Abstract
A multiplex PCR was developed to simultaneously detect Naegleria fowleri and other Naegleria species in the environment. Multiplex PCR was also capable of identifying N. fowleri isolates with internal transcribed spacers of different sizes. In addition, restriction fragment length polymorphism analysis of the PCR product distinguished the main thermophilic Naegleria species from the sampling sites.
The free-living amoebae belonging to the genus Naegleria occur worldwide and inhabit soil and aquatic environments. One member of this genus, Naegleria fowleri, causes primary amoebic meningoencephalitis (PAM) in humans. Although PAM is rare (more than 190 cases reported worldwide [15]), this central nervous system disease is lethal within 1 week in most cases (3). The majority of the fatal infections due to N. fowleri occur in young people exposed to warm water in ponds, swimming pools, and lakes. N. fowleri is thermophilic and generally found in natural and artificially heated water, in particular in the cooling ponds of power plants, in which this species can proliferate intensively (4, 11, 12, 27, 28). Two other thermophilic Naegleria species are currently found in these sites: Naegleria lovaniensis, which is harmless, and Naegleria australiensis, which is pathogenic in mice. The thermophilic species Naegleria italica was also reported to be pathogenic in mice but is rarely encountered (7). These two species could be potentially dangerous for humans.
In a preventive measure, water monitoring was performed regularly in the nuclear power plants of France in order to check for the proliferation of N. fowleri. The identification of Naegleria species has been carried out by an isoenzymatic procedure which allowed simultaneous detection of the three main thermophilic Naegleria species. Other immunological and molecular techniques are now available to specifically detect N. fowleri in environmental sites (14, 25, 26). Recently, ribosomal internal transcribed spacers (ITS) were reported to be useful markers for Naegleria (10, 20), and species-specific primers were defined for N. fowleri (20). In addition, variations in the sequence and size of the ITS were found within this human-pathogenic species, with five different variants which were mostly detected in France. However, the geographic dispersal and the prevalence of these variants were not well established, since too few samples were examined at different sites.
In this study, we applied a simplified ITS PCR procedure to the environmental Naegleria isolates for several reasons: (i) to rapidly and easily analyze a large number of isolates, (ii) to detect the presence of N. fowleri in the potential sites and to further explore the genetic diversity of this species, and (iii) to identify the other thermophilic Naegleria isolates at the sites by using PCR restriction fragment length polymorphism (RFLP) analysis.
Amoeba isolation from the environment.
Five hundred environmental strains were isolated from water of the cooling ponds and downstream of five different nuclear power plants (Table 1). The other free-living amoebae, Nuclearia spp., Acanthamoeba spp., Hartmannella spp., Vahlkampfia spp., and Willaertia spp., were isolated from various sites of France. To isolate and identify the amoebae from the sites, large volumes of water samples (10 100-ml replicates and 30 10-ml replicates) were filtered on 1-μm-pore-size cellulose nitrate membranes (Whatman). The membranes were cut in half and were inverted on nonnutrient agar plates overlaid with a thin pellicle of Escherichia coli as a food source. After 3 to 4 days of incubation at 44°C, the plates were observed under an inverted microscope (×200) to identify the Naegleria species by their morphological characters, i.e., double-walled cysts accompanied by a variable percentage of degenerated empty cysts and trophozoites with eruptive and lobose pseudopodia (17). The identification of the strains as members of the genus Naegleria was confirmed by an enflagellation test. When a Naegleria isolate was identified, two agar squares were cut from the corresponding plate for species identification with biochemical and molecular methods. For the biochemical procedure, subcultures were carried out to prevent fungal overgrowth and to obtain the isolates in sufficient quantity. The environmental Naegleria strains were identified by detection of one of two diagnostic enzymes, malic enzyme or l-threonine dehydrogenase, after isoelectric focusing as previously described (21, 22) with the following simplifications. Each Naegleria strain was harvested by scraping the advancing front at the surface of an agar plate with a sterile loop and was suspended in 22 μl of NaCl (0.017 M)-saccharose (0.22 M) in an Eppendorf microtube. This amoebic suspension was then lysed by a single cycle of freeze-thawing, and after centrifugation, the supernatant (8 to 9 μl) was directly subjected to isoelectric focusing. For the molecular procedure, the agar square was deposited in an Eppendorf tube containing 150 to 200 μl of NaCl (0.017 M)-saccharose (0.22 M) solution. The cell samples were vortexed and transferred into new Eppendorf tubes. The cells were centrifuged and then recovered in a lysis solution containing 2 μl of proteinase K (10 mg/ml) in 28 μl of distilled water. The solution was incubated at 55°C for 20 min and then heated at 95°C for 5 min. Two to three microliters of the solution was used for each PCR amplification.
TABLE 1.
Number of Naegleria isolates for each nuclear power plant examined in this study by ITS-PCR
| Site | Date of isolation | No. of isolates examined
|
|
|---|---|---|---|
| Naegleria | N. fowleri | ||
| Dampierre | June 1998 | 135 | 90 |
| October 1998 | 45 | 3 | |
| November 1999 | 20 | 10 | |
| Golfech | November 1998 | 65 | 2 |
| May 1999 | 130 | 117 | |
| October 1999 | 10 | 0 | |
| Bugey | June 1999 | 20 | 4 |
| June 2001 | 20 | 2 | |
| Chinon | October 1999 | 5 | 1 |
| Nogent | June 2000 | 40 | 0 |
| June 2001 | 10 | 4 | |
PCR amplification.
The ITS region was amplified by PCR with oligonucleotide primers (5′GAACCTGCGTAGGGATCATTT and 5′TTTCTTTTCCTCCCCTTATTA), or the species-specific primers Fw1 and Fw2 (5′GTGAAAACCTTTTTTCCATTTACA and 5′AAATAAAAGATTGACCATTTGAAA), designed to specifically detect N. fowleri (20). Amplification reactions were performed in volumes of 15 μl containing 1× Taq DNA polymerase buffer, a 0.2 mM concentration of each deoxynucleoside triphosphate, a 0.6 μM concentration of each primer, 1.5 mM MgCl2, and 1 to 2 U of Taq DNA polymerase (Appligene, Illkirch, France). For multiplex PCR with the conserved primers and the species-specific primers, the amplification reactions were performed in 15 μl containing 1× Taq DNA polymerase buffer, a 0.2 mM concentration of each deoxynucleoside triphosphate, a 0.4 μM concentration of each conserved primer, a 0.8 μM concentration of each species-specific primer,1.5 mM MgCl, and 1 to 2 U of Taq DNA polymerase. The hot-start PCR method (5) was carried out; more precisely, Taq DNA polymerase was added after a pre-PCR heat cycle.
Amplifications were performed in a GeneAmp model 2400 thermal cycler (Perkin-Elmer-Cetus, Norwalk, Conn.) and programmed as follows: 1 pre-PCR heat cycle at 94°C for 5 min; 35 cycles at 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s; a final cycle at 72°C for 5 min. Eight microliters of each amplification product were analyzed by electrophoresis in 1.2 to 1.4% agarose gels, with 1× TBE buffer (50 mM Tris-HCl, 50 mM boric acid, and 1 mM EDTA). Amplified DNA bands were visualized after staining of the gel with bromide ethidium.
For RFLP analysis, the conserved PCR product (15 μl) was digested with MseI or NlaIV (BioLabs) in a reaction mixture containing 5 μl of the appropriate buffer, 100 μg of bovine serum albumin per ml, 10 U of endonuclease, and sterile water to obtain a final volume of 50 μl. The solution was incubated for 2 h at 37°C. Ten to fifteen microliters of each digestion mixture was separated in a 3% NuSieve gel with 1× TBE buffer and visualized by staining with bromide ethidium.
Molecular identification of N. fowleri from the environment.
Because of the great number of isolates, it was necessary to use a rapid and simple DNA extraction method which must be consistent with the PCR analysis. Preliminary experiments with approximately 100 to 200 N. fowleri cells from pure culture plates showed a positive signal with all the N. fowleri strains. The trophozoites and cysts were tested separately and gave identical results, as already shown (14). The results also showed that the hot-start PCR was needed to obtain reproducible results. In order to test the sensitivity, the amoebae cells harvested from the culture plates by scraping and resuspended in NaCl (0.017 M)-saccharose (0.22 M) and their concentration was determined by counting with a Thoma hemacytometer. The result showed that DNA from as few as five trophozoites or cysts amplified successfully. The PCR fragment was visible with fewer than five cells, but the results were not reproducible.
Multiplex PCR.
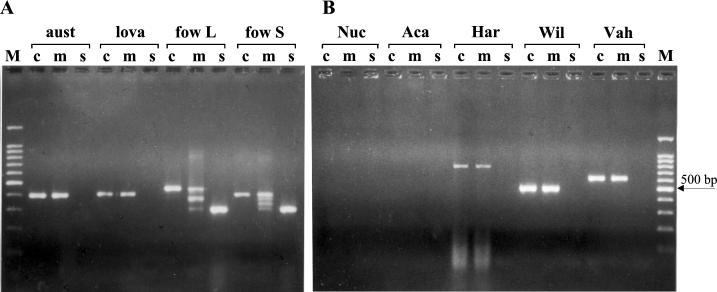
When the conserved and the species-specific primers were combined, four amplified fragments were expected with N. fowleri. The higher and the lower fragments corresponded to the elongation of the conserved primers and the specific primers, respectively; the two intermediate fragments were due to the combination of specific and conserved primers. When the same concentration was used for the conserved and the species-specific primers, the intensity of the smallest stained bands in the gel was very often lower than that of the other ones. In order to obtain equal intensities of amplification products, the species-specific-primer concentrations had to be higher than those of the nonspecific primers. Interestingly, the multiplex PCR could detect the different variants of N. fowleri due to the ITS1 length polymorphism. Particularly, the short-ITS variant, with a 42-bp ITS1, displayed the four expected bands, whereas the long-ITS variant, with an ITS1 length of 86 bp, produced only three bands, due to the comigration of the two intermediate fragments of 388 and 376 bp in the gel (Fig. 1A). As already shown (20), no PCR fragment was observed with the species-specific PCR with the other thermophilic Naegleria strains. The multiplex profile was identical to that obtained with the conserved PCR (Fig. 1A).
FIG. 1.
ITS PCR products generated from the DNA of various free-living amoebae using conserved primers only (c), conserved and species-specific primers (m), and species-specific primers only (s). (A) PCR results for three thermophilic Naegleria species. The N. fowleri long-ITS variant (fow L) and short-ITS variant (fow S) gave single bands of 453 and 409 bp, respectively, with the conserved primers and single bands of 311 bp with only the species-specific primers. With the multiplex PCR, the N. fowleri long-ITS variant displayed three bands of 453, 388 + 376, and 311 bp, whereas the N. fowleri short-ITS variant displayed four bands of 409, 376, 344, and 311 bp. N. australiensis (aust) and N. lovaniensis (lova) gave single fragments of approximately 400 bp with the conserved and the multiplex primers. (B) No PCR fragment was observed with Nuclearia spp. (Nuc) or Acanthamoeba spp. (Aca). Conserved and multiplex PCR gave single fragments of approximately 800, 600, and 500 bp with Hartmannella spp. (Har), Vahlkampfia spp. (Vah), and with Willaertia spp. (Wil), respectively. M, molecular markers (100-bp DNA ladder with a prominent 500-bp fragment).
The presence of fungal contaminants together with N. fowleri did not affect the species specificity of the test. To further evaluate the specificity of the N. fowleri PCR, DNA from Acanthamoeba, Hartmannella, Nuclearia, Vahlkampfia, and Willaertia isolates were tested. Most of them were often found to contaminate the culture plates during monitoring. No species-specific PCR product was observed for all the strains tested (Fig. 1B). By using either the conserved primers or the multiple primers, a single fragment was visible with Hartmannella, Vahlkampfia, and Willaertia isolates and the fragment size was different for each. This indicates that multiplex PCR was also able to detect the most frequent competitors associated with thermotolerant Naegleria species. This result is also interesting in that this method could estimate the other amoebae in the sample sites. Some of them are known to harbor pathogenic bacteria (2, 16, 29). However, to extend the field of investigation, it is necessary to refine the conserved primers, which are very useful for the typing of Naegleria but are inappropriate for distantly related amoebae, particularly Acanthamoeba.
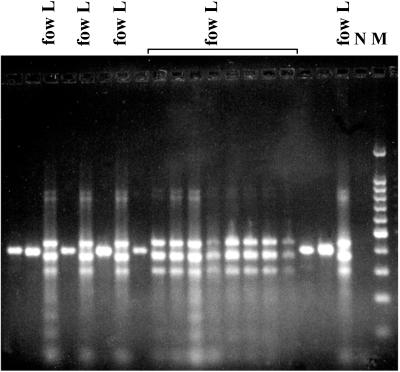
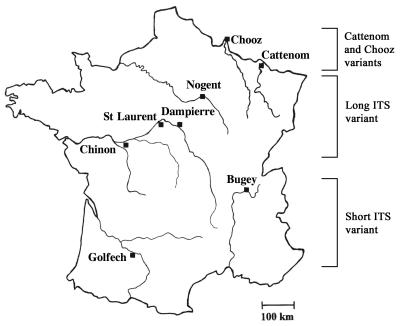
In this study, the nonspecific and the species-specific PCRs were performed separately to identify the pathogenic species from the environment. In a second step, the multiplex PCR was performed with 50% of the isolates, and the results fit perfectly with the two independent PCRs. The molecular results were in accordance with the biochemical ones (more than 99%). All the N. fowleri strains isolated from the Bugey and Golfech sites corresponded to the short-ITS variant and corresponded to the widespread variant previously described by the randomly amplified polymorphic DNA (RAPD) analysis (18, 19). All the isolates from the other sites had the long ITS fragments and exhibited the characteristic profile generated by the multiplex PCR (Fig. 2). Therefore, our results confirmed the predicted geographical distribution of the size variants in France and unveiled some other interesting points. The ITS analysis showed the persistence of the same variants among the sites: the short-ITS variant was always found at Golfech and Bugey, whereas the long-ITS variant was found at the other sites (Fig. 3). Furthermore, only one variant was observed per site, indicating that the N. fowleri variants tend to occupy separate areas. Two cases, however, seem to indicate that certain sites might be colonized by different variants. Among six isolates collected at a Tulsa, Okla., site (13), one isolate was a short-ITS variant and the remaining ones were long-ITS variants (unpublished data). Similarly, two different size variants were found at the Cattenom, France, site (10). It should be noted that particular variants closely related to the South Pacific variant (19, 23) were found in Cattenom and Chooz (eastern France). Until now, RAPD and ITS analyses had not detected these variants elsewhere in France, suggesting that they were restricted to the eastern sites.
FIG. 2.
Multiplex PCR analysis for the simultaneous detection of the human-pathogenic N. fowleri and the non-human-pathogenic isolates from the Dampierre site. Two banding patterns are observed in the gel. The pattern with three bands indicated the presence of the N. fowleri long-ITS variant (fow L). The single-band pattern corresponded to all other thermophilic Naegleria strains. N, negative control amplification in the absence of DNA; M, 100-bp DNA ladder with a prominent 500-bp fragment.
FIG. 3.
Geographical distribution of N. fowleri variants in France. The short-ITS variant was found only at Bugey and Golfech, and the long-ITS variant was found at the other sites analyzed. St Laurent, Cattenom, and Chooz were not examined in this study. Previous RAPD and ITS analyses showed that Cattenom and Chooz were colonized by variants closely related to the South Pacific variant. In addition, Cattenom was the only French site at which different variants were found.
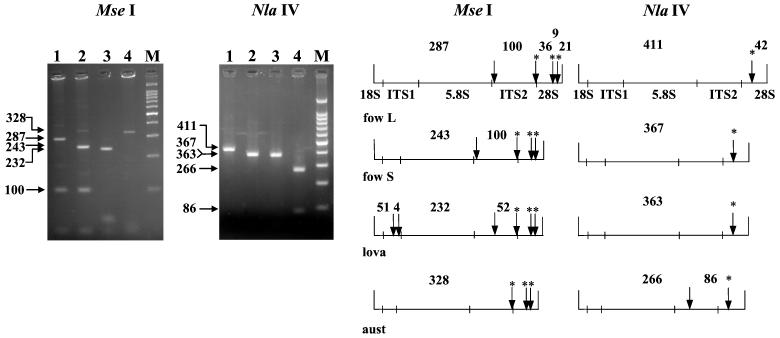
The ITS-RFLP analysis with NlaIV and MseI allowed the identification of the main thermophilic Naegleria species that were collected at the sites (Fig. 4). Digestion with MseI gave major fragments of 287, 243, 232, and 328 bp for the N. fowleri long-ITS variant, the N. fowleri short-ITS variant, N. lovaniensis, and N. australiensis, respectively. An additional specific fragment of 100 bp was visible for N. fowleri and could be used as an additional marker to further identify this species. Digestion with NlaIV allowed us to identify the reference N. australiensis strains LSR 34, pp397, LSR 49, and By33, producing 266- and 86-bp fragments, since one restriction site existed in only the ITS2 region of this species. In addition, some Naegleria gruberi strains thought to be closely related to N. australiensis showed no NlaIV site in the ITS2 region. We also examined some Naegleria isolates that were previously identified as N. australiensis by isoenzymatic typing during the environmental survey. All the isolates examined from various sites (Bugey, Dampierre, Nogent, and Golfech) displayed the characteristic NlaIV profile. However, only a few strains were examined, and further analyses are needed to validate this diagnostic test. The few of N. australiensis and N. lovaniensis samples analyzed could also explain the absence of polymorphism in these two species. Indeed, biochemical and molecular analyses (1, 6, 24) suggest that N. lovaniensis and N. australiensis are more heterogeneous than N. fowleri. RFLP analysis of the ribosomal DNA small subunit distinguished between different Naegleria species (9). However, some of them, such as N. italica, had a group I intron in their small-subunit rRNA (8) and hampered the taxonomic analyses. One particular HinfI restriction site, however, was present only in N. australiensis. This site combined with the NlaIV site could help to further examine N. australiensis in the environment and particularly to identify the undefined environmental isolates thought to be very close to N. australiensis. Restriction analyses with NlaIV and MseI are very helpful for the exploration of the natural populations of N. lovaniensis and N. australiensis. If these restriction sites are found to be species specific, they could be useful for molecular identification. Otherwise, they could detect variability within these two species.
FIG. 4.
MseI and NlaIV digestion patterns of the ITS PCR products for the long-ITS variant of N. fowleri (lane 1), the short-ITS variant of N. fowleri (lane 2), N. lovaniensis (lane 3), and N. australiensis (lane 4). M, 100-bp DNA ladder. On the right are the restriction maps of MseI and NlaIV for the long and the short N. fowleri variants (fow L and fow S), N. lovaniensis (lova), and N. australiensis (aust). The conserved sites are marked with asterisks.
The present work is a first step for fingerprinting of the genus Naegleria and its close relatives. It is now possible to explore the natural habitats and determine the prevalence of the variants of N. fowleri among the sites. Therefore, ITS may help to better understand the significance of the numerous variants of N. fowleri found. Analyses are in progress in order to examine whether similar levels of variability exist elsewhere and to explore the variability in the two other thermophilic species, N. australiensis and N. lovaniensis. From a clinical point of view, the diagnosis of PAM is based on microscopic examination of the cerebrospinal fluid for mobile amoebae. This examination is laborious and can be misleading. The clinical application of ITS PCR for the diagnosis of PAM might give rise to a simple, rapid, and more sensitive clinical test.
Acknowledgments
We thank Leslie Garcia, Rachel JeanGerard, and Aurélien Meyet for the biochemical analyses.
REFERENCES
- 1.Adams, M., R. H. Andrews, B. Robinson, P. Christy, P. R. Baverstok, P. J. Dobson, and S. J. Blackler. 1989. A genetic approach to species criteria in the amoeba genus Naegleria using allozyme electrophoresis. Int. J. Parasitol. 19:823-834. [DOI] [PubMed] [Google Scholar]
- 2.Brown, M. R., and J. Barker. 1999. Unexplored reservoirs of pathogenic bacteria: protozoa and biofilms. Trends Microbiol. 7:46-50. [DOI] [PubMed] [Google Scholar]
- 3.Carter, R. F. 1970. Description of a Naegleria sp. isolated from two cases of primary amoebic meningo-encephalitis, and of the experimental pathological changes induced by it. J. Pathol. 100:217-244. [DOI] [PubMed] [Google Scholar]
- 4.Cerva, L., W. Kasprzak, and T. Mazur. 1982. Naegleria fowleri in cooling waters of power plants. J. Hyg. Epidemiol. Microbiol. Immunol. 26:152-161. [PubMed] [Google Scholar]
- 5.Chou, Q., M. Russell, D. E. Birch, J. Raymond, and W. Bloch. 1992. Prevention of pre-PCR mis-priming and primer dimerization improves low-copy-number amplifications. Nucleic Acids Res. 20:1717-1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark, C. G., G. A. M. Cross, and J. F. De Jonckheere. 1989. Evaluation of evolutionary divergence in the genus Naegleria by analysis of ribosomal DNA plasmid restriction patterns. Mol. Biochem. Parasitol. 34:281-296. [DOI] [PubMed] [Google Scholar]
- 7.De Jonckheere, J. F., P. Pernin, M. Scaglia, and R. Michel. 1984. A comparative study of 14 strains of Naegleria australiensis demonstrates the existence of a highly virulent subspecies: N. australiensis italica n. spp. J. Protozool. 31:324-331. [DOI] [PubMed] [Google Scholar]
- 8.De Jonckheere, J. F. 1993. A group I intron in the SSUrDNA of some Naegleria spp. demonstrated by polymerase chain reaction amplification. J. Eukaryot. Microbiol. 40:179-187. [DOI] [PubMed] [Google Scholar]
- 9.De Jonckheere, J. F. 1994. Riboprinting of Naegleria spp.: small-subunit versus large-subunit rDNA. Parasitol. Res. 80:230-234. [DOI] [PubMed] [Google Scholar]
- 10.De Jonckheere, J. F. 1998. Sequence variation in the ribosomal internal transcribed spacers, including the 5.8S rDNA, of Naegleria spp. Protist 149:221-228. [DOI] [PubMed] [Google Scholar]
- 11.Dive, D. G., H. Leclerc, J. De Jonckheere, and J. M. Delattre. 1981. Isolation of Naegleria fowleri from the cooling pond of an electric power plant in France. Ann. Microbiol. (Paris) 132:97-105. [PubMed] [Google Scholar]
- 12.Huizinga, H. W., and G. L. MacLaughlin. 1990. Thermal ecology of Naegleria fowleri from a power plant cooling reservoir. Appl. Environ. Microbiol. 56:2200-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John, D. T., and M. J. Howard. 1995. Seasonal distribution of pathogenic free-living amebae in Oklahoma waters. Parasitol. Res. 81:193-201. [DOI] [PubMed] [Google Scholar]
- 14.Kilvington, S., and J. Beeching. 1995. Development of a PCR for identification of Naegleria fowleri from the environment. Appl. Environ. Microbiol. 61:3764-3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall, M. M., D. Naumovitz, Y. Ortega, and C. R. Sterling. 1997. Waterborne protozoan pathogens. Clin. Microbiol. Rev. 10:67-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moffat, J. F., and L. S. Tompkins. 1992. A quantitative model of intracellular growth of Legionella pneumophila in Acanthamoeba castellanii. Infect. Immun. 60:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Page, F. C. 1967. Taxonomic criteria for limax amoebae, with descriptions of 3 new species of Hartmannella and 3 of Vahlkampfia. J. Protozool. 14:499-521. [DOI] [PubMed] [Google Scholar]
- 18.Pélandakis, M., S. S. Kaundun, J. F. De Jonckheere, and P. Pernin. 1997. DNA diversity among the free-living amoeba Naegleria fowleri detected by the RAPD method. FEMS Microbiol. Lett. 151:31-39. [Google Scholar]
- 19.Pélandakis, M., J. F. De Jonckheere, and P. Pernin. 1998. Genetic variation in the free-living amoeba Naegleria fowleri. Appl. Environ. Microbiol. 64:2977-2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pélandakis, M., S. Serre, and P. Pernin. 2000. Analysis of the 5.8S rRNA gene and the internal transcribed spacers in Naegleria spp. and in N. fowleri. J. Eukaryot. Microbiol. 47:116-121. [DOI] [PubMed] [Google Scholar]
- 21.Pernin, P., M. L. Cariou, and A. Jacquier. 1985. Biochemical identification and phylogenetic relationships in free-living amoebas of the genus Naegleria. J. Protozool. 32:592-603. [DOI] [PubMed] [Google Scholar]
- 22.Pernin, P., and G. Grelaud. 1989. Application of isoenzymatic typing to the identification of nonaxenic strains of Naegleria (Protozoa, Rhizopoda). Parasitol. Res. 75:595-598. [DOI] [PubMed] [Google Scholar]
- 23.Pernin, P., and J. F. De Jonckheere. 1992. Appearance in Europe of Naegleria fowleri displaying the Australian type of restriction-fragment-length-polymorphism. Parasitol. Res. 78:479-481. [DOI] [PubMed] [Google Scholar]
- 24.Pernin, P., A. Ataya, and M. L. Cariou. 1992. Genetic structure of natural populations of the free-living amoeba Naegleria lovaniensis. Evidence for sexual reproduction. Heredity 68:173-181. [DOI] [PubMed] [Google Scholar]
- 25.Sparagano, O., E. Drouet, R. Brebant, E. Manet, G. A. Denoyel, and P. Pernin. 1993. Use of monoclonal antibodies to distinguish pathogenic Naegleria fowleri (cysts, trophozoites, or flagellate forms) from other Naegleria species. J. Clin. Microbiol. 31:2758-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sparagano, O., and A. Revol. 1995. Isolation of the freshwater Naegleria fowleri (Carter, 1970) from a river using a monoclonal antibody and the polymerase chain reaction. Acta Hydrobiologica 37:29-32. [Google Scholar]
- 27.Sykora, J. L., G. Keleti, and A. J. Martinez. 1983. Occurrence and pathogenicity of Naegleria fowleri in artificially heated waters. Appl. Environ. Microbiol. 45:974-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tyndall, R. L., K. S. Ironside, P. L. Metler, E. L. Tan, T. C. Hazen, and C. B. Fliermans. 1989. Effects of thermal additions on the density and distribution of thermophilic amoebae and pathogenic Naegleria fowleri in a newly created cooling lake. Appl. Environ. Microbiol. 55:722-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wadowsky, R. M., T. M. Wilson, N. J. Kapp, A. J. West, J. M. Kuchta, S. J. States, J. N. Dowling, and R. B. Yee. 1991. Multiplication of Legionella spp. in tap water containing Hartmannella vermiformis. Appl. Environ. Microbiol. 57:1950-1955. [DOI] [PMC free article] [PubMed] [Google Scholar]