Abstract
Listeria monocytogenes is a food-borne bacterial pathogen that is able to grow at refrigeration temperatures. To investigate microbial gene expression associated with cold acclimation, we used a differential cDNA cloning procedure known as selective capture of transcribed sequences (SCOTS) to identify bacterial RNAs that were expressed at elevated levels in bacteria grown at 10°C compared to those grown at 37°C. A total of 24 different cDNA clones corresponding to open reading frames in the L. monocytogenes strain EGD-e genome were obtained by SCOTS. These included cDNAs for L. monocytogenes genes involved in previously described cold-adaptive responses (flaA and flp), regulatory adaptive responses (rpoN, lhkA, yycJ, bglG, adaB, and psr), general microbial stress responses (groEL, clpP, clpB, flp, and trxB), amino acid metabolism (hisJ, trpG, cysS, and aroA), cell surface alterations (fbp, psr, and flaA), and degradative metabolism (eutB, celD, and mleA). Four additional cDNAs were obtained corresponding to genes potentially unique to L. monocytogenes and showing no significant similarity to any other previously described genes. Northern blot analyses confirmed increased steady-state levels of RNA for all members of a subset of genes examined during growth at a low temperature. These results indicated that L. monocytogenes acclimation to growth at 10°C likely involves amino acid starvation, oxidative stress, aberrant protein synthesis, cell surface remodeling, alterations in degradative metabolism, and induction of global regulatory responses.
Listeria monocytogenes is a gram-positive rod-shaped bacterium that causes listeriosis, a potentially life-threatening illness. Pregnant women, infants, and elderly and immunocompromised individuals are at greatest risk (15). L. monocytogenes is a psychrotolerant organism with the ability to grow at refrigeration temperatures and thus to resist the traditional food preservation technology of chilling. When contaminated food is stored for extended periods at low temperatures, L. monocytogenes is able to grow, leading to costly product recalls and posing a significant public health threat (46).
L. monocytogenes has a minimum growth temperature estimated to be just below 2°C (34, 70). Low temperatures have profound effects on the growth of bacteria through influences on the ribosome and cytoplasmic membrane and alterations in protein synthesis and solute uptake (7, 52, 65). A major emphasis of recent studies has been placed on the increased expression of bacterial cold shock proteins (Csps) in response to reduced temperatures (49, 52, 71), rather than long-term adaptive responses to growth at low temperatures. Studies focusing on the Escherichia coli cold shock response have indicated that members of a major class of Csps are small RNA-binding proteins that mediate transcription elongation and message stability (3, 52, 65, 72). The regulation of bacterial RNA levels in response to cold is therefore of interest in other bacterial species where the mechanisms underlying these responses have not yet been examined in detail.
A limited number of studies have examined the molecular mechanisms of psychrotolerance in L. monocytogenes. Annous et al. (2) showed that the cytoplasmic membranes of bacteria grown at low temperatures contain a larger proportion of shorter-chain fatty acids, with a switch in branching from iso to anteiso. Edgcomb et al. (14) and Jones et al. (33) provided evidence that an anteiso branched-chain fatty acid-deficient mutant that was unable to grow at low temperatures had reduced membrane fluidity. Various low-molecular-weight solutes have been shown to stimulate the growth of L. monocytogenes at low temperatures, i.e., act as cryoprotectants (5, 37). Ko et al. (37) described a chill-activated transport system for the osmoprotectant and cryoprotectant glycine betaine. Becker et al. (6) provided evidence for the role of an alternative sigma factor (SigB) in both cryoprotectant uptake and growth of L. monocytogenes at low temperatures.
Two studies used two-dimensional gel electrophoresis to examine the cold shock responses of L. monocytogenes. Bayles et al. (4) observed increased production of 12 Csps in response to a temperature shift from 37 to 5°C. Phan-Thanh and Gormon (53) described significantly increased production of 32 proteins following a similar temperature shift. A major 18-kDa Csp was recently identified as the Flp protein—a ferritin-like, nonheme iron protein (9, 27). Bayles et al. (4) also observed increased production of four proteins in L. monocytogenes cultures growing at 5°C and designated these cold acclimation proteins (Caps).
Based on studies of other bacteria, we expect a considerable number of gene products to have roles in L. monocytogenes cold adaptation responses. No studies have yet examined changes in L. monocytogenes global gene expression patterns associated with decreased temperature. Recently, a novel differential cDNA cloning method (selective capture of transcribed sequences [SCOTS]) was developed for the identification of Mycobacterium tuberculosis genes expressed in response to host interactions (24). This approach has distinct advantages for the current studies in being applicable to any microbe from which total nucleic acids can be obtained and in efficiently identifying differentially expressed genes directly on the basis of the nucleotide sequence rather than promoter regions potentially associated with the expression of downstream genes (42, 67).
Here we used SCOTS to identify L. monocytogenes RNAs differentially expressed by bacteria growing at 10°C relative to those in reference cultures growing at 37°C. We focused on the cold acclimation response, rather than cold shock, because of its potentially greater relevance to L. monocytogenes growth in refrigerated food.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
L. monocytogenes strain 10403S was grown to mid-exponential phase in 100 ml of brain heart infusion (BHI) broth (Difco Laboratories, Detroit, Mich.) in 1-liter Erlenmeyer flasks incubated at either 37 or 10°C with shaking at 240 rpm. E. coli strains DH5α and TOP10F′ were grown in Luria broth (LB) at 37°C. When necessary, ampicillin was added to a final concentration of 100 μg ml−1.
Preparation of genomic DNA.
Harvested cells were lysed by treatment with mutanolysin (Sigma, St. Louis, Mich.) (10 U μl−1) and lysozyme (20 μg μl−1) in 50 mM Tris-HCl (pH 7.2) at 37°C for 20 min. Genomic DNA was prepared by using a Wizard Plus DNA purification system as described by the manufacturer (Promega, Madison, Wis.). Aliquots (100 μg) of genomic DNA in 0.6 ml of H2O were sonicated at 40% maximum intensity by pulsing three times for 10 s each time. Sonicated fragments were photobiotinylated according to the manufacturer's instructions by adding 100 μg of photobiotin (Sigma) and incubation on ice directly under a 250-W reading lamp. Biotin-labeled chromosomal DNA was purified by 2-butanol extraction, precipitated, and resuspended in 0.1× TE buffer (1 mM Tris-Cl, 0.1 mM EDTA [pH 8.0]) at a concentration of 0.5 μg μl−1.
Oligonucleotides.
The oligonucleotides shown in Table 1 were synthesized by Genosys Biotechnology (Woodlands, Tex.).
TABLE 1.
Oligonucleotides used for this study
| Oligonucleotide | Sequence |
|---|---|
| F1 | CTGGCTCAGGACGAACGCTGG |
| B1 | GCTGGCTCCATAAAGGTGACC |
| F2 | TCGGTAGGGTCACCTTTATGG |
| B2 | CTCACACTCACTGCTTGGACG |
| F3 | AATGGAAATGTGCGTCCAAGC |
| B3 | ACTTTTCAAATTGCCCGGCAG |
| K9 | GACACTCTCGAGACATCACCGGTACCNNNNNN |
| F9 | GCCGGAGCTCTGCAGAATTCNNNNNN |
| K-N6 | GACACTCTCGAGACATCACCGGTACC |
| F-N6 | GCCGGAGCTCTGCAGAATTC |
| DapE5′ | GGTTTTGGCATTTTCAGGGC |
| DapE3′ | GATTCTGTGTCCACTCGGTTCG |
Cloning of the L. monocytogenes rDNA operon.
The published sequences for L. monocytogenes 16S, 23S, and 5S ribosomal DNAs (rDNAs) (GenBank accession numbers X98530, X92951, and AF16093, respectively) were used to design primers for the amplification of overlapping DNA fragments corresponding to the entire coding region of the ribosomal operon. Three sets of primers (F1-B1, F2-B2, and F3-B3) were used to amplify three rDNA fragments. PCR was performed with TaqDNA polymerase (Gibco BRL, Rockville, Md.) according to the manufacturer's instructions and with L. monocytogenes 10403S genomic DNA as a template. Amplified DNA fragments were integrated into the pTA cloning vector (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions, and the desired constructs were confirmed by automated sequencing with an ABI Prism 310 apparatus and an ABI Prism dye terminator cycle sequencing ready reaction kit (Perkin-Elmer, Foster City, Calif.). The three cloned rDNA plasmids were purified by using Qiaprep spin columns (Qiagen Inc., Valencia, Calif.).
RNA purification and cDNA synthesis.
L. monocytogenes 10403S was grown to mid-exponential phase (optical density at 600 nm [OD600], 0.6) at 37 and 10°C. Growing cultures were poured directly into prechilled centrifuge bottles on wet ice and centrifuged at 4°C. RNA was isolated from bacterial pellets on ice by using TRI reagent according to the manufacturer's instructions (Molecular Research Center, Inc., Cincinnati, Ohio). RNAs were treated with DNase I according to the manufacturer's directions (Gibco BRL) prior to cDNA synthesis. First-strand cDNA was synthesized as previously described (22) with Superscript II reverse transcriptase (Gibco BRL), 5 μg of each RNA template, and either 1 μg of primer K9 for bacteria grown at 37°C or 1 μg of primer F9 for bacteria grown at 10°C (Table 1). Second-strand synthesis was carried out with the large fragment of DNA polymerase I (Gibco BRL) after denaturation, and the same primers were allowed to reanneal to newly synthesized first-strand cDNA (16). Double-stranded cDNAs were purified with QIAquick PCR purification columns (Qiagen) and PCR amplified as previously described (24) with appropriate primers lacking the 3′-terminal random nucleotides that were used in cDNA synthesis (Table 1). Three standard 50-μl PCRs were performed in parallel for each cDNA before samples were pooled for use in SCOTS.
Normalization of cDNAs and enrichment for cold-induced sequences by SCOTS.
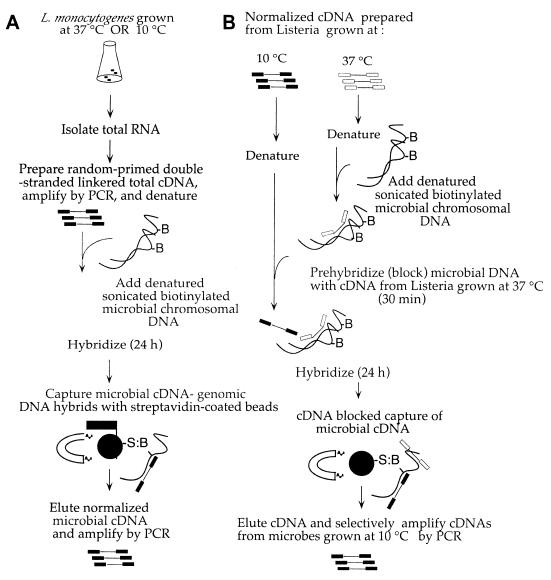
Methods for equalizing the abundance of cDNAs representing bacterial RNAs (normalization) and the reduction of rDNA content have been described elsewhere (24), and a schematic diagram of the entire process is presented in Fig. 1. Briefly, 5 μg of PCR-amplified cDNA from bacteria grown at 10 or 37°C was denatured in 8 μl of hybridization buffer (10 mM EPPS-1 mM EDTA), 2 μl of 1 M NaCl was added, and nucleic acids were allowed to reanneal at 65°C for 30 min. Similarly, 5 μg of rDNA plasmid and 0.3 μg of sonicated biotinylated genomic DNA were denatured in 8 μl of hybridization buffer, 2 μl of 1 M NaCl was added, and rDNA blocking was carried out at 65°C for 30 min. Partially renatured cDNAs were then added to tubes containing rDNA-blocked genomic DNAs (20-μl final volume; oil overlay), and incubation was continued at 65°C for 18 to 24 h. Following blocked-capture hybridization, 100 μl of H2O was added to each 20-μl hybridization reaction. Bacterial cDNA-chromosomal DNA hybrids were bound to streptavidin-coated beads (Dynal, New York, N.Y.), collected, and washed as described by the manufacturer. Captured cDNAs were eluted by treatment with 100 μl of 0.5 M NaOH-0.1 M NaCl at 37°C for 30 min. Samples were concentrated by using ethanol with the addition of 1 μg of glycogen (Ambion, Austin, Tex.) carrier and then were resuspended in 20 μl of H2O. Normalized cDNAs were PCR amplified by using primer F-N6 or K-N6 in 10 parallel 50-μl PCRs, the results of which were pooled for the second round of SCOTS. Eluted normalized cDNAs were amplified in triplicate parallel reactions for the second and all subsequent PCR amplifications as previously described (24) (Fig. 1A). PCR products from the third round of rDNA blocked-capture hybridization were used for three rounds of enrichment by differential hybridization and PCR amplification as previously described (24) (Fig. 1B). Prehybridization with normalized amplified cDNA from bacteria grown at 37°C was used to block the hybridization of commonly expressed cDNAs to biotinylated bacterial genomic DNAs as described above for blocking the capture of rDNA sequences. cDNAs from bacteria grown at 10°C and capable of hybridizing to blocked genomic DNAs were eluted and specifically amplified with DNA primer F-N6, complementary to the terminal sequence of the cDNA from cells grown at 10°C.
FIG. 1.
Schematic diagram outlining the procedure (SCOTS) used to obtain cDNAs for L. monocytogenes RNAs expressed in response to growth at a low temperature. (A) Process used to reduce rDNA sequence content and increase the representation of less abundant RNAs (normalized). (B) Differential hybridization method used for selective amplification of bacterial RNAs expressed in response to a reduced temperature. S:B, streptavidin:biotin.
Sequencing of the SCOTS clones.
PCR-amplified cDNA fragments obtained from bacteria grown at 10°C and enriched for differentially expressed sequences were cloned into the pTA vector according to the manufacturer's instructions. Individual plasmid clones were verified by releasing an EcoRI cDNA insert fragment from the vector. DNA sequences were determined with an ABI Prism 310 apparatus and an ABI Prism dye terminator cycle sequencing ready reaction kit. Sequence analyses were performed by using MacVector software, and National Center for Biotechnology Information BLAST was used to search GenBank for previously described sequences with similarity to SCOTS cDNA clones.
Northern blot analyses.
L. monocytogenes RNAs in 1× morpholinepropanesulfonic acid (MOPS) buffer were separated by electrophoresis in a 1.2% agarose gel with formaldehyde. Gels were stained with ethidium bromide to ensure that equivalent amounts of total RNA were loaded in the lanes. Gel-purified EcoRI fragments from SCOTS clones were radiolabeled by using [α-32P]dCTP (ICN Biochemicals, Irvine, Calif.) with the Prime a Gene labeling system (Promega) according to the manufacturers' instructions and were used as probes for Northern blots. A dapE DNA fragment was PCR amplified from L. monocytogenes 10403S genomic DNA with primers DapE5′ and DapE3′ (Table 1) and was cloned into the pTA vector for use as a template for the synthesis of a housekeeping RNA control probe (57). Approximately 1 × 107 to 5 × 107 cpm was added to each hybridization bag, and hybridizations were performed overnight with formamide at 42°C.
RESULTS
Nucleotide sequence analysis of the cDNA clones obtained by SCOTS.
To identify L. monocytogenes RNAs whose steady-state levels increased during growth at 10°C, a differential cDNA cloning strategy was used to obtain E. coli plasmid cDNA clones for differentially expressed bacterial transcripts (24). Three rounds of hybridization of rDNA-blocked L. monocytogenes genomic DNA fragments with PCR-amplified cDNA pools obtained from bacteria cultured at either 10 or 37°C were used to remove rRNA sequences and to equalize (normalize) the abundance of cDNAs representing the entire RNA populations obtained from L. monocytogenes grown at the two temperatures (Fig. 1A). Three rounds of differential hybridization of normalized cDNAs from bacteria grown at 10 and 37°C to biotinylated genomic DNA fragments were then used to select and amplify specific sequences for bacterial transcripts expressed in response to growth at 10°C (Fig. 1B). These final PCR products were then ligated into a plasmid cloning vector for nucleotide sequence analysis. Most SCOTS plasmid cDNA clones had inserts ranging from 200 to 600 bp, as expected from the appearance of the amplified cDNA on ethidium bromide-stained agarose gels. A total of 90 cDNA sequences were determined for randomly selected SCOTS clones.
Nucleotide sequence similarity searches with the BLAST network service of the National Center for Biotechnology Information indicated that 57 of these 90 sequences corresponded to various regions of the previously described L. monocytogenes flaA gene (Table 2). cDNA inserts corresponding to the L. monocytogenes flaA, flp, rpoN, lhkA, groEL, clpP, and trxB genes and those showing the greatest nucleotide sequence similarity to the adaB, yycJ, bglG, clpB, hisJ, trpG, cysS, aroA, eutB, mleA, psr, celD, and fbp genes of other bacterial species were identified (Table 2). All cDNA clones corresponded to regions within the L. monocytogenes strain EGD-e genome (22; www//genolist.pasteur.fr/ListListindex.html), and their corresponding accession numbers are given in Table 2. Four cDNA sequences (SCOTS 4, 26, 50, and 69) showed no other significant similarity to any previously described genes, as determined by default BLAST search criteria. In summary, 24 different L. monocytogenes RNAs were identified by SCOTS as being potentially differentially expressed during growth at 10°C relative to grown at 37°C (standard cultures).
TABLE 2.
L. monocytogenes cDNA clones identified by SCOTS
| Category | SCOTS clone(s) | Gene | Predicted gene product | GenBank accession no. | Strain EGD-e
|
|
|---|---|---|---|---|---|---|
| Gene | Chromosomal location (kb) | |||||
| Previously described L. monocytogenes cold-adaptive responses | 12 | flaA | Flagellin | X65624 | lmo0690 | 724.9 |
| 25 | flp | Ferritin-like protein | AJ244014 | lmo0943 | 979.1 | |
| Regulatory adaptive responses | 19, 62 | rpoN (sigL) | RNA polymerase sigma factor σ54 | X93169.1 | lmo2461 | 2535.2 |
| 73 | lhkA | Histidine kinase sensor | AJ010495 | lmo1508 | 1538.5 | |
| 6 | yycJ | Two-component signal transduction system | BAA11296 | lmo0291 | 316.9 | |
| 22 | bglG | Similar to transcription antiterminator BglG family | lmo0501 | 536.3 | ||
| 75 | adaB | Similar to methyltransferase | X53399 | lmo0571 | 608.5 | |
| General microbial stress responses | 70 | groEL | Major heat shock protein | AF335323 | lmo2068 | 2149.3 |
| 55 | clpP | Serine protease | AF102775 | lmo2612 | 2627.7 | |
| 7 | trxB | Thioredoxin reductase | AF009622 | lmo2478 | 2554.4 | |
| 60 | clpB | ClpB protease | lmo2206 | 2297.2 | ||
| Alterations in amino acid metabolism | 17, 24, 45 | hisJ | Histidinol phosphate phosphatase | D70002 | lmo0570 | 607.4 |
| 18 | trpG | Anthranilate synthase | S74362 | lmo2749 | 2824.1 | |
| 32 | cysS | Cysteinyl-tRNA synthetase | L14580 | lmo0239 | 259.5 | |
| 65 | aroA | 3-Deoxy-d-arabino-heptulosonate 7-phosphate synthase | X69545 | lmo1600 | 1645.2 | |
| 51 | celD | Endoglucanase D | U07818 | lmo1719 | 178.1 | |
| Alterations in degradative metabolism | 15, 27, 39, 43, 64, 84 | eutB | Ethanolamine ammonia lyase heavy chain | P19635 | lmo1175 | 1203.6 |
| 79 | mleA | Similar to malolactic enzyme | X82326 | lmo1915 | 1988.2 | |
| Alterations of the L. monocytogenes cell surface | 3 | psr | PBP5 synthesis repressor | U42211 | lmo0443 | 472.8 |
| 56 | fbp | Fibronectin-binding protein | AJ132543 | lmo0721 | 751.3 | |
| Proteins with unknown functions | 4 | Unknown conserved hypothetical protein | lmo0599 | lmo0599 | 640.3 | |
| 26 | Unknown protein | lmo0170 | lmo0170 | 168.0 | ||
| 50 | Unknown protein | lmo0719 | lmo0719 | 750.0 | ||
| 69 | Unknown protein | lmo1535 | lmo1535 | 1570.9 | ||
Northern blot analyses of selected SCOTS clones.
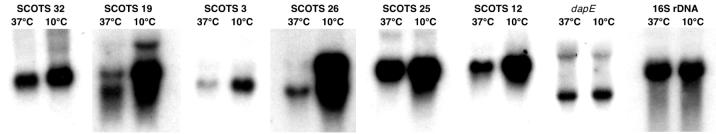
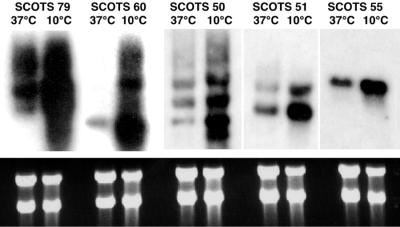
In order to confirm the differential expression of transcripts corresponding to cDNAs cloned by SCOTS, total RNA isolated from bacteria grown at 37 and 10°C was examined directly by Northern hybridization (Fig. 2 and 3). Ethidium bromide staining was used to verify that equivalent amounts of total RNA had been loaded onto agarose gels (Fig. 3). Radiolabeled probes prepared from cloned L. monocytogenes 16S rRNA and dapE DNA fragments were used as housekeeping controls (Fig. 2). Probes prepared from cDNA inserts of individual plasmid clones obtained by SCOTS clearly showed increased hybridization with total RNA prepared from bacteria grown at 10°C compared to those grown at 37°C for all 11 cDNAs examined (Fig. 2 and 3). The extents of differential RNA expression in bacteria grown at the low temperature varied, with SCOTS clone 26 showing the greatest increase in the steady-state level (Fig. 2). Multiple transcripts capable of hybridizing to cDNA probes from SCOTS clones 51 (similar to Bacillus stearothermophilus celD), 50 (lmo0719), 60 (clpB), and 79 (mleA) were detected (Fig. 3), suggesting rapid cellular turnover of these messages.
FIG. 2.
Northern hybridization analyses of RNAs from mid-log-phase L. monocytogenes 10403S cultures grown at 37 and 10°C. EcoRI-digested inserts from plasmid cDNA clones obtained by SCOTS were used as templates to prepare radiolabeled probes. Hybridization was performed as described in Materials and Methods. Nylon filters were subsequently hybridized with a 16S rDNA probe and a housekeeping gene (dapE) probe as controls. Table 2 lists the corresponding genes for SCOTS cDNA clones.
FIG. 3.
Northern hybridization analyses of L. monocytogenes RNAs. Total RNA was isolated from mid-log-phase L. monocytogenes 10403S grown at either 37 or 10°C. RNAs were electrophoresed as described in Materials and Methods and stained with ethidium bromide to ensure that equivalent amounts of total RNA were present in the lanes (bottom panel). EcoRI-digested inserts from plasmid cDNA clones obtained by SCOTS were used as templates to prepare radiolabeled probes (top panel). Table 2 lists the genes corresponding to SCOTS cDNA clones.
DISCUSSION
In this study, we examined the adaptation of L. monocytogenes to growth at a low temperature by identifying bacterial RNAs whose steady-state levels were increased in bacteria growing at 10°C relative to those growing at 37°C. Like many other pathogenic bacteria, L. monocytogenes has an optimal growth temperature of about 37°C. Mid-logarithmic-phase cultures grown at this temperature were used as our standard for the RNA expression studies reported here. This strategy allowed us to compare our findings with prior descriptions of bacteria grown at human body temperature. To study the effects of reduced temperature, a temperature of 10°C was chosen for practical reasons as one at which growth was not excessively slow.
We identified 24 different L. monocytogenes transcripts with the SCOTS differential cDNA cloning approach, and all of those whose levels were directly examined by Northern blot analyses were shown to be differentially expressed. Although we expect that the production of the encoded factors by bacteria is under additional levels of posttranscriptional regulation, it also seems likely that expressing mRNAs to elevated levels is in general intended to increase the production of the encoded factors. We therefore discuss the potential physiological relevance of the encoded proteins in terms of what is known from previous studies of L. monocytogenes and other bacteria, with the understanding that additional studies will be necessary both to show increased production of each corresponding final gene product at a reduced temperature and to establish roles for these products in bacterial psychrotolerance. Several of the mRNAs identified encode proteins with potential roles in general classes of bacterial adaptive responses that are described below.
Previously described L. monocytogenes cold-adaptive responses.
Increased motility and flagellum production have long been known to be associated with the growth of L. monocytogenes at low temperatures (40). flaA encodes the L. monocytogenes major flagellin protein FlaA (13). Western blot analyses indicated that trace amounts of flagellin are present in cells grown at 37°C (35, 51). Dons et al. (13) were able to detect flaA transcripts at 25 but not 37°C by Northern hybridization. flaA transcripts were readily detected in our Northern blot analyses of total RNA isolated from bacteria grown at 10°C; much lower levels of easily detected transcripts were present in bacteria grown at 37°C (Fig. 3). The significance of increased flagellum production during growth at low temperatures is unclear. However, motility is associated with microbial biofilm formation in other species, and reduced temperature is a potential environmental signal associated with natural environmental reservoirs capable of hosting L. monocytogenes (15). It has also been demonstrated that L. monocytogenes bacteria grown at a reduced temperature (4 or 22°C) have greater virulence upon intravenous inoculation than bacteria grown at 37°C (13, 63).
Listeria innocua flp encodes the 18-kDa Flp protein, which forms a multimeric 240-kDa complex (9). Purified Flp functions as an authentic ferritin in oxidizing and chelating readily mobilizable inorganic iron (9). Flp appears to be the major 18-kDa Csp in L. monocytogenes and is translated from a single 0.8-kb transcript (27). Our Northern blot analyses demonstrated that the flp transcript is expressed to higher levels by bacteria growing at 10°C than by those growing at 37°C. Bayles et al. (4) demonstrated a major 18-kDa Csp in cold-shocked cells that was also a Cap in cultures growing at 5°C. The expression of Flp is also known to be induced by low temperatures in Anabaena variabilis (EMBL/GenBank accession number D01016) and to be involved in protection against oxidative stress in Campylobacter jejuni (69).
Regulatory adaptive responses.
Differential production of bacterial factors in response to environmental changes is typically the result of coordinated regulation of gene expression, often via mechanisms that are resistant to specific inhibitory effects exerted by these changes. L. monocytogenes rpoN encodes the alternative sigma factor σ54. σ54 was originally recognized for its role in the regulation of nitrogen metabolism in enteric bacteria (47) but has subsequently been shown to be involved in the regulation of transcription of genes encoding products involved in the utilization of carbon sources, energy metabolism, RNA modification, chemotaxis, flagellation, electron transport, response to heat and phage shock, and expression of other alternative sigma factors (11, 32). A Pseudomonas aeruginosa rpoN mutant lacks motility due to a defect in flagellum and pilus synthesis (66). The L. monocytogenes rpoN gene has been shown to have a role in resistance to the antibacterial peptide mesentericin Y105 (59). Increased expression of rpoN mRNA at low temperatures suggests a previously unrecognized role for this sigma factor in cold acclimation. σ54 may be involved in regulating the expression of L. monocytogenes flaA gene and other genes whose mRNA levels have been found to be increased at low temperatures. Additional studies are necessary to define this potential cold shock regulon.
Bacterial two-component signal transduction systems, consisting of histidine kinase sensors and DNA-binding response regulators, allow bacteria to respond to diverse environmental stimuli. Although no previous studies implicated similar two-component systems in the regulation of L. monocytogenes gene expression during growth at low temperatures, our identification of increased lhkA mRNA levels raises the possibility that the encoded histidine kinase is an important environmental sensor in the cold adaptation response. The histidine kinase Hik33 senses reduced temperatures in the cyanobacterium Synechocystis sp. strain PCC6803 and is involved in the regulation of cold-inducible gene expression (64). The DesK-DesR two-component system regulates the expression of the Bacillus subtilis cold-inducible des gene (1).
The L. monocytogenes bglG gene was also identified by SCOTS. The BglG protein is a transcriptional antiterminator acting within the beta-glucoside operon of E. coli by interacting with the nascent mRNA (31). Transcription attenuation mechanisms have been shown to be central in the autogenous regulation of Csp expression in E. coli (3, 52). Although there have been no reports of bglG being related to low-temperature growth, it is interesting that celD also appears to be induced by low temperatures in L. monocytogenes (see below). CelD is involved in the breakdown of the beta-glucoside polymer cellulose. L. monocytogenes has a large number of transporter genes devoted to carbohydrate transport by the phosphoenolpyruvate-dependent phosphotransferase systems, particularly the beta-glucosides (22). Our results suggest that another environmental signal, reduced temperatures, may modulate the previously described regulation of L. monocytogenes gene expression by beta-glucoside carbohydrate availability (10)
SCOTS cDNA clone 3 shows the greatest similarity to Enterococcus hirae psr and encodes a repressor protein highly similar to those of several other species, including Staphylococcus aureus (msrR) and Streptococcus pyogenes (cpsA) (62). The E. hirae psr gene is located upstream of the structural gene for penicillin-binding protein 5 (PBP5) and may regulate both PBP5 production and its own transcription (41, 58). E. hirae mutants exhibit decreased rhamnose cell wall content (43) and other cell wall alterations (44). Increased expression of L. monocytogenes psr mRNA at 10°C suggests an important regulatory role for the encoded repressor in cold-adaptive responses. Future studies of this repressor and its target(s) are likely to provide important insight into the physiology of L. monocytogenes cold acclimation.
General microbial stress responses.
Increased expression of mRNAs for chaperone proteases (GroEL, ClpP, and ClpB) may reflect the production of improperly folded or otherwise damaged proteins arising during L. monocytogenes growth at low temperatures. The production of the major bacterial heat shock chaperone GroEL is dramatically increased in response to various environmental stimuli, such as heat shock, low pH, ethanol, salt, and bile salts (26, 61). GroEL is a highly conserved protein that functions together with GroES to manage unfolded, misfolded, damaged, or aggregated proteins as well as to produce proper protein folding (28). L. monocytogenes GroEL is expressed at elevated levels during infection and is required for maximum virulence potential (17). Increased groEL expression at reduced temperatures may be involved in the maintenance of protein solubility and function in the cytoplasm or may be part of a more general stress response to adverse environmental conditions signaled by multiple stimuli.
Caseinolytic proteins (Clps) form large protein complexes that have both proteolytic and chaperone activities (23). These energy-dependent bacterial proteases are involved in the degradation of damaged polypeptides and the salvage of amino acids. ClpP is a serine protease that is able to cleave polypeptides of six amino acids or less (45), forming a more processive enzyme complex upon association with ClpA or ClpX ATPases. B. subtilis clpP mRNA levels have been shown to increase in response to heat shock, salt, and ethanol stress (19). In the cyanobacterium Synechococcus, clpP has been shown to be essential for acclimation and growth at 25°C. L. monocytogenes clpP encodes a 21.6-kDa protein and is necessary for survival in host macrophages (18). A 21-kDa protein was identified as both a Csp and a Cap in studies of L. monocytogenes by Bayles et al. (4). However, the inactivation of clpP did not alter L. monocytogenes growth at 4°C, although no data were presented (18). Increased clpP mRNA levels in cells grown at 10°C suggests that the encoded enzyme may be involved in the degradation of abnormal polypeptides or truncated peptides that arise during low-temperature growth. Studies on enteric bacteria have shown that bacterial ribosome function is severely impaired upon cold shock and that cold-induced accessory translation factors are necessary to resume normal peptide synthesis (52, 71). Alternatively, increased clpP mRNA levels may result from the activation of more general stress response pathways. We also identified mRNA for another Clp protease, ClpB (SCOTS clone 60), as expressed in response to a reduced temperature. ClpB induction plays a critical role in cold acclimation in the cyanobacterium Synechococcus sp. strain PCC7942 (54).
Increased levels of transcripts encoding thioredoxin reductase (trxB) suggest that L. monocytogenes experiences increased oxidative stress at 10°C. As described above, increased expression of flp mRNA, which encodes a ferritin-like protein, may also be related to an alteration of the oxidation-reduction status of the cell. Thioredoxin reductase, along with thioredoxin and NADPH, constitutes a thiol-dependent oxidation-reduction system that catalyzes the reduction of certain proteins (30). The thioredoxin system, along with the glutaredoxin system, maintains a reducing environment in the bacterial cytoplasm, reducing disulfide bonds that form spontaneously in aerobically grown cultures (56).
Alterations in amino acid metabolism.
SCOTS clone 32 was identified as cysteinyl-tRNA synthetase. Cysteinyl-tRNA synthetase charges cysteinyl-tRNA with cysteine and, like genes for several other aminoacyl-tRNA synthetases and amino acid biosynthetic genes (including trpG and aroA), contains an untranslated upstream sequence that allows the regulation of expression by transcription antitermination mechanisms (25, 73). Such transcription attenuation mechanisms are central in the autogenous regulation of Csp expression in E. coli (3, 52).
Three other identified mRNAs encode products related to amino acid biosynthesis. These are aroA, trpG, and hisJ, encoding products involved in the biosynthesis of aromatic compounds, tryptophan, and histidine, respectively. O'Donovan and Ingraham (48) showed that the first enzyme in histidine biosynthesis was more sensitive to feedback inhibition by histidine at low temperatures. It is possible that L. monocytogenes cells become starved for certain amino acids at least partially through feedback inhibition and respond by inducing biosynthetic enzymes for histidine and aromatic amino acids. A cold-sensitive Salmonella enterica serovar Typhimurium mutant required tryptophan for growth at 20°C (29). Studies with yeast cells have indicated that transport of the amino acid tryptophan is a rate-limiting step during growth at reduced temperatures (36). L. monocytogenes is a multiple-amino-acid auxotroph requiring several amino acids for growth and as nitrogen sources. Peptides appear to be an important source of amino acids for L. monocytogenes (68), especially in laboratory media, such as brain heart infusion broth, which contain large amounts of peptides. Borezee et al. (8) reported that OppA, an oligopeptide-binding protein from L. monocytogenes, is required for bacterial growth at low temperatures. An oppA deletion mutant was unable to grow at 5°C. At low temperatures, peptide transport may be inhibited, imposing further limitations on amino acids and nitrogen (55).
Alterations of the L. monocytogenes cell surface.
In addition to mRNA capable of encoding an L. monocytogenes peptide similar to the E. hirae psr repressor and the FlaA protein, we also identified mRNA for a novel L. monocytogenes fibronectin-binding protein expressed in response to reduced temperatures. L. monocytogenes fbp encodes a 24.6-kDa peptide (21) which has a PrfA-like box that appears to play a regulatory role in the expression of virulence genes (38). L. monocytogenes Fbp is unique in that it shares no similarity with previously described bacterial fibronectin-binding proteins and was identified by its ability to bind human fibronectin (20). The L. monocytogenes fibronectin-binding protein may facilitate the entry of bacteria into mammalian cells (20). Any relationship between fbp mRNA expression and low-temperature growth of L. monocytogenes remains to be elucidated.
Alterations in degradative metabolism.
Six SCOTS cDNA clones showed the greatest similarity to E. coli eutB, which encodes a heavy chain of ethanolamine ammonia lyase, the first enzyme involved in the degradation of ethanolamine to acetaldehyde and ammonia (60). SCOTS cDNA clone 51 showed significant similarity (58% identity over 343 bp) to celD, which encodes endoglucanase D of B. stearothermophilus. CelD is part of a multienzyme complex known as the cellulosome that is able to break down cellulose. The significance of ethanolamine and cellulose degradation in the low-temperature metabolism of L. monocytogenes is unclear. Ethanolamine catabolism may serve as a source of ammonia. L. monocytogenes is ubiquitous in the environment and is often found associated with decaying vegetation. Park and Kroll (50) have shown that the environmentally ubiquitous molecule cellobiose represses the expression of L. monocytogenes factors involved in host colonization. Temperatures below that of mammalian hosts are potential environmental signals that could trigger metabolic changes important for the growth of L. monocytogenes as free-living bacteria in natural reservoirs.
mleA encodes the malolactic enzyme, which is involved in malolactate fermentation in some lactic acid bacteria, where l-malate is converted to l-lactate (39). This fermentation may provide metabolic energy, and the induction of this enzyme is not incompatible with the microaerophilic and facultatively anaerobic life style of L. monocytogenes (22).
Adaptations potentially unique to L. monocytogenes.
Several genes identified by SCOTS are so far unique to L. monocytogenes. These include SCOTS clones 4, 26, 50, and 69 (Table 2). BLAST analyses with nucleotide sequences from these clones failed to reveal any similarity to previously described sequences in GenBank. The entire L. monocytogenes strain EGD-e genome sequence was published (22) during revision of this article, enabling us to identify the corresponding full open reading frame sequences for all SCOTS cDNA clones (Table 2). Northern blot analyses with cDNA inserts from plasmid clones 26 and 50 as probes indicated that steady-state levels of these Listeria-specific RNAs were increased during growth at 10°C (Fig. 2 and 3). The genes for several RNAs expressed by the organism in response to growth at a low temperature are potentially uniquely present in this species.
Concluding remarks.
Members of the CspA family, proposed RNA chaperones, have received considerable attention in studies of the bacterial cold shock response. It appears that there is a transitory requirement for high levels of these proteins and that other factors are required for cold acclimation. We have identified a group of bacterial genes showing increased RNA expression levels during sustained growth at 10°C compared to 37°C. Allele inactivation studies addressing the roles of several of these genes during low-temperature growth are now under way. These studies will provide further insight into the basic physiology of bacterial cold adaptation responses and should guide the development of novel strategies to control L. monocytogenes growth at refrigeration temperatures.
Acknowledgments
This work was supported by award 99-35201-8607 from the National Research Initiative competitive grants program of the U.S. Department of Agriculture and a grant from the Illinois Council on Food and Agriculture Research.
REFERENCES
- 1.Aguilar, P. S., A. M. Hernandez-Arriaga, L. E. Cybulski, A. C. Erazo, and D. Mendoza. 2001. Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 20:1681-1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Annous, B. A., L. A. Becker, D. O. Bayles, D. P. Labeda, and B. J. Wilkinson. 1997. Critical role of anteiso-C15:0 fatty acid in the growth of Listeria monocytogenes at low temperatures. Appl. Environ. Microbiol. 63:3887-3894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bae, W., B. Xia, M. Inouye, and K. Severinov. 2000. Escherichia coli CspA-family RNA chaperones are transcription antiterminators. Proc. Natl. Acad. Sci. USA 97:7784-7789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayles, D. O., B. A. Annous, and B. J. Wilkinson. 1996. Cold stress proteins induced in Listeria monocytogenes in response to temperature down shock and growth at low temperatures. Appl. Environ. Microbiol. 62:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayles, D. O., and B. J. Wilkinson. 2000. Osmoprotectants and cryoprotectants for Listeria monocytogenes. Lett. Appl. Microbiol. 30:23-27. [DOI] [PubMed] [Google Scholar]
- 6.Becker, L. A., S. N. Evans, R. W. Hutkins, and A. K. Benson. 2000. Role of σB in adaptation of Listeria monocytogenes to growth at low temperature. J. Bacteriol. 182:7083-7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry, E. D., and P. M. Foegeding. 1997. Cold temperature adaptation and growth of microorganisms. J. Food Prot. 60:1583-1594. [DOI] [PubMed] [Google Scholar]
- 8.Borezee, E., E. Pellegrini, and P. Berche. 2000. OppA of Listeria monocytogenes, an oligopeptide-binding protein required for bacterial growth at low temperature and involved in intracellular survival. Infect. Immun. 68:7069-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bozzi, M., G. Mignogna, S. Stefanini, D. Barra, C. Longhi, P. Valenti, and E. Chiancone. 1997. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Listeria innocua. J. Biol. Chem. 272:3259-3265. [DOI] [PubMed] [Google Scholar]
- 10.Brehm, K., M. T. Ripio, J. Kreft, and J. A. Vazquez-Boland. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by beta-glucosides. J. Bacteriol. 181:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck, M., M.-T. Gallegos, D. J. Studholme, Y. Guo, and J. D. Gralla. 2000. The bacterial enhancer-dependent σ54 transcription factor. J. Bacteriol. 182:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czuprynski, C. J., J. F. Brown, and J. T. Roll. 1989. Growth at reduced temperature increases the virulence of Listeria monocytogenes for intravenously but not intragastrically inoculated mice. Microb. Pathog. 7:213-223. [DOI] [PubMed] [Google Scholar]
- 13.Dons, L., O. F. Rasmussen, and J. E. Olsen. 1992. Cloning and characterization of a gene encoding flagellin of Listeria monocytogenes. Mol. Microbiol. 6:2919-2929. [DOI] [PubMed] [Google Scholar]
- 14.Edgcomb, M. R., S. Sirimanne, B. J. Wilkinson, P. Drouin, and R. P. D. Morse II. 2000. Electron paramagnetic resonance studies of the membrane fluidity of the foodborne pathogenic psychrotroph Listeria monocytogenes. Biochim. Biophys. Acta 1463:31-42. [DOI] [PubMed] [Google Scholar]
- 15.Farber, J. M., and P. I. Peterkin. 1991. Listeria monocytogenes, a food-borne pathogen. Microbiol. Rev. 55:476-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Froussard, P. 1993. rPCR: a powerful tool for random amplification of whole RNA sequences. PCR Methods Appl. 2:185-190. [DOI] [PubMed] [Google Scholar]
- 17.Gahan, C. G., J. O'Mahony, and C. Hill. 2001. Characterization of the groESL operon in Listeria monocytogenes: utilization of two reporter systems (gfp and hly) for evaluating in vivo expression. Infect. Immun. 69:3924-3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaillot, O., E. Pellegrini, S. Bregenholt, S. Nair, and P. Berche. 2000. The ClpP serine protease is essential for the intracellular parasitism and virulence of Listeria monocytogenes. Mol. Microbiol. 35:1286-1294. [DOI] [PubMed] [Google Scholar]
- 19.Gerth, U., E. Kruger, I. Derre, T. Msadek, and M. Hecker. 1998. Stress induction of the Bacillus subtilis clpP gene encoding a homologue of the proteolytic component of the Clp protease and the involvement of ClpP and ClpX in stress tolerance. Mol. Microbiol. 28:787-802. [DOI] [PubMed] [Google Scholar]
- 20.Gilot, P., P. Andre, and J. Content. 1999. Listeria monocytogenes possesses adhesins for fibronectin. Infect. Immun. 67:6698-6701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilot, P., Y. Jossin, and J. Content. 2000. Cloning, sequencing and characterisation of a Listeria monocytogenes gene encoding a fibronectin-binding protein. J. Med. Microbiol. 49:887-896. [DOI] [PubMed] [Google Scholar]
- 22.Glaser, P., L. Frangeul, C. Buchrieser, C. Rusniok, A. Amend, F. Baquero, P. Berche, H. Bloecker, P. Brandt, T. Chakraborty, A. Charbit, F. Chetouani, E. Couve, A. de Daruvar, P. Dehoux, E. Domann, G. Dominguez-Bernal, E. Duchaud, L. Durant, O. Dussurget, K. D. Entian, H. Fsihi, F. G. Portillo, P. Garrido, L. Gautier, W. Goebel, N. Gomez-Lopez, T. Hain, J. Hauf, D. Jackson, L. M. Jones, U. Kaerst, J. Kreft, M. Kuhn, F. Kunst, G. Kurapkat, E. Madueno, A. Maitournam, J. M. Vicente, E. Ng, H. Nedjari, G. Nordsiek, S. Novella, B. de Pablos, J. C. Perez-Diaz, R. Purcell, B. Remmel, M. Rose, T. Schlueter, N. Simoes, A. Tierrez, J. A. Vazquez-Boland, H. Voss, J. Wehland, and P. Cossart. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 23.Gottesman, S., and M. R. Maurizi. 1992. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol. Rev. 56:592-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grundy, F. J., and T. M. Henkin. 1993. tRNA as a positive regulator of transcription antitermination in Bacillus subtilis. Cell 74:475-482. [DOI] [PubMed] [Google Scholar]
- 26.Hartke, A., J. Frere, P. Boutibonnes, and Y. Auffray. 1997. Differential induction of the chaperonin GroEL and the co-chaperonin GroES by heat, acid, and UV-irradiation in Lactococcus lactis subsp. lactis. Curr. Microbiol. 34:23-26. [DOI] [PubMed] [Google Scholar]
- 27.Hebraud, M., and J. Guzzo. 2000. The main cold shock protein of Listeria monocytogenes belongs to the family of ferritin-like proteins. FEMS Microbiol. Lett. 190:29-34. [DOI] [PubMed] [Google Scholar]
- 28.Hendrick, J. P., and F. U. Hartl. 1993. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 62:349-384. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann, B., and J. L. Ingraham. 1970. A cold sensitive mutant of Salmonella typhimurium which requires tryptophan for growth at 20°C. Biochim. Biophys. Acta 201:970-972. [DOI] [PubMed] [Google Scholar]
- 30.Holmgren, A. 1985. Thioredoxin. Annu. Rev. Biochem. 54:237-271. [DOI] [PubMed] [Google Scholar]
- 31.Houman, F., M. R. Diaz-Torres, and A. Wright. 1990. Transcriptional antitermination in the bgl operon of E. coli is modulated by a specific RNA binding protein. Cell 62:1153-1163. [DOI] [PubMed] [Google Scholar]
- 32.Jagannathan, A., C. Constantinidou, and C. W. Penn. 2001. Roles of rpoN, fliA, and flgR in expression of flagella in Campylobacter jejuni. J. Bacteriol. 183:2937-2942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones, S. L., P. Drouin, B. J. Wilkinson, and P. D. Morse II. 2002.. Correlation of long-range membrane order with temperature-dependent growth characteristics of parent and a cold-sensitive, branched-chain-fatty acid deficient mutant of Listeria monocytogenes. Arch. Microbiol. 177:217-222. [DOI] [PubMed] [Google Scholar]
- 34.Junttila, J. R., S. I. Niemela, and J. Hirn. 1988. Minimum growth temperatures of Listeria monocytogenes and non-haemolytic Listeria. J. Appl. Bacteriol. 65:321-327. [DOI] [PubMed] [Google Scholar]
- 35.Kathariou, S., R. Kanenaka, R. D. Allen, A. K. Fok, and C. Mizumoto. 1995. Repression of motility and flagellin production at 37°C is stronger in Listeria monocytogenes than in the nonpathogenic species Listeria innocua. Can. J. Microbiol. 41:572-577. [DOI] [PubMed] [Google Scholar]
- 36.Kawamura, D., I. Yamashita, O. Nimi, and A. Toh-e. 1994. Cloning and nucleotide sequence of a gene conferring ability to grow at low temperature on Saccharomyces cerevisiae tryptophan autotrophs. J. Ferment. Bioeng. 77:1-9. [Google Scholar]
- 37.Ko, R., L. T. Smith, and G. M. Smith. 1994. Glycine betaine confers enhanced osmotolerance and cryotolerance on Listeria monocytogenes. J. Bacteriol. 176:426-431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kreft, J., and J. A. Vazquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 39.Labarre, C., J. Guzzo, J. F. Cavin, and C. Divies. 1996. Cloning and characterization of the genes encoding the malolactic enzyme and the malate permease of Leuconostoc oenos. Appl. Environ. Microbiol. 62:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leifson, E., and M. I. Palen. 1955. Variations and spontaneous mutations in the genus Listeria in respect to flagellation and motility. J. Bacteriol. 70:233-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ligozzi, M., F. Pittaluga, and R. Fontana. 1993. Identification of a genetic element (psr) which negatively controls expression of Enterococcus hirae penicillin-binding protein 5. J. Bacteriol. 175:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mahan, M. J., J. M. Slauch, and J. J. Mekalanos. 1993. Selection of bacterial virulence genes that are specifically induced in host tissues. Science 259:686-688. [DOI] [PubMed] [Google Scholar]
- 43.Massidda, O., R. Kariyama, L. Daneo-Moore, and G. D. Shockman. 1996. Evidence that the penicillin-binding protein 5 synthesis repressor (psr) of Enterococcus hirae is also involved in the regulation of cell wall composition and other cell wall-related properties. J. Bacteriol. 178:5272-5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Massidda, O., O. Dardenne, M. B. Whalen, W. Zorzi, J. Coyette, G. D. Shockman, and L. Daneo-Moore. 1998. The PBP 5 synthesis repressor (psr) gene of Enterococcus hirae ATCC 9790 is substantially longer than previously reported. FEMS Microbiol. Lett. 166:355-360. [DOI] [PubMed] [Google Scholar]
- 45.Maurizi, M. R., W. P. Clark, Y. Katayama, S. Rudikoff, J. Pumpherey, B. Bowers, and S. Gottesman. 1990. Sequence and structure of ClpP, the proteolytic component of the ATP-dependent Clp protease of Escherichia coli. J. Biol. Chem. 265:12536-12545. [PubMed] [Google Scholar]
- 46.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Merrick, M. J. 1993. In a class of its own-the RNA polymerase sigma factor σ54 (σN). Mol. Microbiol. 10:903-909. [DOI] [PubMed] [Google Scholar]
- 48.O'Donovan, G. A., and J. L. Ingraham. 1965. Cold sensitive mutants of Escherichia coli resulting from increased feedback inhibition. Proc. Natl. Acad. Sci. USA 54:451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Panoff, J. M., B. Thammavongs, M. Gueguen, and P. Boutibonnes. 1998. Cold stress responses in mesophilic bacteria. Cryobiology 36:75-83. [DOI] [PubMed] [Google Scholar]
- 50.Park, S. F., and R. G. Kroll. 1993. Expression of listeriolysin and phosphatidylinositol-specific phospholipase C is repressed by the plant-derived molecule cellobiose in Listeria monocytogenes. Mol. Microbiol. 8:653-661. [DOI] [PubMed] [Google Scholar]
- 51.Peel, M., W. Donachie, and A. Shaw. 1988. Temperature-dependent expression of flagella of Listeria monocytogenes studied by electron microscopy, SDS-PAGE and Western blotting. J. Gen. Microbiol. 134:2171-2178. [DOI] [PubMed] [Google Scholar]
- 52.Phadtare, S., K. Yamanaka, and M. Inouye. 2000. The cold shock response, p. 33-45. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 53.Phan-Thanh, L., and T. Gormon. 1995. Analysis of heat and cold shock proteins in Listeria by two-dimensional electrophoresis. Electrophoresis 16:444-450. [DOI] [PubMed] [Google Scholar]
- 54.Porankiewicz, J., and A. K. Clarke. 1997. Induction of the heat shock protein ClpB affects cold acclimation in the cyanobacterium Synechococcus sp. strain PCC 7942. J. Bacteriol. 179:5111-5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Premaratne, R. J., W. J. Lin, and E. A. Johnson. 1991. Development of an improved chemically defined minimal medium for Listeria monocytogenes. Appl. Environ. Microbiol. 57:3046-3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Prinz, W. A., F. Aslund, A. Holmgren, and J. Beckwith. 1997. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in the Escherichia coli cytoplasm. J. Biol. Chem. 272:15661-15667. [DOI] [PubMed] [Google Scholar]
- 57.Raffelsbauer, D., A. Bubert, F. Engelbrecht, J. Scheinpflug, A. Simm, J. Hess, S. H. Kaufmann, and W. Goebel. 1998. The gene cluster inlC2DE of Listeria monocytogenes contains additional new internalin genes and is important for virulence in mice. Mol. Gen. Genet. 260:144-158. [DOI] [PubMed] [Google Scholar]
- 58.Rice, L. B., L. L. Carias, R. Hutton-Thomas, F. Sifaoui, L. Gutmann, and S. D. Rudin. 2001. Penicillin-binding protein 5 and expression of ampicillin resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 45:1480-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robichon, D., E. Gouin, M. Debarbouille, P. Cossart, Y. Cenatiempo, and Y. Hechard. 1997. The rpoN (σ54) gene from Listeria monocytogenes is involved in resistance to mesentericin Y105, an antibacterial peptide from Leuconostoc mesenteroides. J. Bacteriol. 179:7591-7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roof, D. M., and J. R. Roth. 1989. Functions required for vitamin B12-dependent ethanolamine utilization in Salmonella typhimurium. J. Bacteriol. 171:3316-3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salotra, P., D. K. Singh, K. P. Seal, N. Krishna, H. Jaffe, and R. Bhatnagar. 1995. Expression of DnaK and GroEL homologs in Leuconostoc mesenteroides in response to heat shock, cold shock or chemical stress. FEMS Microbiol. Lett. 131:57-62. [DOI] [PubMed] [Google Scholar]
- 62.Singh, V. K., R. K. Jayaswal, and B. J. Wilkinson. 2001. Cell wall-active antibiotic induced proteins of Staphylococcus aureus identified using a proteomic approach. FEMS Microbiol. Lett. 199:79-84. [DOI] [PubMed] [Google Scholar]
- 63.Stephens, J. C., I. S. Roberts, D. Jones, and P. W. Andrew. 1991. Effect of growth temperature on virulence of strains of Listeria monocytogenes in the mouse: evidence for a dose dependence. J. Appl. Bacteriol. 70:239-244. [DOI] [PubMed] [Google Scholar]
- 64.Suzuki, I., Y. Kanesaki, K. Mikami, M. Kanehisa, and N. Murata. 2001. Cold-regulated genes under control of the cold sensor Hik33 in Synechocystis. Mol. Microbiol. 40:235-244. [DOI] [PubMed] [Google Scholar]
- 65.Thieringer, H. A., P. G. Jones, and M. Inouye. 1998. Cold shock and adaptation. Bioessays 20:49-57. [DOI] [PubMed] [Google Scholar]
- 66.Totten, P. A., J. C. Lara, and S. Lory. 1990. The rpoN gene product of Pseudomonas aeruginosa is required for expression of diverse genes, including the flagellin gene. J. Bacteriol. 172:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valdivia, R. H., and S. Falkow. 1996. Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22:367-378. [DOI] [PubMed] [Google Scholar]
- 68.Verheul, A., A. Hagting, M. R. Amezaga, I. R. Booth, F. M. Rombouts, and T. Abee. 1995. A di- and tripeptide transport system can supply Listeria monocytogenes Scott A with amino acids essential for growth. Appl. Environ. Microbiol. 61:226-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wai, S. N., K. Nakayama, K. Umene, T. Moriya, and K. Amako. 1996. Construction of a ferritin-deficient mutant of Campylobacter jejuni: contribution of ferritin to iron storage and protection against oxidative stress. Mol. Microbiol. 20:1127-1134. [DOI] [PubMed] [Google Scholar]
- 70.Walker, S. J., P. Archer, and J. G. Banks. 1990. Growth of Listeria monocytogenes at refrigeration temperatures. J. Appl. Bacteriol. 68:157-162. [DOI] [PubMed] [Google Scholar]
- 71.Wouters, J. A., F. M. Rombouts, O. P. Kuipers, W. M. De Vos, and T. Abee. 2000. The role of cold-shock proteins in low-temperature adaptation of food-related bacteria. Syst. Appl. Microbiol. 23:165-173. [DOI] [PubMed] [Google Scholar]
- 72.Yamanaka, K., and M. Inouye. 2001. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 183:2808-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yanofsky, C. 2001. Advancing our knowledge in biochemistry, genetics, and microbiology through studies on tryptophan metabolism. Annu. Rev. Biochem. 70:1-37. [DOI] [PubMed] [Google Scholar]