for about 2 min until the color (annatto) indicated that a homogeneous mass was formed (note: if the mix is stirred before cheese has melted completely, phase separation may occur). The fluid cheese was pressed to obtain a uniform layer in the pan, allowed to solidify at 4°C, vacuum packaged, and stored at −20°C.
Aqueous extracts for HPLC were prepared from 10-g samples of spiked cheese sample mixed with 90 ml of double-deionized water and homogenized in a Stomacher 400 (Seward Medical, Ltd., London, United Kingdom) for 6 min at 25°C. Approximately 15 ml of the homogenate was centrifuged at 4,400 × g for 30 min at 4°C, and then 3 ml of the aqueous fraction was collected and passed through a 0.2-μm low-protein-binding syringe-mounted filter (Gelman Sciences, Ann Arbor, Mich.). Next, 0.4 ml of 1 M NaCl was mixed with 1.6 ml of the filtered extract, and the salted solution was size fractioned through a 30,000-molecular-weight cutoff membrane filter (Amicon, Inc., Beverly, Mass.) by centrifugation at 4,900 × g for 1 h at 4°C. The filtrate was collected and stored at −20°C until needed. Standard solutions of purified synthetic peptides were prepared as described by Strickland et al. (36), and then reverse-phase HPLC was performed as described previously (4) on spiked cheese extracts and peptide standard solutions. The extraction efficiency of each peptide from cheese was calculated by dividing the peptide peak area from an HPLC chromatogram of spiked cheese by the area obtained from the same concentration of peptide in water. Extraction efficiency values for β-CN (f 193-209) and αS1-CN (f 1-9) represent the mean obtained from at least 3 independent experiments.
Assembly of trained bitter sensory panel.
Approximately 100 potential panelists were screened for the ability to taste bitterness using aqueous quinine sulfate (QS) solutions with duotrio testing (1). A labeled reference of distilled water was presented to each panelist followed by a series of paired samples. Within each pair, one sample contained distilled water and the other held increasingly concentrated levels of USP grade QS (Goldline Laboratories, Fort Lauderdale, Fla.). Panelists were asked to identify the sample in the pair that was different from the labeled reference water and to describe its taste. Individual taste thresholds were defined as the lowest concentration perceived with no mistakes in higher concentration pairs. Panelists able to taste ≤0.05 mg of QS per liter in water were then screened for their ability to detect and rate bitter flavor intensity in model cheese that had been spiked (as described for peptides) with 1 to 75 mg of QS per 400 g of cheese. Solutions of lactic acid were also included to identify panelists able to differentiate bitterness from sourness. Apple slices and mouth rinse water were used between cheese taste samples throughout the study. After approximately 10 training sessions, 14 panelists were chosen based on taste sensitivity and ability to differentiate samples.
Sensory evaluation.
Permission to perform sensory analysis on cheeses manufactured with isogenic L. lactis starter bacteria was obtained from the Utah State University Biosafety Committee and Institutional Review Board. After 2, 4, and 6 months of ripening, 3-g samples of experimental cheeses were coded and presented in a randomized order to trained panelists, who scored each sample for bitter flavor intensity (where 0 = not bitter, 1 = just perceivable bitterness, 3 = slightly bitter, 5 = moderately bitter, 7 = very bitter, 9 = extremely bitter). Statistical analysis of variance of sensory data was performed with SAS software (SAS Institute, Inc., Cary, N.C.).
RESULTS
Construction and characterization of lactococcal lactocepin isogens.
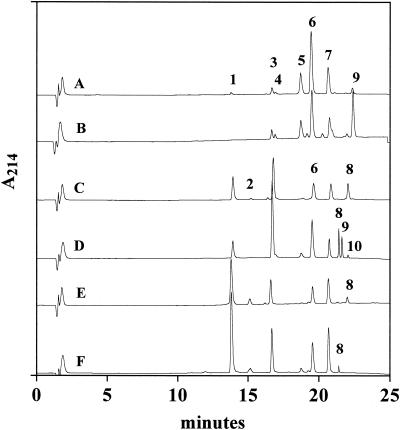
In L. lactis, a FMC+ phenotype requires the ability to ferment lactose, produce lactocepin, and take up oligopeptides (43). Because L. lactis strain MG1363ΔacmA cannot ferment lactose or produce lactocepin, the bacterium was first transformed with a naturally occurring L. lactis lactose plasmid to generate the LCP− host strain L. lactis PH. Independent transformation of L. lactis PH with pNZ521, pGKV552, and pMBS3 subsequently yielded FMC+ CFU whose representative isolates were designated PHa (group a lactocepin), PHe (group e lactocepin), and PHh (group h lactocepin), respectively (Table 1). As expected, incubations of whole cells with αS1-CN (f 1-23) at pH 5.2 in the presence of 4% NaCl showed that the lactocepin specificity of individual constructs matched the respective enzyme from wild-type cells and differed from each other. The group a lactocepin of L. lactis PHa or SK11, for example, had the strongest affinity for the Leu16-Asn17 and Asn17-Glu18 bonds of αS1-CN (f 1-23) but also produced αS1-CN (f 1-13) and low levels of αS1-CN (f 1-9) (Fig. 1A and B). The group e lactocepin of L. lactis PHe or S1 preferentially hydrolyzed αS1-CN (f 1-23) at the Gln13-Glu14 position but also formed αS1-CN (f 1-16) and αS1-CN (f 1-9) (Fig. 1C and D). Finally, incubations with L. lactis PHh or S3 showed that the group h lactocepin of these strains produced high levels of αS1-CN (f 1-9) and also cleaved αS1-CN (f 1-23) at the Leu16-Asn17 and Gln13-Glu14 positions (Fig. 1E and F).
FIG. 1.
Reversed-phase HPLC of the products of αS1-CN (f 1-23) after incubation with whole cells of L. lactis strains PHa (A), SK11 (B), PHe (C), S1 (D), PHh (E), or S3 (F). Incubations were performed in 25 mM Tris-NaH2PO4-Na acetate (pH 5.2) with 4% NaCl. Peptides identified in the chromatograms include peak 1, αS1-CN (f 1-9); peak 2, αS1-CN (f 7-13) and αS1-CN (f 1-6); peak 3, αS1-CN (f 1-13); peak 4, αS1-CN (f 1-14); peak 5, αS1-CN (f 1-17); peak 6, αS1-CN (f 10-16) and αS1-CN (f 1-16); peak 7, αS1-CN (f 17-23) and αS1-CN (f 18-23); peak 8, αS1-CN (f 14-23); peak 9, αS1-CN (f 1-23); and peak 10, αS1-CN (f 10-23). Chromatograms D and F employed a faster flow rate at the end of the run, so peptides in peaks 8, 9, and 10 eluted more rapidly. A214, absorbance at 214-nm wavelength.
Composition of cheeses made with lactocepin isogens.
Percentages of fat, moisture, and salt in moisture contents were very similar among reduced-fat Cheddar cheeses manufactured with L. lactis PH, PHe, PHa, or PHh single-strain starters (13.4 ± 1.0, 49.3 ± 1.0, and 2.9 ± 0.2, respectively). Cheese pH at 1 month was also very similar (pH 5.07 ± 0.09).
As is shown in Table 2, reduced-fat Cheddar cheese made with isogenic single-strain starters contained approximately 109 starter CFU per gram at 2 days, and the numbers of these bacteria declined by 1 to 3 orders of magnitude over 6 months of ripening. Cheeses made with L. lactis PHe or PHh showed the greatest decline in viable starter counts, while cheeses made with PHa or the LCP− isogen PH showed little change. Experimental cheeses also contained 102 to 104 nonstarter lactic acid bacteria per g at day 1, and levels of these bacteria exceeded 107 CFU per g by 4 months in all cheeses (Table 2).
TABLE 2.
Mean number of viable L. lactis starter and nonstarter lactic acid bacteria in experimental Cheddar cheeses during ripeninga
| Time point | No. of bacteria for starter isogen used in cheese manufactureb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| PH
|
PHa
|
PHe
|
PHh
|
|||||
| Starter | NSLAB | Starter | NSLAB | Starter | NSLAB | Starter | NSLAB | |
| d 1 | 8.4 × 108 | 1.8 × 102 | 5.8 × 108 | <10 | 9.9 × 108 | 2.4 × 101 | 3.4 × 109 | 1.1 × 104 |
| 2 mo | 1.4 × 108 | 6.2 × 107 | 2.6 × 108 | 6.7 × 105 | 6.9 × 107 | 5.4 × 106 | 1.3 × 108 | 3.7 × 105 |
| 4 mo | 1.1 × 108 | 4.7 × 108 | 4.3 × 107 | 2.2 × 107 | 1.7 × 106 | 1.0 × 107 | 5.5 × 107 | 3.5 × 107 |
| 6 mo | 7.0 × 107 | 6.1 × 107 | 1.7 × 107 | 1.5 × 107 | 6.6 × 104 | 2.2 × 107 | 5.1 × 106 | 9.1 × 107 |
NSLAB, nonstarter lactic acid bacteria; d, day.
Results are given in CFU/gram.
Effect of starter on peptide accumulation in cheese.
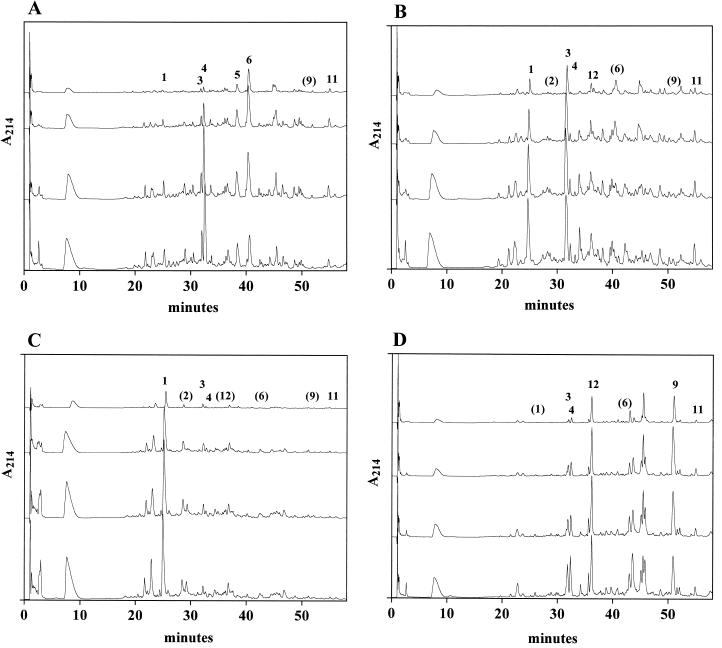
As shown in Fig. 2, aqueous extracts from cheese made with each L. lactis isogen had a characteristic HPLC profile after 2, 4, and 6 months of ripening. Coinjection studies with known standards showed that many of the most prominent peaks contained peptides that were derived from αS1-CN (f 1-23) by lactocepin (Fig. 1) and that the relative abundance of these peptides in cheese differed in a manner that was clearly influenced by starter lactocepin specificity (Table 3). A peak that comigrated with αS2-CN (f 1-21) was also detected in all cheeses, but concentrations were highest in Cheddar made with the LCP− isogen L. lactis PH (Fig. 2 and Table 3). This observation supports our previous suggestion that αS2-CN (f 1-21) is degraded by lactocepin in cheese (4).
FIG. 2.
Reversed-phase HPLC chromatograms of size-fractionated aqueous extracts of reduced-fat Cheddar cheese manufactured with L. lactis strain PHa (A), PHe (B), PHh (C), or PH (D) isogenic single-strain starters. Lines in each panel show, from the top, data collected after pressing (time 0), and after 2, 4, and 6 months of ripening at 7°C. Peptides identified in the chromatogram include peaks 1, αS1-CN (f 1-9); 2, αS1-CN (f 7-13) and αS1-CN (f 1-6); 3, αS1-CN (f 1-13); 4, αS1-CN (f 1-14); 5, αS1-CN (f 1-17); 6, αS1-CN (f 10-16) and αS1-CN (f 1-16); 7, αS1-CN (f 17-23) and αS1-CN (f 18-23); 8, αS1-CN (f 14-23); 9, αS1-CN (f 1-23); 10, αS1-CN (f 10-23); 11, β-CN (f 193-209); and 12, αS2-CN (f 1-21). Numbers in parenthesis indicate the deduced position of that peptide in the chromatogram. A214, absorbance at 214-nm wavelength.
TABLE 3.
Relative abundance of casein-derived peptides in size-fractionated aqueous extracts of reduced-fat Cheddar cheese made with isogenic L. lactis single-strain startersa
| Peptide | Results with strain:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
L. lactis PH
|
L. lactis PHa
|
L. lactis PHe
|
L. lactis PHh
|
|||||||||
| 2 mo | 4 mo | 6 mo | 2 mo | 4 mo | 6 mo | 2 mo | 4 mo | 6 mo | 2 mo | 4 mo | 6 mo | |
| αS1 f 1-9 | NDb | ND | ND | 1.9 | 2.2 | 2.6 | 5.9 | 7.4 | 7.3 | 15.0 | 14.8 | 14.7 |
| αS1 f 1-13 | 2.6 | 3.1 | 4.5 | 2.1 | 2.8 | 3.6 | 10.3 | 10.0 | 8.4 | 2.7 | 2.4 | 0.6 |
| αS1 f 1-14 | 3.0 | 3.5 | 5.5 | 4.6 | 8.3 | 10.6 | 1.5 | 2.0 | 2.2 | 1.4 | 1.0 | 0.3 |
| αS1 f 1-16 | ND | ND | ND | 14.1 | 7.8 | 5.2 | 3.7 | 2.2 | ND | 0.3 | ND | ND |
| αS1 f 1-17 | ND | ND | ND | 4.3 | 4.2 | 3.0 | 2.5 | 2.3 | 1.7 | ND | ND | ND |
| αS1 f 1-23 | 17.2 | 11.0 | 7.0 | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| β-CN f 193-209 | 1.8 | 1.3 | 1.2 | 1.9 | 1.5 | 1.0 | 1.8 | 1.9 | 0.9 | 0.4 | 0.2 | 0.4 |
| αS2 f 1-21 | 9.9 | 8.4 | 6.8 | 1.2 | 1.5 | 1.3 | 3.6 | 2.2 | 2.3 | 0.5 | ND | ND |
Numbers depict the percentage of the total peak area on respective HPLC chromatograms represented by the peak area for each peptide. The latter areas were obtained from the HPLC chromatogram of a representative cheese after 2, 4, and 6 months of ripening and divided by the total peak area for that chromatogram at the respective ripening time.
ND, not detected.
Because αS1-CN (f 1-9) and the chymosin-derived peptide β-CN (f 193-209) have been associated with bitterness in cheese (4, 28), experiments were performed to quantify levels of these peptides in experimental cheeses. To accomplish this, we first determined that the mean extraction of efficiency of αS1-CN (f 1-9) from spiked model cheese was 0.29 ± 0.02, while that of β-CN (f 193-209) was 0.51 ± 0.01. Extrapolation of HPLC peak areas with these data allowed us to estimate that levels of β-CN (f 193-209) in experimental cheeses ranged from 0.3 to 0.5 mg/g at press and increased slightly during aging (Table 4). Statistical analysis of variance (ANOVA) showed that time (P < 0.0001) but not starter choice (P > 0.5) significantly affected β-CN (f 193-209) concentrations in experimental cheeses. In contrast, starter choice (P < 0.009), time (P < 0.0001), and the starter × time interaction (P < 0.0001) each had a significant effect on the concentration of αS1-CN (f 1-9) in cheese. As shown in Table 4, levels of this lactocepin-derived peptide also increased over time, but the magnitude of change differed substantially in cheese made with different isogens.
TABLE 4.
Mean concentrations of αS1-CN (f 1-9) and β-CN (f 193-209) in reduced-fat Cheddar cheese made with single-strain isogenic L. lactis starter bacteriaa
| Starter strain | Concn (mg/g) of:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| αS1-CN (f 1-9)
|
β-CN (f 193-209)
|
|||||||
| 2 d | 2 mo | 4 mo | 6 mo | 2 d | 2 mo | 4 mo | 6 mo | |
| L. lactis PH | 0.1 ± 0.0 | 0.8 ± 0.1 | 1.6 ± 0.2 | 2.9 ± 0.2 | 0.3 ± 0.0 | 0.5 ± 0.1 | 0.6 ± 0.1 | 0.5 ± 0.0 |
| L. lactis PHa | 0.5 ± 0.1 | 1.2 ± 0.4 | 1.9 ± 0.6 | 2.0 ± 1.1 | 0.4 ± 0.0 | 0.7 ± 0.1 | 0.9 ± 0.0 | 0.9 ± 0.2 |
| L. lactis PHe | 1.1 ± 0.5 | 2.7 ± 0.8 | 4.5 ± 1.7 | 4.4 ± 1.6 | 0.5 ± 0.1 | 0.7 ± 0.2 | 1.0 ± 0.4 | 0.8 ± 0.5 |
| L. lactis PHh | 2.8 ± 1.5 | 6.9 ± 1.0 | 10.6 ± 0.2 | 12.1 ± 0.1 | 0.4 ± 0.1 | 0.5 ± 0.2 | 0.5 ± 0.3 | 0.6 ± 0.3 |
Peptide levels in experimental cheeses were quantified by HPLC after determination of the extraction efficiency for each peptide from a model cheese spiked with known peptide concentrations. d, days.
Regression analysis of bitter flavor scores from the trained sensory panel and individual concentrations of αS1-CN (f 1-9) or β-CN (f 193-209) showed that both peptides had a positive correlation with bitterness, but correlation coefficients were only 0.56 and 0.58, respectively. Since bitterness scores likely represent the combined contribution of these and other peptides, we attempted to ascertain the combined effect of αS1-CN (f 1-9) or β-CN (f 193-209). However, simple summation of each peptide's concentration was not informative due to the 10-fold difference in the relative abundance of αS1-CN (f 1-9) versus β-CN (f 193-209). In an effort to adjust for concentration differences, peptide data were transformed into a ratio by dividing the concentration of αS1-CN (f 1-9) or β-CN (f 193-209) in a given cheese by the highest concentration of that peptide in the data set. The ratios for each peptide were then combined, and this value was used for regression analysis with sensory bitterness scores. As shown in Fig. 3, this approach produced a correlation coefficient of 0.84.
FIG. 3.
Regression analysis of bitter flavor scores from the trained sensory panel and the combined effect of αS1-CN (f 1-9) and β-CN (f 193-209) concentrations. Peptide concentration data for each cheese were transformed into a ratio by dividing the concentration of αS1-CN (f 1-9) or β-CN (f 193-209) in that cheese by the highest concentration of respective peptide in the data set. The ratios for each peptide were then combined, and this value was used for regression analysis with sensory bitterness scores.
Bitter flavor intensity.
Evaluation by the trained bitterness panel showed that cheese made with the LCP− isogen, L. lactis PH, did not develop bitter flavor during 6 months of ripening, whereas cheeses made with L. lactis PHa, PHe, or PHh isogens had slight to moderate bitterness, respectively (Table 5). Statistical ANOVA revealed that time and starter choice had a significant effect on bitterness in experimental cheeses (α = 0.05; P < 0.0001). Bitter scores were significantly higher in 6-month-old cheeses than in 2-month-old cheeses, and Cheddar made with L. lactis PHa, PHe, or PHh was significantly more bitter than cheese made with the LCP− isogen, PH (Table 5). In addition, 2-month-old cheese made with L. lactis PHh was significantly (P < 0.05) more bitter than cheese made with PHe or PH, and cheese made with PHh was the only product that was significantly (P < 0.05) more bitter than PH control cheese at all sampling times (Table 5).
TABLE 5.
Bitterness of reduced-fat Cheddar cheese made with isogenic single-strain L. lactis starter bacteria as scored by a trained, screened sensory panela
| Starter strain | Mean score for bitter intensityb
|
|||
|---|---|---|---|---|
| 2 mo | 4 mo | 6 mo | Sum strain means | |
| L. lactis PH | 1.4A | 1.4A | 1.8A | 1.5A |
| L. lactis PHa | 2.5AB | 3.2B | 3.5AB | 3.1B |
| L. lactis PHe | 2.2A | 3.3B | 4.2B | 3.2B |
| L. lactis PHh | 3.5B | 3.6B | 4.7B | 3.9B |
| Sum age means | 2.4X | 2.9X | 3.6Y | |
Starter strain and cheese age were individually significant (P < 0.0001). Means with the same superscript letter in the same column (A to C) or row (X and Y) were not significantly different from each other (α = 0.05).
Scoring: 1 = no perceptible bitterness; 3 = slightly bitter; 5 = moderately bitter; 7 = very bitter; 9 = extremely bitter.
DISCUSSION
Bitterness in Cheddar cheese is a serious economic concern, and several studies have implicated lactocepin specificity in the production of bitter peptides in cheese (4, 25, 38). However, research to define the contribution of lactocepin specificity to bitterness has been hampered by strain-to-strain variability in autolysis and intracellular peptidase activity among L. lactis. This study sought to overcome these limitations via construction of isogenic starter bacteria that resisted autolysis and which differed only in lactocepin specificity. The host bacterium selected for this work, L. lactis MG1363ΔacmA, contains a 700-bp deletion in the gene encoding the major peptidoglycan hydrolase, AcmA (6). Since AcmA is the only peptidoglycan hydrolase in strain MG1363 (6), and because lactocepin specificity affects autolysis via differential degradation of AcmA (7), the absence of AcmA activity in strain MG1363ΔacmA should minimize confounding effects from intracellular enzymes on peptide accumulation and bitterness. As shown in Table 2, numbers of viable (i.e., culturable) starter bacteria in experimental cheeses remained relatively stable over the first 2 months of ripening but decreased more rapidly afterward in cheeses made with L. lactis PHe or PHh. As a result, the degree to which autolysis was controlled in experimental cheeses is unclear, particularly in the later months of ripening. However, indirect evidence suggesting that isogenic starters did not undergo autolysis is available from cheese peptide data. The bitter peptide β-CN (f 193-209), for example, is produced by chymosin (17) and cannot be hydrolyzed by lactocepin (14). It is, however, efficiently hydrolyzed by the lactococcal endopeptidase PepO, an intracellular enzyme released into the cheese matrix by starter autolysis (3, 11). Thus, starter autolysis should reduce β-CN (f 193-209) levels in cheese (3, 4, 13), but ANOVA showed that concentrations of this peptide in experimental cheeses were not significantly affected by starter choice (P > 0.5) and actually increased slightly over time in all cheeses (Table 4). This observation suggests that even though the number of viable starter cells differed over time, extensive autolysis did not occur in any of the experimental cheeses.
Further evidence to suggest that autolysis was controlled comes from αS1-CN (f 1-14) peptide data. The αS1-CN (f 1-14) peptide is not a major product of lactocepin action on αS1-CN (f 1-23) (Fig. 1), and production of this peptide in cheese has also been attributed to lactococcal PepO activity (3, 4, 13). A peptide that comigrated on HPLC chromatograms with αS1-CN (f 1-14) accumulated in all cheeses during ripening, but levels of this peptide were highest in cheeses that showed the smallest change in starter numbers during ripening; i.e., cheese made with L. lactis PHa and PH (Tables 2 and 3). These observations raise new questions regarding the possible origin of αS1-CN (f 1-14) in cheese. Possible alternative sources for this peptide may include nonstarter lactic acid bacteria or another L. lactis cell surface proteinase (e.g., HtrA [32]).
As expected, HPLC data also showed that lactocepin specificity had a marked effect on the peptide pool that accumulated in cheese made with each starter during ripening. Although Cheddar cheeses manufactured with single-strain isogenic LCP− and LCP+ starters had similar compositions, the peptides αS1-CN (f 1-9), αS1-CN (f 1-13), αS1-CN (f 1-16), and αS1-CN (f 1-17) accumulated only in cheeses made with proteinase-producing isogenic starters (Fig. 2 and Table 3), and the incidence and relative abundance of these peptides clearly reflected the in vitro specificity of starter lactocepin (Fig. 1). This observation is significant because each of these lactocepin-derived peptides is small and relatively hydrophobic (mass less than 6,000 and average hydrophobicity greater than 1,300) (28), and some have been implicated in bitterness (4, 24, 26). As was noted previously, bitterness develops when levels of a constituent bitter peptide exceed its taste threshold, and most bitter peptides are small to medium-sized hydrophobic molecules (28).
A direct role for lactocepin-derived peptides in the development of the bitter flavor defect was established by statistical analysis of trained sensory data. Cheddar cheese made with the isogenic starter L. lactis PHa, PHe, or PHh was significantly (P < 0.0001) more bitter than cheese made with the LCP− isogen, L. lactis PH. Moreover, the highest propensity for bitterness was seen in cheese made with starter cells that produced group h lactocepin (Table 5), and this effect was especially pronounced in the early months of ripening, where bitterness is a more significant economic concern. Because all of the starter bacteria used in this study were isogenic variants of a common strain, these data indicate that the propensity for bitterness in some industrial strains might be attenuated by proteinase gene exchange or gene replacement. It is noteworthy that characterization of bitter industrial strains by our group has shown that the group h lactocepin is relatively common among these bacteria (unpublished data). Finally, although other peptides certainly contribute to bitterness, data presented in Fig. 3 suggest that determination of αS1-CN (f 1-9) and β-CN (f 193-209) levels in cheese may be a useful way to predict the propensity of a given cheese to develop bitter flavor defect.
Acknowledgments
We express our appreciation to Willem M. de Vos for supplying the plasmid pNZ521, Jan Kok and Gerard Venema for pGK12, pGKV552, and L. lactis MG1363 acmAΔ1, and Larry L. McKay for L. lactis LM0230. We also thank Urmila Duhan, John Jaeggi, Bill Hoesly, Marianne Smukowksi, Bill Tricomi, Steve Wright, and Rhodia, Inc. (Madison, Wis.) for technical assistance and the Utah State University Biotechnology Center for peptide synthesis and DNA sequencing support.
This work was funded by Dairy Management, Inc., through the Western Dairy Center and the Wisconsin Center for Dairy Research and supported by the Utah Agricultural Experiment Station, Utah State University, Logan, Utah 84322-4810.
Footnotes
Approved as Journal Paper no. 7405 by the Utah Agricultural Experiment Station.
REFERENCES
- 1.American Society for Testing and Materials. 1968. Manual on sensory testing methods, STP 434. American Society for Testing and Materials, Philadelphia, Pa.
- 2.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baankreis, R., S. van Schalkijk, A. C. Alting, and F. A. Exterkate. 1995. The occurrence of two intracellular oligoendopeptidases in Lactococcus lactis and their significance for peptide conversion in cheese. Appl. Microbiol. Biotechnol. 44:386-392. [DOI] [PubMed] [Google Scholar]
- 4.Broadbent, J. R., M. Strickland, B. Weimer, M. E. Johnson, and J. L. Steele. 1998. Peptide accumulation and bitterness in Cheddar cheese made using single-strain Lactococcus lactis starters with distinct proteinase specificities. J. Dairy Sci. 81:327-337. [Google Scholar]
- 5.Brown, J. H., and P. E. Howe. 1922. Transparent milk as a bacteriological medium. J. Bacteriol. 7:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buist, G., J. Kok, J. Leenhouts, M. Dabrowska, G. Venema, and A. J. Haandrikman. 1995. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J. Bacteriol. 177:1554-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buist, G., G. Venema, and J. Kok. 1998. Autolysis of Lactococcus lactis is influenced by proteolysis. J. Bacteriol. 180:5947-5953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caldwell, S., D. J. McMahon, C. J. Oberg, and J. R. Broadbent. 1996. Development and characterization of lactose-positive Pediococcus species for milk fermentation. Appl. Environ. Microbiol. 62:936-941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Efstathiou, J. D., and L. L. McKay. 1977. Inorganic salts resistance associated with a lactose fermenting plasmid in Streptococcus lactis. J. Bacteriol. 30:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Exterkate, F. A. 1987. On the possibility of accelerating the ripening of Gouda cheese; a comment. Neth. Milk Dairy J. 41:189-194. [Google Scholar]
- 11.Exterkate, F. A. 1995. The lactococcal cell envelope proteinases: differences, calcium-binding effects and role in cheese ripening. Int. Dairy J. 5:995-1018. [Google Scholar]
- 12.Exterkate, F. A., and A. C. Alting. 1993. The conversion of the αS1-casein-(1-23)-fragment by the free and bound forms of the cell envelope proteinase of Lactococcus lactis subsp. cremoris under conditions prevailing in cheese. Syst. Appl. Microbiol. 16:1-8. [Google Scholar]
- 13.Exterkate, F. A., and A. C. Alting. 1995. The role of starter peptidases in the initial proteolytic events leading to amino acids in Gouda cheese. Int. Dairy J. 5:15-28. [Google Scholar]
- 14.Exterkate, F. A., A. C. Alting, and C. J. Slangen. 1995. Conversion of the αS1-casein-(24-199)-fragment and β-casein under cheese conditions by chymosin and starter peptidases. Syst. Appl. Microbiol. 18:7-12. [Google Scholar]
- 15.Flambard, B., and V. Julliard. 2000. The autoproteolysis of Lactococcus lactis lactocepin III affects its specificity towards β-casein. Appl. Environ. Microbiol. 66:5134-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fox, P. F., J. Law, P. L. H. McSweeney, and J. Wallace. 1993. Biochemistry of cheese ripening, p. 389-438. In P. F. Fox (ed.), Cheese: chemistry, physics and microbiology, vol. 1, 2nd ed. Chapman and Hall, London, United Kingdom. [Google Scholar]
- 17.Fox, P. F., T. K. Singh, and P. L. H. McSweeney. 1994. Proteolysis in cheese during ripening, p. 1-31. In A. T. Andrews and J. Varley (ed.), Biochemistry of milk products. Royal Society of Chemistry, Cambridge, England.
- 18.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiwaki, M., H. Ikemura, M. Shimuzu-Kadota, and A. Hirashima. 1989. Molecular characterization of a cell wall-associated proteinase gene from Streptococcus lactis NCDO763. Mol. Microbiol. 3:359-369. [DOI] [PubMed] [Google Scholar]
- 20.Kok, J., K. J. Leenhouts, A. J. Haandrikman, A. M. Ledeboer, and G. Venema. 1988. Nucleotide sequence of the cell wall proteinase gene of Streptococcus cremoris Wg2. Appl. Environ. Microbiol. 54:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kok, J., J. M. B. M. van der Vossen, and G. Venema. 1984. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 48:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kok, J., J. M. van Dijl, J. M. B. M. van der Vossen, J. Hugenholtz, and G. Venema. 1985. Cloning and expression of a Streptococcus cremoris proteinase in Bacillus subtilis and Streptococcus lactis. Appl. Environ. Microbiol. 50:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunji, E. R. S., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic system of lactic acid bacteria. Antonie Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 24.Law, B. A., and F. Mulholland. 1995. Enzymology of lactococci in relation to flavour development from milk proteins. Int. Dairy J. 5:833-854. [Google Scholar]
- 25.Law, J., G. F. Fitzgerald, C. Daly, P. F. Fox, and N. Y. Farye. 1992. Proteolysis and flavor development in Cheddar cheese made with the single starter strains Lactococcus lactis ssp. lactis UC317 or Lactococcus lactis ssp. cremoris HP. J. Dairy Sci. 75:1173-1185. [Google Scholar]
- 26.Lee, K. D., C. G. Lo, and J. J. Wartheson. 1996. Removal of bitterness from the bitter peptides extracted from Cheddar cheese with peptidases from Lactococcus lactis ssp. cremoris SK11. J. Dairy Sci. 79:1521-1528. [Google Scholar]
- 27.Lemieux, L., and R. E. Simard. 1991. Bitter flavor in dairy products. I. A review of the factors likely to influence its development, mainly in cheese manufacture. Lait 71:599-636. [Google Scholar]
- 28.Lemieux, L., and R. E. Simard. 1992. Bitter flavor in dairy products. II. A review of bitter peptides from caseins: their formation, isolation and identification, structure masking and inhibition. Lait 72:335-382. [Google Scholar]
- 29.Low, D., J. Ahlgren, D. Horne, D. J. McMahon, C. J. Oberg, and J. R. Broadbent. 1998. Role of Streptococcus thermophilus MR-1C capsular exopolysaccharide in cheese moisture retention. Appl. Environ. Microbiol. 64:2147-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McKay, L. L., K. A. Baldwin, and E. A. Zottola. 1972. Loss of lactose metabolism in lactic streptococci. Appl. Microbiol. 23:1090-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Olson, N. F., and M. E. Johnson. 1990. Light cheese products: characteristics and economics. Food Technol. 44:93-96. [Google Scholar]
- 32.Pouquet, I., V. Saint, E. Seznec, N. Simoes, A. Bolotin, and A. Gruss. 2000. HtrA is the unique surface housekeeping protease in Lactococcus lactis and is required for natural protein processing. Mol. Microbiol. 35:1042-1051. [DOI] [PubMed] [Google Scholar]
- 33.Reid, J. R., and T. Coolbear. 1998. The altered specificity of lactococcal proteinase PI (lactocepin I) in humectant systems giving the water activity and salt content of cheddar cheese. Appl. Environ. Microbiol. 64:588-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reid, J. R., and T. Coolbear. 1999. Specificity of Lactococcus lactis subsp. cremoris SK11 proteinase, lactocepin III, in low-water-activity, high-salt-concentration humectant systems and its stability compared with that of lactocepin I. Appl. Environ. Microbiol. 65:2947-2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stadhouders, J., L. Toepoel, and J. M. T. M. Wouters. 1988. Cheese making with prt− and prt+ variants of N-streptococci and their mixtures. Phage sensitivity, proteolysis and flavour development during ripening. Neth. Milk Dairy J. 42:183-193. [Google Scholar]
- 36.Strickland, M., M. E. Johnson, and J. R. Broadbent. 2001. Qualitative and quantitative analysis of proteins and peptides in milk products by capillary electrophoresis. Electrophoresis 22:1510-1517. [DOI] [PubMed] [Google Scholar]
- 37.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Microbiol. 50:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Visser, S., G. Hup, F. A. Exterkate, and J. Stadhouders. 1983. Bitter flavour in cheese. 2. Model studies on the formation and degradation of bitter peptides by proteolytic enzymes from calf rennet, starter cells and starter cell fractions. Neth. Milk Dairy J. 37:169-180. [Google Scholar]
- 39.Vos, P., G. Simons, R. J. Siezen, and W. M. de Vos. 1989. Primary structure and organization of the gene for a procaryotic, cell envelope-located serine proteinase. J. Biol. Chem. 264:13579-13585. [PubMed] [Google Scholar]
- 40.Vos, P., M. van Asseldonk, F. van Jeveren, R. Seizen, G. Simons, and W. M. de Vos. 1989. A maturation protein is essential for production of active forms of Lactococcus lactis SK11 serine proteinase located in or secreted from the cell envelope. J. Bacteriol. 171:2795-2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weimer, B., B. Dias, U. Madhavi, J. Broadbent, C. Brennand, J. Jaegi, M. Johnson, F. Milani, J. Steele, and D. V. Sisson. 1997. Influence of NaCl and pH on intracellular enzymes that influence Cheddar cheese ripening. Lait 77:383-398. [Google Scholar]
- 42.Willrett, D. 1997. A market survey of dairy processor needs, p. 60-68. In National Cheese Technology Forum, proceedings '97. Dairy Management, Inc., Rosemont, Ill.
- 43.Yu, W., K. Gillies, J. K. Kondo, J. R. Broadbent, and L. L. McKay. 1996. Loss of plasmid-mediated oligopeptide transport system in lactococci: another reason for slow milk coagulation. Plasmid 35:145-155. [DOI] [PubMed] [Google Scholar]