Abstract
A strain of a thermophilic bacterium, tentatively designated Bacillus thermodenitrificans TS-3, with arabinan-degrading activity was isolated. It produced an endo-arabinase (ABN) (EC 3.2.1.99) and two arabinofuranosidases (EC 3.2.1.55) extracellularly when grown at 60°C on a medium containing sugar beet arabinan. The ABN (tentatively called an ABN-TS) was purified 7,417-fold by anion-exchange, hydrophobic, size exclusion, and hydroxyapatite chromatographies. The molecular mass of ABN-TS was 35 kDa as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, and the isoelectric point was pH 4.5. The enzyme was observed to be more thermostable than known ABNs; it had a half-life of 4 h at 75°C. The enzyme had optimal activity at 70°C and pH 6.0. The enzyme had apparent Km values of 8.5 and 45 mg/ml and apparent Vmax values of 1.6 and 1.1 mmol/min/mg of protein against debranched arabinan (α-1,5-arabinan) and arabinan, respectively. The enzyme had no pectin-releasing activity (protopectinase activity) from sugar beet protopectin, differing from an ABN (protopectinase-C) from mesophilic Bacillus subtilis IFO 3134. The pattern of degradation of debranched arabinan by ABN-TS indicated that the enzyme was an endo-acting enzyme and the main end products were arabinobiose and arabinose. The results of preliminary experiments indicated that the culture filtrate of strain TS-3 is suitable for l-arabinose production from sugar beet pulp at high temperature.
Hemicelluloses compose a large fraction of plant cell walls and are a heterogeneous mixture of polysaccharides that include xylans, glucans, mannans, galactans, and arabinans. Arabinans consists of a backbone of α-1,5-linked l-arabinofuranosyl residues, some of which are substituted with α-1,2- and α-1,3-linked side chains of l-arabinose in the furanose conformation (1). There is some evidence to suggest that in plant cell walls arabinan is generally linked to the rhamnopyranosyl units of rhamnogalacturonan (13, 14). Sakai and Sakamoto described a protopectinase (protopectinase-C [PPase-C]) from Bacillus subtilis IFO 3134 that did not catalyze polygalacturonic acid degradation (11). By endo-1,5-α-l-arabinase (ABN) activity, PPase-C splits the α-1,5-l-arabinofuranoside linkage of the arabinan region in arabinogalactan, which attaches pectin to the cell wall constituents, so that PPase-C releases pectin (12). In addition to endo-ABN, α-l-arabinofuranosidase (α-l-ABF) also is involved in degradation of arabinan. The enzyme cleaves the arabinose side chains, allowing endo-ABN to attack the arabinan backbone. These enzymes act synergistically in degrading branched arabinan to generate l-arabinose (21).
It has been proved that l-arabinose selectively inhibits intestinal sucrase in a noncompetitive manner and reduces the glycemic response after sucrose ingestion in animals (16, 17). Based on these observations, l-arabinose can be used as a physiologically functional sugar that inhibits sucrose digestion. Effective l-arabinose production is, therefore, important in the food industry.
Mesophilic microbial endo-1,5-α-l-ABNs have been described for species of Bacillus (5, 11, 22, 23), Aspergillus (9, 21), and Pseudomonas (8). All of these endo-ABNs have molecular masses of about 30 to 35 kDa and have optimal temperatures of between 50 and 60°C. To our knowledge, there has been no report on thermoactive and thermostable endo-1,5-α-l-ABN from thermophilic bacteria.
To develop an effective process of l-arabinose production from hemicellulose, we have attempted isolation of bacteria producing thermostable arabinan-degrading enzymes. This paper deals with the screening of thermophilic bacteria producing thermostable endo-ABN and the purification and characterization of the enzyme.
MATERIALS AND METHODS
Substrate and chemicals.
Arabinan (from sugar beet pulp), debranched arabinan (from sugar beet pulp), arabinobiose, arabinotriose, and arabinotetraose were purchased from Megazyme (Bray, County Wicklow, Ireland). p-Nitrophenyl-α-l-arabinofuranoside was purchased from Sigma-Aldrich Japan (Tokyo, Japan). Unless otherwise specified, all chemicals were of certified reagent grade.
Screening of thermophilic bacilli with thermostable ABN.
A thermophilic bacterium isolated in this research, Bacillus thermodenitrificans TS-3, was used. The strain was isolated from a soil sample collected in Nara City, Japan, as follows. AG medium, containing 0.5% arabinogalactan from sugar beet pulp as the sole carbon source, 0.2% ammonium sulfate, 0.6% KH2PO4, 1.4% K2HPO4, and 0.02% yeast extract (Difco, Detroit, Mich.) (separately autoclaved at pH 7.0), was used for the isolation of organisms. In a test tube (15.5-mm diameter), 3 ml of the medium was inoculated with a soil sample and incubated at 60°C for 48 h on a shaker (120 rpm; 7-cm stroke). An aliquot of the culture broth was transferred into fresh medium and incubated under the same conditions, and then an aliquot of the culture broth in which the organisms had grown was spread out onto the AG medium agar plate. The plates were placed in plastic bags and incubated at 60°C, and microbial colonies were individually picked up and were inoculated into the AG medium. After incubation at 60°C for 24 h, the ABN activity (activity on debranched arabinan) and α-l-ABF activity of the culture filtrate were measured.
Identification of the isolate.
The isolate was identified by methods described previously (3).
Cloning of 16S rRNA gene and nucleotide sequencing.
For 16S ribosomal DNA analysis, genomic DNA of strain TS-3 was prepared by the method of Saito and Miura (10). The 16S ribosomal DNA was amplified for 25 cycles (15 s of denaturation at 98°C, 10 s of annealing at 55°C, and 30 s of elongation at 74°C) by using a bacterium-specific forward primer (5F, 5′-GAAGAGTTTGATCCTGGCTCAGA-3′) and a reverse primer (1506R, 5′-TACCTTGTTACGACTTCACCCCAG-3′). The composition of the PCR mixture (50 μl) was 120 mM Tris-HCl (pH 8.0), 10 mM KCl, 6 mM (NH4)2SO4, 0.1% Triton X-100, 10 μg of bovine serum albumin per ml, 1 mM MgCl2, 200 mM deoxynucleoside triphosphates, 250 ng of genomic DNA, 50 pmol of each primer, and 2.5 units of DNA polymerase from Pyrococcus kodakaraensis strain KOD1 (Toyobo, Tokyo, Japan). The amplified DNA (approximately 1,500 bp) was phosphorylated with T4 DNA kinase, ligated into the SmaI site of pUC18 (Amersham Pharmacia Biotec, Buckinghamshire, United Kingdom), and transformed into Escherichia coli JM109 cells by the method of Hanahan (4). Plasmids of randomly selected clones with inserts of the correct size were then recovered with a plasmid Midi kit (Qiagen, GmbH, Hilden, Germany). The nucleotide sequences were analyzed by the dideoxy chain termination method using a Thermo Sequenase fluorescently labeled primer cycle sequencing kit (Amersham Pharmacia Biotec) on an A.L.F. DNA sequencer (Pharmacia LKB Biotechnology, Uppsala, Sweden). Both strands of the DNA were sequenced, and the resulting sequence was compared with known sequences by using the FASTA program.
Enzyme assays.
ABN activity was assayed in a mixture containing 200 μl of 0.5% arabinan in 100 mM sodium acetate buffer (pH 6.0) and 10 μl of appropriately diluted enzyme solution, with incubation at 70°C for 30 min. Activities on arabinan and debranched arabinan were assayed by measuring the release of reducing groups by the Somogyi method (18). One unit of the activity was defined as the activity that liberates reducing groups corresponding to 1 μmol of l-arabinose per min per ml of reaction mixture at 70°C.
α-l-ABF activity was determined with p-nitrophenyl-α-l-arabinofuranoside as a substrate. The reaction mixture, containing 190 μl of 1 mM p-nitrophenyl-α-l-arabinofuranoside in 100 mM sodium acetate buffer (pH 7.0) and 10 μl of appropriately diluted enzyme solution, was incubated at 70°C for 30 min. The reaction was terminated by the addition of 100 μl of 500 mM Na2CO3, and the released p-nitrophenol was determined by measuring the absorbance at 405 nm. One unit of enzyme activity is defined as the amount of enzyme that releases 1 μmol of p-nitrophenol per min from p-nitrophenyl-α-l-arabinofuranoside at 70°C. The enzymatic activities on p-nitrophenyl-α-l-arabinopyranoside, p-nitrophenyl-α- and -β-d-galactopyranoside, p-nitrophenyl-α-d-xylopyranoside, and p-nitrophenyl-β-d-cellobioside were measured under the same conditions as used for the ABF reaction.
Protopectinase activity (pectin-releasing activity) was assayed in a mixture containing 20 mg of sugar beet protopectin, 990 μl of 100 mM sodium acetate buffer (pH 6.0), and 10 μl of enzyme solution at 70°C for 30 min. The pectic substances liberated from protopectin were measured by the carbazole-H2SO4 method (15). One unit of enzyme activity was defined as the activity that liberates pectic substances corresponding to 1 μmol of d-galacturonic acid under the above-described reaction conditions. Sugar beet protopectin used as a substrate was prepared by the following procedure: sugar beet pulp was homogenized, washed with 2% sodium hexametaphosphate solution (pH 4.0) until the soluble substances that reacted with carbazole-H2SO4 were washed off, and then freeze-dried.
Effect of metals.
The effects of metal ions on the ABN activity were determined by adding the appropriate salts (final concentration, 1 mM) to the standard enzyme reaction mixture, and the activity was then assayed by the standard method.
ABN preparation.
B. thermodenitrificans TS-3 was cultivated in a 300-ml Sakaguchi flask containing 50 ml of a medium composed of 0.5% arabinogalactan, 0.5% Casamino Acids, 0.5% yeast extract, and 0.5% NaCl (pH 7.0) at 60°C in a reciprocating shaker for 16 h. The whole culture broth was then transferred to a 10-liter jar fermentor containing 5 liters of the same medium and cultured at 55°C for 20 h with aeration (1 vol/vol/min) and agitation (400 rpm).
Purification of thermostable ABN.
Purification of the ABN (tentatively called ABN-TS) from B. thermodenitrificans TS-3 was performed as follows.
(i) Preparation of starting material.
For purification of the enzyme from culture filtrate of B. thermodenitrificans TS-3, the culture filtrate (5 liters) was concentrated under reduced pressure to about 500 ml and dialyzed against 20 mM Tris-HCl buffer (pH 8.0). This was used as the starting material for enzyme purification.
(ii) Q-Sepharose column chromatography.
The enzyme solution obtained in step 1 was put on a Q-Sepharose column (5 by 12 cm; Amersham Pharmacia Biotec) equilibrated with 20 mM Tris-HCl buffer, pH 8.0. The column was washed extensively with the same buffer and eluted with a linear gradient of 0 to 500 mM NaCl in the same buffer (pH 8.0) at a flow rate of 4 ml/min, and 10-ml fractions were collected. The highly active fractions were pooled and made 30% saturated with ammonium sulfate by addition of granular ammonium sulfate.
(iii) Phenyl-Sepharose HP column chromatography.
The enzyme solution obtained in step 2 was put on a phenyl-Sepharose HP column (2.6 by 10 cm; Amersham Pharmacia Biotec) equilibrated with 20 mM Tris-HCl buffer (pH 7.5) containing ammonium sulfate at 30% saturation. The column was washed extensively with the same buffer and then eluted with a liner gradient of 30 to 0% ammonium sulfate saturation in 20 mM Tris-HCl buffer (pH 7.5) at a flow rate of 4 ml/min. The highly active ABN fractions were pooled (10 ml) and concentrated to about 2 ml by ultrafiltration with a Centriprep-10 instrument (Millipore, Bedford, Mass.).
(iv) Superose 12 column chromatography.
The concentrated enzyme solution was put on a Superose 12 column (1.6 by 30 cm; Amersham Pharmacia Biotec) equilibrated with 20 mM Tris-HCl buffer containing 100 mM NaCl, pH 7.0. The elution was done with the same buffer at a flow rate of 0.5 ml/min, and fractions of 1 ml were collected. The fractions containing ABN activity were pooled and dialyzed against 50 mM Tris-HCl buffer containing 1 mM KH2PO4, pH 7.5.
(v) Hydroxyapatite column chromatography.
The dialysate was applied to a Biogel HTP column (0.6 by 10 cm; Japan Bio-Rad Laboratories, Tokyo, Japan) equilibrated with the dialysis buffer. The column was washed with the same buffer and eluted with a linear gradient of 1 to 100 mM K2HPO4 in 50 mM Tris-HCl buffer (pH 7.5) at a flow rate of 0.4 ml/min. The fractions containing ABN activity were pooled and dialyzed against 20 mM Tris-HCl buffer, pH 8.0.
(vi) Mono Q column chromatography.
The enzyme solution was loaded onto a Mono Q 5/5 column (Amersham Pharmacia Biotec) equilibrated with the dialysis buffer. The column was washed with the same buffer and eluted with a linear gradient of 0 to 500 mM NaCl in 20 mM Tris-HCl buffer (pH 8.0) at a flow rate of 0.5 ml/min.
(vii) Superdex 75 column chromatography.
The enzyme solution obtained in step 6 was concentrated to about 0.25 ml by ultrafiltration with a Centricon-10 instrument (Millipore) and applied to a Superdex 75 HR10/30 column (1 by 30 cm; Amersham Pharmacia Biotec) equilibrated with 20 mM Tris-HCl buffer containing 100 mM NaCl, pH 7.0. The enzyme was eluted with the same buffer at a flow rate of 0.25 ml/min. The fractions containing ABN activity were pooled and dialyzed against 20 mM Tris-HCl buffer, pH 7.0.
Purification of PPase-C.
An endo-ABN (PPase-C), having protopectinase activity, produced from B. subtilis IFO 3134 was purified by a method described previously (11).
Electrophoresis of proteins.
The homogeneity and molecular mass of the enzyme were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) by the method of Laemmli on 12.5% (wt/vol) polyacrylamide gels (6), and the protein bands were stained with Coomassie brilliant blue R-250. The molecular mass standards used were from Japan Bio-Rad Laboratories and included phosphorylase b (97.4 kDa), bovine serum albumin (66.7 kDa), ovalbumin (45.0 kDa), carbonic anhydrase (31.0 kDa), trypsin inhibitor (21.5 kDa), and lysozyme (14.0 kDa).
Molecular mass measurement.
The apparent molecular mass of the native enzyme was determined by gel filtration on a Superdex 75 HR10/30 column with tyroglobulin (670 kDa), bovine gamma globulin (158 kDa), chicken ovalbumin (44 kDa), and equine myoglobin (17 kDa) as standard proteins.
Isoelectric focusing.
Isoelectric focusing was performed using a model III mini-IEF cell with a 5% (wt/vol) acrylamide gel and Bio-Lyte 3/10 (Japan Bio-Rad Laboratories) according to the manufacturer's instructions. After electrofocusing, protein bands were stained with a silver stain kit (Japan Bio-Rad Laboratories). The pI of the enzyme was estimated from the relationship between the mobilities of the standard proteins (Isoelectric Focusing Calibration kit; Japan Bio-Rad Laboratories) and their pIs.
N-terminal amino acid sequence.
Around 100 pmol of purified enzyme was subjected to SDS-10% PAGE and electrophoretically transferred onto a polyvinylidene difluoride membrane (Immobilon-Psq; Millipore) in 100 mM N-cyclohexyl-3-aminopropanesulfonic acid (CAPS)-NaOH buffer (pH 11.0) at 50 V for 1.5 h. The enzyme-blotted membrane was put into a protein sequencer (ABI model 475; Applied Biosystems, Foster City, Calif.).
Effects of pH and temperature on ABN-TS activity.
The optimum pH of the activity of ABN-TS was found by determining at the activity at 70°C for 30 min with following buffer systems (debranched arabinan was used as the substrate): 100 mM sodium acetate buffer (pH 4.0 to 7.0), 100 mM phosphate buffer (pH 6.0 to 8.0), and 100 mM Tris-HCl buffer (pH 7.0 to 9.0). The enzyme stability at different pHs was measured by determining the residual activities after incubating the enzyme samples (0.69 U/ml) at 37°C for 16 h at pH 3.0 to 11.0. The effect of temperature on the ABN-TS activity was measured by determining the enzyme activity at temperatures ranging from 40 to 90°C.
Thermostability.
Microcentrifuge tubes (1.5 ml) containing 100 μl of purified ABN-TS solution (0.69 U/ml) in 50 mM Tris-HCl buffer (pH 7.0) containing 50 μg of bovine serum albumin per ml were incubated at 70 and 75°C. At regular incubation intervals, the tubes were withdrawn and then kept at 4°C. The residual ABN activity in each tube was determined under the standard assay conditions with debranched arabinan as the substrate.
Kinetic parameters.
The apparent Km and Vmax values against debranched arabinan and arabinan were determined by the double-reciprocal plot method of Lineweaver and Burk (7). The activities against debranched arabinan and arabinan were assayed at concentrations of between 1.25 and 10 mg/ml and 10 and 100 mg/ml, respectively.
Enzymatic hydrolysis of debranched arabinan.
High-pressure liquid chromatography (HPLC) was used identify the products generated by ABN-TS and PPase-C from debranched arabinan. Debranched arabinan (0.1%) was incubated with ABN-TS (1.26 U) and PPase-C (1.18 U) in 1 ml of 20 mM sodium acetate buffer (pH 6.0) at 70 and 50°C, respectively. At regular time intervals, samples were withdrawn and heated in a boiling water bath for 5 min to stop the reaction. The reaction mixtures were then loaded on an Ultron PS-80P column (0.8 by 30 cm; Shinwa Chemical Industries, Kyoto, Japan), and the oligosaccharides were eluted with Milli-Q water at a flow rate of 1 ml/min. The sugars were detected with a refractive index detector (model RID-10A; Shimadzu, Kyoto, Japan) and were identified and quantified by comparing their elution times with those of standard amounts of arabinose and arabino-oligosaccharides, up to arabinotetraose.
l-Arabinose production from sugar beet pulp using ABN-TS.
Sugar beet pulp (30 g) was autoclaved at 121°C for 40 min in 1 liter of 20 mM sodium acetate buffer, pH 6.0. Culture filtrate of strain TS-3, containing 4,100 U of ABN-TS and 380 U of α-l-ABFs, was added to the suspension and incubated at 60°C for 24 h with shaking. To identify the released arabinose, the arabinose in the reaction mixture was purified by crystallization; after the incubation, the reaction mixtures were filtered through a glass filter. Polysaccharides in the filtrate were precipitated by adding ethanol to a final concentration of 75% and discarded by centrifugation at 11,000 × g for 30 min. The resultant supernatant was deionized with mixed bed resin AG 501-X8 (Japan Bio-Rad Laboratories) and concentrated to 5 ml under reduced pressure. Arabinose in the concentrate was crystallized by adding ethanol to a final concentration of 95%. The resultant crystals were analyzed with infrared (IR) spectra by using an FT-IR-300E instrument (JASCO, Tokyo, Japan) and specific rotation by using a SEPA-200 instrument (Horiba Ltd., Kyoto, Japan).
Protein assay.
The protein concentration was estimated by using a Micro-BCA protein assay regent kit (Pierce, Rockford, Ill.) with bovine serum albumin as a standard. The protein concentration in the column effluents was monitored by measuring absorbance at 280 nm.
Nucleotide sequence accession number.
The nucleotide sequence of the 16S rRNA gene of B. thermodenitrificans TS-3 has been submitted to the GenBank/EMBL/DDBJ databases under accession number AB063312.
RESULTS
Isolation and identification of microorganisms which produce thermostable arabinan-degrading enzymes.
Microorganisms which produce thermostable arabinan-degrading enzymes were isolated from soil samples by an enrichment technique using the medium described in Materials and Methods. In Table 1, the endo-ABN (activity on debranched arabinan), α-l-ABF, and protopectinase activities of culture filtrates from the isolated strains are shown. All strains produced optimal activity at 70°C and pH 6.0 on debranched arabinan (ABN activity) and ABF activity; however, these ABN activities varied in thermostability. Pectin-releasing activity on sugar beet pulp was not detected in culture filtrates from any of the strains. The thermostability of the ABN activity of strain TS-3 was the highest (up to 70°C for 30 min) among these strains, and strain TS-3 was selected and used throughout this experiment.
TABLE 1.
Arabinan-degrading activities of culture filtrates from isolates
| Strain | ABN
|
ABF activity (U/ml) | PPase activity (U/ml) | |||
|---|---|---|---|---|---|---|
| Activity (U/ml) | Optimal temp (°C) | Optimal pH | Thermostabilitya (°C) | |||
| TS-2 | 0.06 | 70 | 6.0 | ∼65 | 7.0 | NDb |
| TS-3 | 0.14 | 70 | 6.0 | ∼70 | 3.4 | ND |
| TS-6 | 0.06 | 70 | 6.0 | ∼70 | 3.6 | ND |
| TS-17 | 0.10 | 70 | 6.0 | ∼60 | 1.6 | ND |
| TS-40 | 0.18 | 70 | 6.0 | ∼60 | 8.5 | ND |
| TS-41 | 0.04 | 70 | 6.0 | ∼65 | 8.0 | ND |
Thermostability was assessed by incubating culture filtrate at pH 7.5 for 30 min and assaying remaining activity at 70°C and pH 6.0. Stable means the remaining activity is up to 80%.
ND, not detected.
The properties of strain TS-3 were investigated. The microorganism was gram positive, spore forming, and rod shaped (0.5 to 1.0 by 3.0 to 6.0 μm). It grew well under aerobic conditions. The isolate was positive for reduction of nitrate, hydrolysis of starch, utilization of citrate, and production of catalase and cytochrome oxidase and negative for acetoin formation (Voges-Proskauer test), formation of indole from tryptophan, and production of gas from glucose. It grew over a temperature range of from 45 to 65°C and at initial pH values of between 7.0 and 9.0. The nucleotide sequence of the 16S rRNA gene from TS-3 showed 99.8% identity with that of B. thermodenitrificans (accession no. AB028234), 99.5% with that of Bacillus denitrificans (Z26927), and 99.1% with that of Geobacillus subterraneus strain K (AF2763), suggesting the bacterium is affiliated with the thermophilic Bacillus groups. Based on these results, strain TS-3 was defined as a strain of B. thermodenitrificans, belonging to genus Bacillus, and was tentatively designated B. thermodenitrificans strain TS-3.
Purification of ABN.
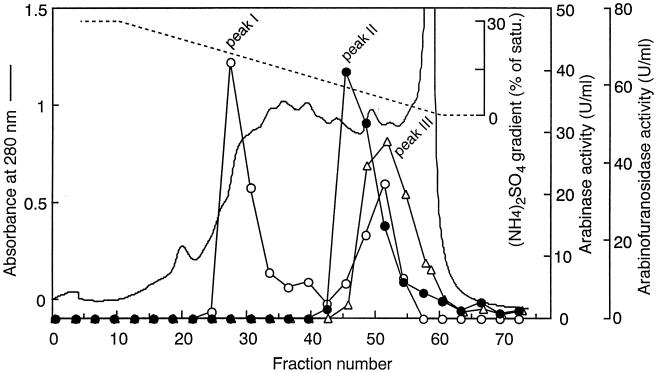
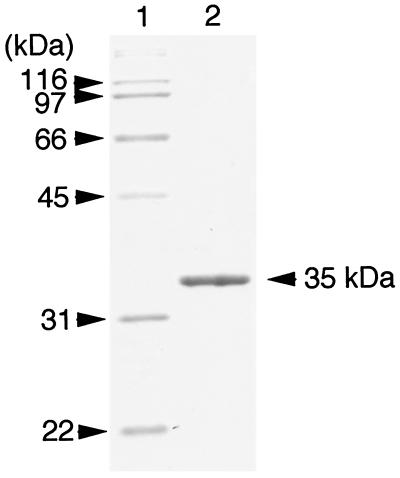
The procedure for purification of the ABN is summarized in Table 2. As shown in Fig. 1, debranched-arabinan-degrading activity (peak II) was separated from two peaks of ABF activity (peaks I and III) by phenyl-Sepharose HP column chromatography, indicating that this activity was endo-ABN. Peak I showed strong activity on p-nitrophenyl-α-l-arabinofuranoside but no activity on arabinan. On the other hand, peak III showed activities on both p-nitrophenyl-α-l-arabinofuranoside and arabinan. As peak II seemed to be endo-ABN, it was used for further purification steps. In the final purification step, which involved Superdex 75, the enzyme activity was eluted as a single peak in parallel to the protein concentration (data not shown). As shown in Table 2, the procedure described here gave 2.2 mg of the purified enzyme from 5 liters of the culture filtrate and a 28.8% recovery of total ABN activity in the crude enzyme. The purified enzyme gave a single sharp protein band on SDS-PAGE (Fig. 2). The enzyme was tentatively designated ABN-TS, and used in the following studies.
TABLE 2.
Purification procedure for arabinase from B. thermodenitrificans TS-3
| Purification step | Vol (ml) | Protein (mg) | ABN activity (U) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Culture filtrate | 5,200 | 54,202 | 3,400 | 0.06 | 1.0 | 100 |
| Q-Sepharose | 500 | 1,927 | 2,522 | 1.31 | 21.8 | 74.2 |
| Phenyl-Sepharose | 70 | 185 | 2,278 | 12.3 | 205 | 67.0 |
| Superose 12 | 11 | 41.3 | 1,263 | 30.6 | 510 | 37.1 |
| Hydroxyapatite | 16 | 6.1 | 1,203 | 197 | 3,283 | 35.4 |
| Mono Q | 2 | 2.5 | 1,023 | 409 | 6,817 | 30.1 |
| Superdex 75 | 1.5 | 2.2 | 978 | 445 | 7,417 | 28.8 |
FIG. 1.
Elution profile of ABN and ABF from a phenyl-Sepharose column. The conditions for chromatography are described in Materials and Methods. ○, ABF activity on p-nitrophenyl-α-l-arabinofuranoside; •, ABN activity on debranched arabinan; ▵, ABN activity on arabinan.
FIG. 2.
SDS-PAGE of purified ABN-TS. Lane 1, marker proteins; lane 2, purified ABN-TS (1 μg).
Enzymatic properties of ABN-TS.
The molecular mass of ABN-TS was estimated by permeation chromatography on Superdex 75 and SDS-PAGE. From the result of permeation chromatography, the molecular mass of the enzyme was estimated to be around 23 kDa, and from SDS-PAGE, it was estimated to be 35 kDa (Fig. 2). These results indicated that the enzyme was a monomeric structure. The isoelectric point of the enzyme was determined to be around pH 4.5 by using Bio-Lyte-PAGE (data not shown).
The N-terminal amino acid sequence obtained from the purified ABN-TS was Val-His-Phe-His-Pro-Phe-Gly-Asn-Val-Asn-Phe-Tyr-Glu-Met-Asp-Trp-Ser-Leu-Lys-Gly. The sequences were analyzed against the SWISS-PROT protein sequence database with the BLASTP program, but no similar sequence was found.
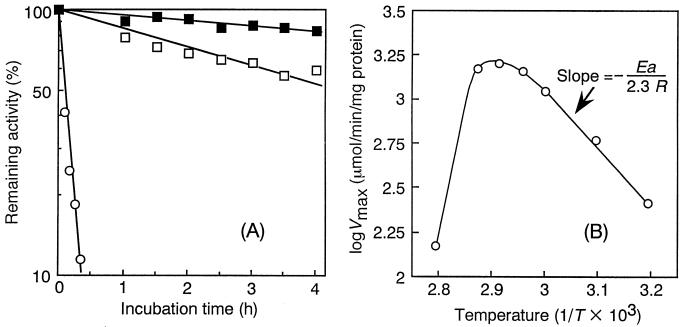
The effects of pH and temperature on the activity and stability of ABN-TS were investigated. The enzyme was most active at pH 6.0 and was quite stable between pH 6.0 and 11.0 when incubated at 37°C for 16 h, as shown in Table 3. The optimal temperature for the activity was around 70°C, as shown in Table 3. The thermostability of ABN-TS was determined in detail by incubation at various temperatures and measurement of residual activities at intervals of incubation, comparing with that of PPase-C (an endo-ABN from mesophilic B. subtilis). The inactivation kinetics of the enzyme at pH 7.0 were determined. They fitted a first-order reaction, and the half-lives of ABN-TS were 11 and 4 h at 70 and 75°C, respectively (Fig. 3A). On the other hand, the thermostability of PPase-C was very low compared with that of ABN-TS (the half-life of inactivation at 65°C was 5 min). The Arrhenius plots for calculating activation energy for the ABN-TS reaction are shown in Fig. 3B. Here, Vmax values were observed to increase with the reaction temperature up to 70°C, and the activation energy for the ABN-TS reaction was determined to be 8.6 kcal/mol at up to 70°C and to decrease at higher temperatures. The Q10 value was calculated to be 1.45 in the temperature range of between 60 and 70°C.
TABLE 3.
Comparison of some physicochemical and enzymatic properties of endo-arabinases from various microorganisms
| Species or strain | Molecular mass (kDa)a | pI | Optimal pH | pH stability | Optimal temp (°C) | End product of reaction | Reference |
|---|---|---|---|---|---|---|---|
| TS-3 | 35 | 4.5 | 6.0 | 6-11 | 70 | Arabinobiose | This paper |
| B. subtilis | 30 | 9.0 | 6.0 | 6-9 | 60 | Arabinobiose | 11 |
| A. nidulans | 40 | 3.25 | 5.5 | NDb | 68 | ND | 9 |
| A. niger | 45 | 3.0 | 4.6 | ND | 51 | Arabinobiose | 21 |
Determined by SDS-PAGE.
ND, not determined.
FIG. 3.
Effect of temperature on stability (A) and reaction rate (B) of ABN-TS. (A) Thermostabilities of ABN-TS and PPase-C. Residual ABN activities were measured at 70°C and pH 6.0 after incubation at 70°C (ABN-TS) (▪), 75°C (ABN-TS) (□), or 65°C (PPase-C) (○), at various times. (B) Relationship between temperature and activity of ABN-TS (Arrhenius plots).
The effects of different metal ions on the activity of ABN-TS were determined. No effect on activity was detected with Ba2+, Mg2+, and Mn2+, whereas Hg2+ and Ag2+ completely inhibited the activity. Hg2+ is known to react with protein sulfhydryl groups as well as with histidine and tryptophan residues. As the addition of EDTA did not affect the activity, the enzyme seemed to not need any metal ion for the enzymatic reaction.
Substrate specificity and kinetic analysis.
The enzyme was active on arabinan (from sugar beet pulp) and debranched arabinan (from sugar beet pulp). The enzyme was inactive on arabinogalactan (from larch wood), carboxymethyl cellulose, xylan (from oat spelt), polygalacturonate (from orange), pectin (from apple), galactan (from lupin), p-nitrophenyl-α-l-arabinofuranoside, p-nitrophenyl-α-l-arabinopyranoside, p-nitrophenyl-α- and -β-d-galactopyranoside, p-nitrophenyl-α-d-xylopyranoside, and p-nitrophenyl-β-d-cellobioside at pH 6.0.
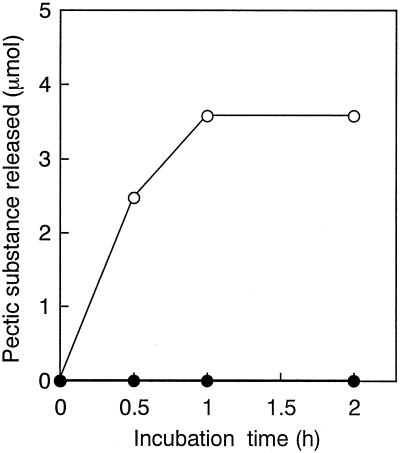
Endo-ABN (PPase-C) from B. subtilis IFO 3134 has pectin-releasing activity (protopectinase activity) with sugar beet pulp (Fig. 4). We examined the protopectinase activity of ABN-TS against sugar beet protopectin; however, the enzyme was not active on sugar beet protopectin even when the reaction was performed using activities (as arabinan-degrading activity on debranched arabinan) 10 times greater than that of PPase-C (Fig. 4).
FIG. 4.
Protopectinase activity of PPase-C and ABN-TS. Sugar beet protopectin (20 mg) was incubated with ABN-TS (5.9 U) and PPase-C (0.59 U) in 1 ml of 100 mM sodium acetate buffer (pH 6.0) at 50°C. Samples were taken out and filtered, and released pectic substance was measured by the carbazole-sulfuric acid method. •, ABN-TS; ○, PPase-C.
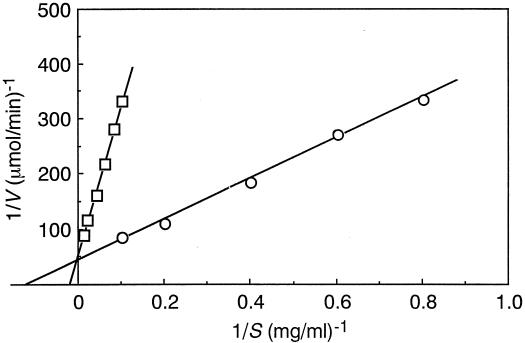
The influence of the extent of branching of the arabinan substrates on the apparent Km and Vmax values of ABN-TS is shown in Fig. 5. The apparent Km values for degradation of debranched arabinan and arabinan were 8.5 and 45 mg/ml (at pH 6.0), and the apparent Vmax values were 1.6 and 1.1 mmol/min/mg of protein (at pH 6.0), respectively.
FIG. 5.
Determination of apparent Km and Vmax values of ABN-TS at 70°C from a Lineweaver-Burk plots. ○, debranched arabinan; □, arabinan.
Mode of action of the enzyme.
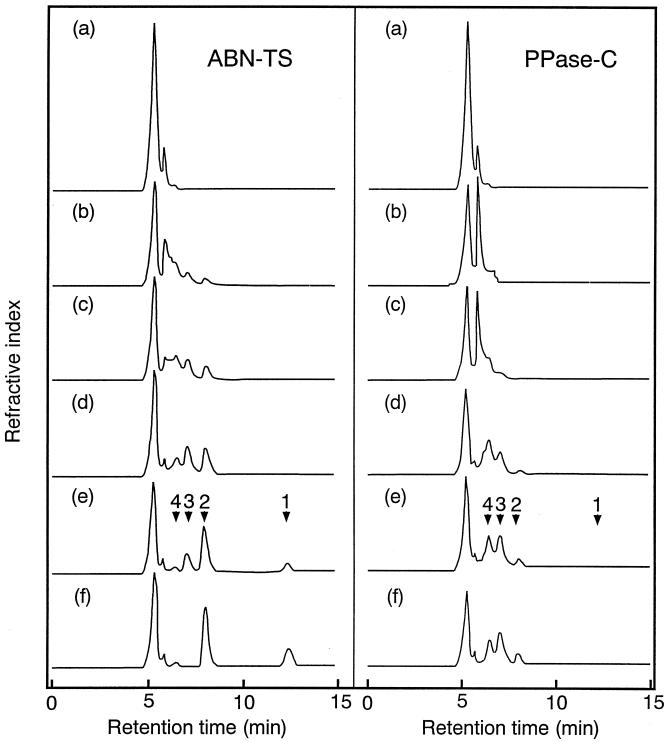
To investigate the reaction pattern of ABN-TS with debranched arabinan, HPLC analyses were done on the reaction products. Figure 6 shows the high-pressure liquid chromatograms of the reaction products of ABN-TS compared with those of PPase-C, an ABN produced by B. subtilis IFO 3134. At an early stage of the PPase-C reaction, a mixture of arabino-oligosaccharides was released, and the primary end product was arabinotriose. These facts indicate that PPase-C is a typical endo-acting enzyme. On the other hand, ABN-TS degraded the debranched arabinan by an endo mode, the same as PPase-C, but the end products were arabinobiose and arabinose, different from those with PPase-C.
FIG. 6.
HPLC analysis of products generated from debranched arabinan by ABN-TS and PPase-C. Debranched arabinan (0.1%) was incubated with ABN-TS (1.26 U) and PPase-C (1.18 U) in 1 ml of 20 mM sodium acetate buffer (pH 6.0) at 70 and 50°C, respectively. At 0 min (a), 1 min (b), 2 min (c), 5 min (d), 30 min (e), and 2 h (f), reaction mixtures were removed and boiled for 5 min to stop the reaction. Twenty microliters of each samples was subjected to HPLC analysis as described in Materials and Methods. The elution times of the standards arabinose (1), arabinobiose (2), arabinotriose (3), and arabinotetraose (4) are indicated.
Preliminary experiments for l-arabinose production using ABN-TS.
To confirm that our enzyme is available in l-arabinose produced from a biomass resource, sugar beet pulp (3%, wt/vol) was autoclaved and incubated with culture filtrate of strain TS-3, containing 4.1 U of ABN-TS per ml and 0.38 U of α-l-ABFs per ml, at 60°C for 24 h with shaking. After the incubation, the arabinose generated from sugar beet pulp by the enzymes was estimated by HPLC. Only arabinose, as a monosaccharide, was detected in the reaction product. Approximately 80 mg of arabinose per g of sugar beet pulp was released, corresponding to 100% of the arabinose initially present in the sugar beet pulp. Arabinose released in the reaction mixture was crystallized by adding ethanol. The IR spectra in FT-IR and the specific rotation ([α] D14.5 = +102 to 104°) of the resulting crystals were identical to those of authentic l-arabinose. Thus, the crystals were confirmed to be l-arabinose.
DISCUSSION
This is the first report regarding a thermostable endo-ABN. We isolated a strain, tentatively designated B. thermodenitrificans TS-3, producing thermostable ABN. Our enzyme, ABN-TS, from the isolate is most active at around 70°C (at pH 6.0). This is the highest temperature for optimal activity among known ABNs; the ABNs of B. subtilis (11) and Aspergillus niger (21) are reported to be highly active at 60 and 50°C, respectively. ABN-TS is highly thermostable, and it has a half-life of 4 h at 75°C, whereas the ABN (PPase-C) from the mesophilic bacterium B. subtilis IFO 3134 is inactivated completely by incubation at 65°C within 30 min. In Table 3, some properties of known ABNs are listed. An ABN showing an optimal reaction temperature of 68°C, which is close to that of our enzyme, was isolated from Aspergillus nidulans (9). However, details on its thermostability were not reported. Thus, ABN-TS is the most thermoactive and thermostable ABN reported to date.
The molecular mass of ABN-TS is 35 kDa as determined by SDS-PAGE. This value is similar to those for ABNs from other microorganisms: the molecular masses of ABNs of B. subtilis, A. niger, and A. nidulans are reported to be 30, 43, and 40 kDa, respectively. The pI of ABN-TS (4.5) is much more acidic than that of PPase-C (9.0) from B. subtilis IFO 3134 (11), and it is similar to those of ABNs from fungi such as A. niger (3.0) and A. nidulans (3.25) (9). The N-terminal amino acid sequence of ABN-TS has no similarity to those of other ABNs. From these facts, ABN-TS seems to be unique in molecular characteristics among ABNs. We have isolated and characterized thermostable pectate lyase (PL 47, a pectin-degrading enzyme) from a thermophilic bacterium, Bacillus sp. strain TS 47 (19, 20). We have found that the pI of PL 47 (5.3) is much more acidic than those of less-thermostable pectate lyases from other species of the bacilli, and we have speculated that acidic amino acids in the enzyme contribute to its thermostability. In order to elucidate the peculiarity of thermostable enzymes, we are studying the molecular structure of ABN-TS, and the results will be published elsewhere.
The rate of arabinan hydrolysis of the ABN-TS reaction is very low compared to that of debranched arabinan at low substrate concentrations. The Lineweaver-Burk plots show that branching of arabinan does not affect the apparent Vmax values but does affect substrate affinity: the apparent Km values for the degradation of debranched arabinan and arabinan are 8.5 and 45 mg/ml, respectively (Fig. 5). From these facts, it appears that removal of arabinose residues on the side chain of the 1,5-α-l-arabinan backbone increases the affinity (1/Km) of ABN-TS for arabinan.
Since one of the objectives of this study is to develop an efficient system for production of l-arabinose from biomass resources, accelerating the rate of arabinan degradation by ABN is very important, as is the thermostability of the enzymes. In the culture filtrate of our strain, at least two α-l-ABF activities, which may serve to remove side-chain-containing arabinoses on the 1,5-α-l-arabinan backbone, were detected by phenyl-Sepharose HP column chromatography (Fig. 1).
One ABF (in peak III in Fig. 1, tentatively named ABF II) shows activity on sugar beet arabinan but not on debranched arabinan; thus, ABF II is thought to be classified as a family 54 ABF, in accordance with the classification of Beldman et al. (2), which acts by cleaving arabinofuranosyl linkages attached on side chains of such arabinose-containing polysaccharides as arabinan, arabinogalactan, and arabinoxylan. ABF II may take part with ABN-TS in degrading arabinan by removing side-chain-containing arabinose from arabinan: the ABF II works to make debranched arabinan, and then the ABN-TS degrades the resultant debranched arabinan to produce, effectively, arabino-oligosaccharides. Another ABF (in peak I in Fig. 1, tentatively named ABF I) is inactive on arabinan but active on arabino-oligosaccharides, and the enzyme is assumed to be an enzyme classified as a family 51 ABF, in accordance with the classification by Beldman et al. (2), which degrades arabino-oligosaccharides produced by the ABN-TS reaction to form the end product l-arabinose (unpublished data). Thus, arabinan is degraded to form arabinose by the cooperation of the above three enzymes in strain TS-3. The two ABFs are thermostable, as is ABN-TS: their half-lives at 75°C (at pH 7.0) are around 4 h (unpublished data). A biochemical reaction at high temperature is favorable in terms of not only preventing microbial infection of the reaction mixture but also accelerating the reaction rate. Thus, our strain seems to be favorable for producing l-arabinose from biomass materials, especially from sugar beet pulp.
Sakai and Sakamoto have described that PPase-C (from B. subtilis IFO 3134) has ABN activity and catalyzes the splitting of the α-1,5-l-arabinofuranoside linkages of the arabinan region in arabinogalactan of sugar beet protopectin, which attaches pectin molecules to the cell wall constituents, so that it releases pectin molecules (12). On the other hand, ABN-TS does not show pectin-releasing activity on sugar beet protopectin, although it shows the same level of activity on soluble arabinan as PPase-C. Based on these data, ABNs could be divided in two groups: one that has protopectinase activity and the other which does not. Although we have little knowledge as to why ABN-TS does not attack the arabinan region in the sugar beet protopectin while the enzyme degrades soluble arabinan from sugar beet, we assume that the molecular structure of the enzyme is unsuitable to react on insoluble substances.
With ABN-TS, to elucidate the relationship between molecular structure and protopectinase activity, we are currently working on its molecular structure. The results obtained in that study will be published elsewhere.
REFERENCES
- 1.Bacic, A., P. J. Harris, and B. A. Stone. 1988. Structure and function of plant cell walls, p. 297-371. In J. Preiss (ed.), The biochemistry of plants, vol. 14. Academic Press, San Diego, Calif. [Google Scholar]
- 2.Beldman, G., H. A. Schols, S. M. Piston, M. J. F. Searl-van Leeuwen, and A. G. J. Voragen. 1997. Arabinans and arabinan degrading enzymes. Adv. Macromol. Carbohydr. Res. 1:1-64. [Google Scholar]
- 3.Gibson, T., and R. E. Gordon. 1974. Genus I. Bacillus, p. 529-550. In R. E. Buchanan and N. E. Gibbons (ed.), Bergey's manual of determinative bacteriology, 8th ed. The Williams & Wilkins Co., Baltimore, Md.
- 4.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 5.Kaji, A., and T. Saheki. 1975. Endo-arabinanase from Bacillus subtilis F-11. Biochim. Biophys. Acta 410:354-360. [DOI] [PubMed] [Google Scholar]
- 6.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 7.Lineweaver, H., and D. Burk. 1934. The determination of enzyme dissociation constants. J. Am. Chem. Soc. 56:658-666. [Google Scholar]
- 8.McKie, V. A., G. W. Black, S. J. Millward-Sadler, G. P. Hazlewood, J. I. Laurie, and H. J. Gilbert. 1997. Arabinanase A from Pseudomonas fluorescens subsp. cellulosa exhibits both an endo- and an exo- mode of action. Biochem. J. 323:547-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramón, D., P. van der Veen, and J. Visser. 1993. Arabinan degrading enzymes from Aspergillus nidulans: induction and purification. FEMS Microbiol. Lett. 113:15-22. [DOI] [PubMed] [Google Scholar]
- 10.Saito, H., and K. Miura. 1963. Preparation of transforming deoxyribonucleic acid by phenol treatment. Biochim. Biophys. Acta 72:619-629. [PubMed] [Google Scholar]
- 11.Sakai, T., and T. Sakamoto. 1990. Purification and some properties of a protopectin-solubilizing enzyme that has potent activity on sugar beet protopectin. Agric. Biol. Chem. 54:879-889. [Google Scholar]
- 12.Sakamoto, T., J. Yoshinaga, T. Shogaki, and T. Sakai. 1993. Studies on protopectinase-C mode of action: analysis of the chemical structure of the specific substrate in sugar beet protopectin and characterization of the enzyme activity. Biosci. Biotechnol. Biochem. 57:1832-1837. [Google Scholar]
- 13.Sakamoto, T., and T. Sakai. 1994. Protopectinase-T: a rhamnogalacturonase able to solubilize protopectin from sugar beet. Carbohydr. Res. 259:77-91. [DOI] [PubMed] [Google Scholar]
- 14.Sakamoto, T., and T. Sakai. 1995. Analysis of structure of sugar-beet pectin by enzymatic methods. Phytochemistry 39:821-823. [DOI] [PubMed] [Google Scholar]
- 15.Seibert, F. B., and J. Anto. 1946. Determination of polysaccharide in serum. J. Biol. Chem. 163:511-522. [PubMed] [Google Scholar]
- 16.Semenza, G., and A.-K. von Balthazar. 1974. Steady-state kinetics of rabbit-intestinal sucrase. Kinetic mechanism, Na+ activation, inhibition by Tris(hydroxymethyl)aminomethane at the glucose subsite. Eur. J. Biochem. 41:149-162. [DOI] [PubMed] [Google Scholar]
- 17.Seri, K., K. Sanai, N. Matsuo, K. Kawakubo, C.-Y. Xue, and S. Inoue. 1996. l-Arabinose selectively inhibits intestinal sucrase in an uncompetitive manner and reduces glycaemic response after sucrose ingestion in animals. Metabolism 45:1368-1374. [DOI] [PubMed] [Google Scholar]
- 18.Somogyi, M. 1952. Note on sugar determination. J. Biol. Chem. 195:19-23. [PubMed] [Google Scholar]
- 19.Takao, M., T. Nakaniwa, K. Yoshikawa, T. Terashita, and T. Sakai. 2000. Purification and characterization of thermostable pectate lyase with protopectinase activity from Thermophilic Bacillus sp. TS 47. Biosci. Biotechnol. Biochem. 64:2360-2367. [DOI] [PubMed] [Google Scholar]
- 20.Takao, M., T. Nakaniwa, K. Yoshikawa, T. Terashita, and T. Sakai. 2001. Molecular cloning, DNA sequence, and expression of the gene encoding for thermostable pectate lyase of thermophilic Bacillus sp. TS 47. Biosci. Biotechnol. Biochem. 65:322-329. [DOI] [PubMed] [Google Scholar]
- 21.Voragen, A. G. J., F. M. Rombouts, M. F. Searle-van Leeuwen, H. A. Schols, and W. Pilnik. 1987. The degradation of arabinans by endo-arabinase and arabinofuranosidases purified from Aspergillus niger. Food Hydrocolloides 1:423-437. [Google Scholar]
- 22.Weinstein, L., and P. Albersheim. 1979. Structure of plant cell walls. Purification and partial characterization of a wall-degrading endo-arabinase and an arabinosidase from Bacillus subtilis. Plant Physiol. 63:425-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yoshihara, O., and A. Kaji. 1983. An endo-1,5-α-l-arabinase, which can disintegrate potato tissue. Agric. Biol. Chem. 47:1935-1940. [Google Scholar]