Abstract
The gastric fluid and feces of three belugas from the Mystic Aquarium were assessed for the presence of Helicobacter spp. Gastric fluid and feces from the two clinically healthy belugas were negative for helicobacter, and endoscopy performed on these animals revealed no lesions. However, a helicobacter isolate and PCR product similar to helicobacter strains previously recovered from dolphins were identified, respectively, from the feces and gastric fluid of a beluga manifesting intermittent inappetence and lethargy. Esophageal and forestomach ulcers were noted on endoscopy. This is the first report of novel Helicobacter spp. being identified from whales.
Belugas are a circumpolar species with an endangered relic population living in the Saint Lawrence River Estuary (17, 18, 23). Three belugas (animals 1, 2, and 3) cohoused at the Mystic Aquarium were assessed for helicobacter infection. Animal 1 manifested clinical signs, including intermittent inappetence and lethargy, and esophageal and forestomach ulcers were observed at endoscopic examination. Animals 2 and 3 showed no clinical signs, nor were esophageal and forestomach ulcers diagnosed endoscopically.
Rectal swabs and gastric fluid were obtained from all three belugas for culture and PCR. A sterile swab (Becton Dickinson and Company, Sparks, Md.) was inserted through the anus to a depth sufficient to coat the swab with feces. The swab was then removed and placed in vials containing 20% glycerol in brucella broth. The gastric fluid was collected by endoscopy (Olympus 210-cm Videoscope) and aliquoted in individual one-dram vials. The endoscope and its channels were rinsed sequentially in dilute chlorhexidine, 70% alcohol, and water between each sampling. A Massachusetts Institute of Technology (MIT) accession number was assigned to each fecal and gastric fluid sample collected: samples from animal 1 were MIT 00-7128 and MIT 00-7129, those from animal 2 were MIT 00-7125 and MIT 00-7126, and those from animal 3 were MIT 00-7131 and MIT 00-7132. These sample numbers are used throughout this work.
The fecal and gastric fluid samples used for microaerobic culture were placed in individual vials with 3 ml of 20% glycerol in brucella broth. The samples were placed on dry ice and then stored at −70°C prior to culture. The media used for culture were Trypticase soy agar with 5% sheep blood and TVP (trimethoprim, vancomycin, and polymyxin) and CVA (cefoperazone, vancomycin, and amphotericin B) antibiotic-impregnated media (Remel Laboratories, Lenexa, Kans.). In addition, selective antibiotic medium (ABM) was prepared with blood agar base (Oxoid; Remel), 5% horse blood (Remel), amphotericin B (50 μg/ml), vancomycin (100 μg/ml), polymyxin B (3.3 μg/ml), bacitracin (200 μg/ml), and nalidixic acid (10.7 μ/ml) (all antibiotics from Sigma Chemical Co., St. Louis, Mo.). Approximately 0.5 g of feces was homogenized in 1 ml of brucella broth (Difco Laboratories, Detroit, Mich.) containing 5% fetal calf serum (Summit Technologies, Fort Collins, Colo.) in a glass tissue grinder. Approximately 100 μl of each sample was applied directly to TVP, CVA, and ABM media. In order to remove larger bacterial contaminants, half of the remaining portion of the sample was filtered through a 0.45-μm-pore-size filter onto a blood agar plate. The plates were incubated at 37°C under microaerobic conditions for 2 to 4 weeks in vented jars containing N2, H2, and CO2 (80:10:10). Biochemical and morphological analyses following a previously described protocol were performed on isolated bacteria (13, 20).
DNA was extracted from the feces and gastric fluid using a modified Mini QIAamp DNA kit (Qiagen Inc., Valencia, Calif.). Helicobacter sp.-specific primers C97 and C05 were used to generate 16S rRNA amplicons of 1,200 bp (Table 1) (8). Approximately 40 ng (10 μl) of the DNA preparation from the feces and from the gastric fluid was used for PCR. The PCR mixture contained 1× Expand High Fidelity PCR System buffer, a 0.5 μM concentration of each of the two primers, a 200 μM concentration of each deoxynucleoside triphosphate, and 200 μg of bovine serum albumin per ml. The samples were heated at 94°C for 4 min, centrifuged briefly, and cooled to 58°C, and 2.6 U of High Fidelity PCR System enzyme mix (Roche Molecular Biochemicals, Indianapolis, Ind.) was added. Amplification conditions were as follows: denaturation at 94°C for 1 min, annealing at 58°C for 2 min, and elongation at 72°C for 3 min. Thirty-five cycles were completed before a final elongation step at 72°C for 8 min. A 15-μl aliquot of the PCR product was electrophoresed through a 1% agarose gel separation matrix prior to ethidium bromide staining and viewing under a UV light.
TABLE 1.
PCR primers used in this study
| Primera | Position | Orientation | PCR specificity | Sequence |
|---|---|---|---|---|
| F24 | 9-27 | Forward | Universal | GAGTTTGATYMTGGCTCAG |
| F25 | 1525-1541 | Reverse | Universal | AAGGAGGTGWTCCARCC |
| C97 | 276-291 | Forward | Helicobacter | GCTATGACGGGTATCC |
| C05 | 1477-1495 | Reverse | Helicobacter | ACTTCACCCCAGTCGCTG |
Primers F24 and F25 were used for PCR of genomic DNA for cycle sequencing. Primers C97 and C05 were used to amplify a 1.2-kb product from fecal samples for cloning.
A TOPO TA cloning kit vector was used for cloning the PCR products (Invitrogen, Carlsbad, Calif.). Four microliters of PCR product was ligated with 1 μl of vector at room temperature for 30 min and then transferred into Top 10 competent cells. Ampicillin plates with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) were used to select positive clones. Plasmid DNA was isolated from Escherichia coli with a QIAprep Spin Mini Prep kit (Qiagen). The 1,200-bp sequence of one PCR product using Helicobacter sp. genus-specific primers was obtained by cycle sequencing (8).
The 16S rRNA cistrons from a fecal culture isolate (MIT 00-7128) were amplified by PCR, purified, and sequenced using our protocol previously described (8, 15).
Sequence data were entered into RNA, a program set written in Microsoft QuickBasic for data entry, editing, sequence alignment, secondary structure comparison, similarity matrix generation, and dendrogram construction for 16S rRNA sequences (22). Sequences were aligned as previously described (22). Our database contains over 1,000 sequences obtained in our laboratory and over 500 retrieved from GenBank. Dendrograms were constructed by the neighbor-joining method.
Helicobacter species-specific 1.2-kb PCR products were subjected to restriction fragment length polymorphism (RFLP) analysis. DNA digestion was accomplished by adding 10 U each of restriction endonucleases AluI and HhaI (New England Biolabs, Beverly, Mass.) and 2 μl of restriction buffer (New England Biolabs) to 16 μl of DNA and incubating at 37°C for 2 h. The samples were then electrophoresed through a 6% Visigel separation matrix followed by ethidium bromide staining and were viewed by UV illumination.
The culture results were positive for Helicobacter sp. from the feces (sample MIT 00-7128) of the clinically ill beluga, animal 1. Helicobacter spp. were not isolated by culture from the feces of animals 2 and 3, nor were they isolated from the gastric fluid of any of the three animals. The Helicobacter sp. isolated from the fecal culture was oxidase, catalase, and urease positive but negative for nitrate reduction, alkaline phosphatase hydrolysis, and indoxyl acetate hydrolysis. The isolate grew at 37 and 42°C and was susceptible to cephalothin and resistant to nalidixic acid.
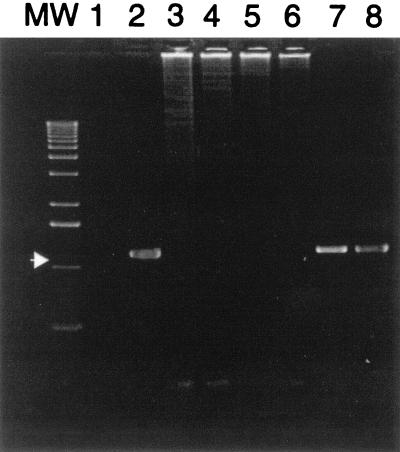
The feces and gastric fluid from the clinically ill beluga (animal 1) were positive for the presence of a 1,200-bp PCR-specific Helicobacter sp. product, whereas the feces and gastric fluid from animal 2 (samples MIT 00-7125 and MIT 00-7126) and animal 3 (sample MIT 00-7131 and MIT 00-7132) were negative (Fig. 1).
FIG. 1.
Gel electrophoresis with ethidium bromide staining demonstrating 1,200-bp PCR target sequence using Helicobacter-specific primers (arrow). Lane MW, molecular weight standards; lane 1, reagent control; lane 2, Helicobacter-positive control (H. hepaticus); lanes 3 and 4, MIT 00-7125 and MIT 00-7126 (animal 2), representing the beluga feces and gastric fluid, respectively; lanes 5 and 6, MIT 00-7131 and MIT 00-7132 (animal 3), representing the beluga feces and gastric fluid, respectively; lanes 7 and 8, MIT 00-7128 and MIT 00-7129, representing the DNA from beluga (animal 1) feces and gastric fluid, respectively.
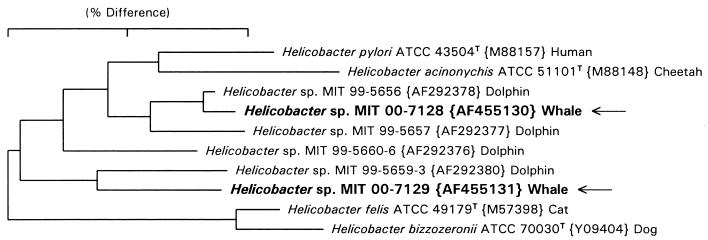
A full 16S rRNA sequence of about 1,490 bp was obtained from the fecal culture bacterial isolate MIT 00-7128, and a 1,200-bp Helicobacter species-specific PCR product was sequenced from the gastric fluid specimen MIT 00-7129. A dendrogram illustrating the alignment of these sequences is shown in Fig. 2. The beluga helicobacter sequences clustered with two novel dolphin helicobacter isolates previously described by us as well as with other recognized species of gastric helicobacters. The sequence for the novel helicobacter beluga fecal culture isolate MIT 00-7128 (ATCC BAA-429) differs from that of helicobacter dolphin isolate MIT 99-5656 by only 4 bp (99.7% similar) and thus appears to represent the same species (15). The 1,200-bp sequence from the beluga gastric PCR product, MIT 00-7129, is most closely related to a 1,200-bp PCR product from the stomach of another dolphin, sample MIT 99-5659-3 (15). However, these sequences differ by 28 bp (97.7% similar) and may represent a different taxon.
FIG. 2.
Dendrogram depicting the phylogenetic location of the Helicobacter spp. constructed on the basis of 16S rRNA sequence similarity values. The sequences from the beluga whale (isolates MIT 00-7128 and MIT 00-7129) are identified with arrows. The number in parentheses following the MIT accession number is the GenBank accession number. The scale bare is equal to a 2% difference in nucleotide sequences as determined by measuring the lengths of the horizontal lines connecting two species.
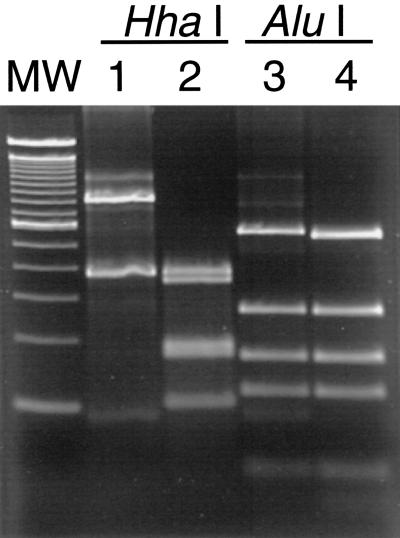
Two RFLP patterns (one from feces and the other from gastric fluid) were observed from the two 1,200-bp Helicobacter sp.-specific PCR products obtained from animal 1. Two patterns were observed in the HhaI digests; one pattern was observed in the AluI digests. Thus, the two RFLP patterns agree with the 16S rRNA sequence data indicating that the two beluga helicobacters are distinct from each other (Fig. 3). Although the RFLP patterns of samples MIT 00-7128 (feces) and MIT 00-7129 (gastric fluid) differed from each other, each was identical to that of its closest neighbor in the 16S rRNA tree: Atlantic white-sided dolphin isolates MIT 99-5656 and MIT 99-5659-3, respectively (Fig. 3) (15).
FIG. 3.
PCR-RFLP patterns of the 1,200-bp species-specific Helicobacter PCR product from beluga gastric fluid and feces. Lane MW, molecular weight standards; lane 1, MIT 00-7128 PCR product from beluga fecal isolate digested by HhaI; lane 2, MIT 00-7129 PCR product from beluga gastric fluid digested by HhaI; lane 3, MIT 00-7128 PCR product from beluga fecal isolate digested by AluI; lane 4, MIT 00-7129 PCR product from beluga gastric fluid digested by AluI.
Cetaceans have a three-chambered stomach: the nonglandular forestomach, the glandular main stomach, and the pyloric stomach (5, 16, 25, 26). Gastric helicobacters colonize only the glandular tissue and do not colonize the nonglandular stomach. In cetaceans, ulcers are visualized in both the nonglandular and glandular stomach (1, 7, 14, 28, 29). Due to the size of the animal and the length of the endoscope, it is not known if the helicobacter-positive beluga had lesions in the glandular stomach. Previous studies indicated that helicobacters colonize the main and pyloric stomachs in dolphins and did not colonize the forestomach (15). Interestingly, pigs, like cetaceans, have glandular and nonglandular portions of their stomachs (6, 9, 24, 27). In swine, gastric ulcers are identified in the nonglandular pars esophagea (6). Although the etiopathogenesis of gastric ulcer disease in pigs remains unclear, investigators have recently suggested that gastric lesions in the nonglandular stomach may be related to the presence of helicobacters in the glandular stomach (3, 6, 27).
The majority of the studies on the epizootiology of helicobacter infections in animals have been conducted with H. mustelae in ferrets (11, 12). The data indicated that transmission of H. mustelae is fecal-oral (4, 11, 12). H. pylori also has been cultured from feces and may survive in water in a nonculturable but viable coccoid form (10, 30, 31). The other proposed route of transmission is oral-oral (2, 19, 21, 31). H. pylori has also been cultured from saliva and dental plaques from humans, which argues for an oral-oral transmission (2, 19). Helicobacter spp. were identified by PCR in the gastric fluid and feces of the clinically affected beluga which manifested esophageal and forestomach ulcers on endoscopy. Interestingly, Helicobacter spp. were not identified in the other two whales, which had no clinical signs. In addition, gastric lesions were not visualized in the two clinically normal belugas during endoscopy despite the fact that all three belugas had been housed together since 1984. Although whale feces usually dissipates within a few seconds and disinfection of the water in the aquarium is attempted with a combination of ozone and chlorine, the ability to culture a gastric helicobacter from the feces of a beluga suggests that fecal-oral transmission may be important in the epizootiology of this infection in cetaceans. This seems possible given that the whales previously resided in an enclosed aquatic ecosystem that contained at least one bottlenose dolphin with confirmed gastric ulcers. Alternatively, helicobacter infection acquired prior to capture is supported by the apparent lack of helicobacter infection in the other two belugas sharing the same environment.
In this study, a beluga with clinical signs was infected with a helicobacter identical to a novel helicobacter isolated from the inflamed main stomachs of stranded dolphins (15). The Helicobacter sp. (specimen MIT 00-7128) isolated from fecal culture was urease, catalase, and oxidase positive, which is biochemically consistent with the three gastric helicobacters isolated from dolphins (15). While divergence in the 16S rRNA sequences of up to 4% would suggest that the dolphin and whale isolates could represent up to five species, the phenotypic consistency and limited number of strains would suggest grouping them in a single cetacean taxon until additional isolates are obtained and studied. To our knowledge this is the first report of a Helicobacter sp. being isolated and characterized from a whale. Identifying Helicobacter spp. from the feces and gastric fluid provides a noninvasive method to diagnose helicobacter infection in marine mammals. Further studies are required to define the role of Helicobacter spp. in the etiopathogenesis of gastric ulcers in cetaceans.
Acknowledgments
This work was supported in part by NIH grants R01-AI37750, T32-RR07036, and R01-DE10374.
We thank Gayle Sirpenski, David St. Aubin, and Carrie Goertz of the Mystic Aquarium marine mammal team and Elaine Robbins for their help and expertise.
Footnotes
This work constitutes contribution no. 133 of the Sea Research Foundation.
REFERENCES
- 1.Abollo, E. 1998. Long-term recording of gastric ulcers in cetaceans stranded in the Galician (NW Spain) coast. Dis. Aquat. Org. 32:71-73. [DOI] [PubMed] [Google Scholar]
- 2.Banatvala, N., C. Romero Lopez, and R. J. Owen. 1994. Use of the polymerase chain reaction to detect Helicobacter pylori in the dental plaque of healthy and symptomatic individuals. Microb. Ecol. Health Dis. 7:1-8. [Google Scholar]
- 3.Barbosa, A. J., J. C. Silva, A. M. Nogueira, E. Paulino, and C. R. Miranda. 1995. Higher incidence of Gastrospirillum sp. in swine with gastric ulcer of the pars oesophagea. Vet. Pathol. 32:134-139. [DOI] [PubMed] [Google Scholar]
- 4.Batchelder, M., J. G. Fox, and A. Hayward. 1996. Natural and experimental Helicobacter mustelae re-infection following successful antimicrobial eradication in ferrets. Helicobacter 1:34-42. [DOI] [PubMed] [Google Scholar]
- 5.Coffey, D. 1977. Dolphins, whales and porpoises and encyclopedia of sea mammals, p. 39. MacMillan Publishing Co. Inc., New York, N.Y.
- 6.De Groote, D., R. Ducatelle, L. J. van Doorn, D. Tilmant, A. Verschuuren, and F. Haesebrouck. 2000. Detection of “Candidatus Helicobacter suis” in gastric samples of pigs by PCR: comparison with other invasive diagnostic techniques. J. Clin. Microbiol. 38:1131-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Guise, S., A. Lagace, P. Beland, C. Girard, and R. Higgins. 1995. Non-neoplastic lesions in beluga whales (Delphinapterus leucas) and other marine mammals from the St. Lawrence Estuary. J. Comp. Pathol. 112:257-271. [DOI] [PubMed] [Google Scholar]
- 8.Dewhirst, F. E., C. C. Chien, B. J. Paster, R. L. Paster, R. Orcutt, D. B. Schauer, and J. G. Fox. 1999. Phylogeny of the defined murine microbiota: altered Schaedler flora. Appl. Environ. Microbiol. 65:3287-3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doster, A. R. 2000. Porcine gastric ulcer. Vet. Clin. N. Am. Food Anim. Pract. 16:163-174. [DOI] [PubMed] [Google Scholar]
- 10.Engstrand, L. 2001. Helicobacter in water and waterborne routes of transmission. Symp. Ser. Soc. Appl. Microbiol. 30:80S-84S. [DOI] [PubMed] [Google Scholar]
- 11.Fox, J. G., B. J. Paster, and F. E. Dewhirst. 1992. Helicobacter mustelae isolation from feces of ferrets: evidence to support fecal-oral transmission of gastric Helicobacter. Infect. Immun. 60:606-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fox, J. G., M. Blanco, and L. Lyan. 1993. Role of gastric pH in isolation of Helicobacter mustelae from the feces of ferrets. Gastroenterology 104:86-92. [DOI] [PubMed] [Google Scholar]
- 13.Fox, J. G., F. E. Dewhirst, and J. C. Tully. 1994. Helicobacter hepaticus sp. nov., a microaerophilic bacterium isolated from livers and intestinal mucosal scrapings from mice. J. Clin. Microbiol. 32:1238-1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geraci, J. R., and K. Gerstmann. 1966. Relationship of dietary histamine to gastric ulcers in the dolphin. J. Am. Vet. Med. Assoc. 149:884-890. [PubMed] [Google Scholar]
- 15.Harper, C. M., C. Dangler, S. Xu, Y. Feng, Z. Shen, B. Sheppard, A. Stamper, F. E. Dewhirst, B. J. Paster, and J. G. Fox. 2000. Isolation and characterization of Helicobacter sp. from the gastric mucosa of dolphins, Lagenorhynchus acutus and Delphinus delphis. Appl. Environ. Microbiol. 66:4751-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harrison, R. J., F. R. Johnson, and B. A. Young. 1970. The esophagus and stomach of dolphins. (Tursiops, Delphinus, Stenella). J. Zool. Lond. 160:377-390. [Google Scholar]
- 17.Lesage, V., and M. Kingsley. 1998. Updated status of the St. Lawrence River population of the beluga. Delphinapterus leucas. Can. Field-Nat. 112:98-113. [Google Scholar]
- 18.Marine Mammal Commission. 2000. Marine Mammal Commission Annual Report to Congress, p. 33. Marine Mammal Commission, Bethesda, Md.
- 19.Megraud, F. 1995. Transmission of Helicobacter pylori: fecal-oral versus oral-oral route. Aliment. Pharmacol. Ther. 9:85-91. [PubMed] [Google Scholar]
- 20.Mendes, E. N., D. M. Quieroz, F. E. Dewhirst, B. J. Paster, S. B. Moura, and J. G. Fox. 1996. Helicobacter trogontum sp. nov., isolated from the rat intestine. Int. J. Syst. Bacteriol. 46:916-921. [DOI] [PubMed] [Google Scholar]
- 21.Parsonnet, J., H. Shmuely, and T. Haggerty. 1999. Fecal and oral shedding of Helicobacter pylori from healthy infected adults. JAMA 282:2240-2245. [DOI] [PubMed] [Google Scholar]
- 22.Paster, B. J., and F. E. Dewhirst. 1988. Phylogeny of Campylobacter, wolinellas, Bacteroides gracilis, and Bacteroides ureolyticus by 16S ribosomal ribonucleic acid sequencing. Int. J. Syst. Bacteriol. 38:56-62. [Google Scholar]
- 23.Pippard, L. 1985. Status of the St. Lawrence River population of beluga (Delphinapterus leucas). Can. Field-Nat. 99:438-450. [Google Scholar]
- 24.Queiroz, D. M., G. Rocha, E. Mendes, S. de-Moura, A. De-Oliveira, and D. Miranda. 1996. Association between Helicobacter and gastric ulcers disease of the pars esophagea in swine. Gastroenterology 111:19-27. [DOI] [PubMed] [Google Scholar]
- 25.Ridgway, S. H. 1968. The bottlenose dolphin in biomedical research, p. 435. In W. I. Gay (ed.), Methods of animal experimentation, vol. III. Academic Press, New York, N.Y. [Google Scholar]
- 26.Ridgway, S. H. 1972. Mammals of the sea: biology and medicine, p. 52. Charles C. Thomas Publisher, Springfield, Ill.
- 27.Roosendaal, R., J. Vos, T. Roumen, R. van Vugt, A. Bart, H. Klaasen, E. Kuipers, C. Vanderbroucke-Grauls, and J. Kusters. 2000. Slaughter pigs are commonly infected by closely related but distinct gastric ulcerative lesion-inducing gastrospirilla. J. Clin. Microbiol. 38:2661-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sweeney, J. 1978. Marine mammals, p. 600. In M. E. Fowler (ed.), Zoo and wild animal medicine. Saunders, Philadelphia, Pa.
- 29.Sweeney, J. C., and S. H. Ridgway. 1975. Common diseases of small cetaceans. J. Am. Vet. Med. Assoc. 167:533-540. [PubMed] [Google Scholar]
- 30.Versalovic, J., and J. G. Fox. 1999. Helicobacter, p. 727-738. In P. R. Murray et al. (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 31.West, A. P., M. R. Miller, and D. S. Tompkins. 1990. Survival of Helicobacter pylori in water and saline. J. Clin. Pathol. 43:609. [DOI] [PMC free article] [PubMed] [Google Scholar]