Abstract
Temporal changes of the bacterioplankton from a meromictic lake (Lake Vilar, Banyoles, Spain) were analyzed with four culture-independent techniques: epifluorescence microscopy, PCR-denaturing gradient gel electrophoresis (DGGE) fingerprinting, fluorescence in situ whole-cell hybridization and flow cytometry sorting. Microscopically, blooms of one cyanobacterium (Synechococcus sp.-like), one green sulfur bacterium (Chlorobium phaeobacteroides-like), and one purple sulfur bacterium (Thiocystis minor-like) were observed at different depths and times. DGGE retrieved these populations and, additionally, populations related to the Cytophaga-Flavobacterium-Bacteroides phylum as predominant community members. The analyses of partial 16S ribosomal DNA sequences from the DGGE fingerprints (550 bp analyzed) revealed higher genetic diversity than expected from microscopic observation for most of these groups. Thus, the sequences of two Synechococcus spp. (both had a similarity of 97% to Synechococcus sp. strain PCC6307 in 16S rRNA), two Thiocystis spp. (similarities to Thiocystis minor of 93 and 94%, respectively), and three Cytophaga spp. (similarities to Cytophaga fermentans of 88 and 89% and to Cytophaga sp. of 93%, respectively) were obtained. The two populations of Synechococcus exhibited different pigment compositions and temporal distributions and their 16S rRNA sequences were 97.3% similar. The two Thiocystis populations differed neither in pigment composition nor in morphology, but their 16S rRNA sequences were only 92.3% similar and they also showed different distributions over time. Finally, two of the Cytophaga spp. showed 96.2% similarity between the 16S rRNA sequences, but one of them was found to be mostly attached to particles and only in winter. Thus, the identity of the main populations changed over time, but the function of the microbial guilds was maintained. Our data showed that temporal shifts in the identity of the predominant population is a new explanation for the environmental 16S rRNA microdiversity retrieved from microbial assemblages and support the hypothesis that clusters of closely related 16S rRNA environmental sequences may actually represent numerous closely related, yet ecologically distinct, populations.
The number of prokaryotes on earth is estimated to be 4 × 1030 to 6 × 1030 cells (56). They constitute, by far, the largest reservoir of life and encompass the major part of earth's physiological and phylogenetic diversity (58). Characterization of the diversity hidden behind this tremendous population size has been, however, chronically limited by the small cell size, cryptic morphology, and low cultivability of microorganisms (39, 41). Fortunately, our perception of microbial diversity has dramatically changed in the last decade by retrieving rRNA genes (mostly 16S ribosomal DNA [rDNA] and 18S rDNA) from the environment, without the need to have pure cultures to examine (for a review, see references 3 and 34). As a result, new rDNA sequences, substantially different from any previously known sequences, have led to the discovery of new phyla of still uncultured microorganisms (26, 31, 38). However, groups of closely related rRNA sequences (with similarities ranging from 90 to 99.9%) also indicate high diversity but at a much smaller scale (microdiversity) (18, 32). Such microheterogeneity in the ribosomal sequences has been observed in a wide range of environments and in genetic libraries obtained from bacteria, archaea, and eukaryotic microorganisms (2, 5, 6, 10, 12, 16, 19, 25, 29, 31, 33). Therefore, this might be a widespread characteristic of microbial populations. However, only a few recent studies focused on the marine cyanobacterium Prochlorococcus and on freshwater Synechococcus provide some clues about the ecological significance of microdiversity in natural populations (32, 43, 44, 54, 55).
The ecological relevance of this small-scale diversity is difficult to establish essentially for two reasons. First, because methodological difficulties that arise in the long process from natural samples to sequences may give a biased view of the microbial assemblage (57). In the worst case, the small-scale diversity in the ribosomal sequences could be due to PCR artifacts or to rRNA multioperons belonging to a single population. In order to circumvent this problem, fluorescence in situ hybridization (FISH) and flow cytometry can be used to “visualize” cells in situ (2, 3, 32). The second caveat is that most of the sequences deposited in databases have been obtained from single samples and, therefore, do not reflect time-depth dynamics of microbial assemblages. In order to compare microbial assemblages and to assess temporal and spatial changes, fingerprinting techniques, such as denaturing gradient gel electrophoresis (DGGE) (36), offer the best alternative. Finally, testing the ecological significance of environmental rDNA-defined populations requires the selection of an appropriate environment to study (see, for example, reference 52).
In the present work, we selected a small, well-known, stratified, and sulfide-rich lake (Lake Vilar) for a survey of the temporal changes of microbial populations with limited 16S rRNA sequence variability. Detailed and frequent long-term sampling can be easily carried out in lakes (42), and many studies have taken place in Lake Vilar over many years by traditional (4, 22-24, 30) and, recently, by molecular RNA-based methods (7-9). These studies have shown that microbial populations are finely adapted to vertical and temporal gradients of physicochemical conditions. These fine gradients are very difficult to mimic in the laboratory, and microorganisms inhabiting these systems are difficult to culture (8, 9). In addition, closely related 16S rDNA sequences had been previously detected in Lake Vilar, providing us with an appropriate natural model system for this study.
MATERIALS AND METHODS
Site and sampling.
Lake Vilar is a small freshwater meromictic lake in the Banyoles karstic system in Girona in northeastern Spain (42°8′N, 2°45′E). The lake consists of two basins with a total surface area of ca. 11,000 m2 and a maximum depth of 9 m. Sulfide (up to 1.5 mM) is present during the whole year, although it is restricted to the deeper, higher conductivity waters. The chemocline is found at 4.5 m. Here, dense populations of photosynthetic sulfur bacteria can develop, provided that sufficient light penetrates. Blooms of phototrophic populations (both oxygenic and anoxygenic) are segregated in depth, along gradients of light and sulfide, and in time. Autofluorescent cyanobacteria (Synechococcus sp.-like), green sulfur bacteria (Chlorobium phaeobacteroides-like), and purple sulfur bacteria (Thiocystis minor-like) were counted by epifluorescence microscopy. Total cells were counted by using DAPI (4′,6′-diamidino-2-phenylindole) staining. In each case, the standard deviation was <10% of the cell count. The lake was sampled fortnightly from 19 February to 5 June 1996, and physicochemical analyses were carried out as described previously (9). Samples for molecular biological analyses were kept in the dark, on ice, for processing in the laboratory 2 to 4 h later.
Nucleic acid analyses, DGGE, and sequencing.
One to five liters of lake water were concentrated by using a refrigerated centrifuge (Sorvall Instruments, DuPont, Del.) at 8,000 × g for 20 min and then stored at −80°C. This method recovered most of the cells (9). Total DNA was extracted and purified by using the hot phenol-sodium dodecyl sulfate method as described previously (9) and was used as target in the PCR to amplify bacterial 16S rRNA genes. Bacterial 550-bp fragments suitable for subsequent DGGE analysis were obtained with the primer combination of 341f with a GC clamp (40-nucleotide GC-rich sequence, 5′-CCT ACG GGA GGC AGC AG-3′) and 907r (5′-CCG TCA ATT CMT TTG AGT TT-3′) (35). Specific primers targeting cyanobacteria and chloroplasts from algae were also used with DGGE conditions reported elsewhere (37). DGGE for bacteria was run as described before (9) for 3.5 h at a constant voltage of 200 V and at 60°C in a 20 to 80% vertical denaturant gradient (the 100% denaturant agent is 7 M urea and 40% deionized formamide). Gels were photographed with UV transillumination after ethidium bromide staining. The pictures were digitalized and analyzed by using the gel plotting macro tool of the NIH Image software package version 1.62 (National Institutes of Health, Bethesda, Md.). After background subtraction, the intensity of each band was measured integrating the area under the peak and was expressed as a percentage of the total area in the profile. The error among replicates was <4%. Several bands were excised from the denaturing gradient gels, reamplified, and purified for sequencing as reported earlier (9). Partial sequences were evaluated by using the basic local alignment search tool (BLAST) (1) on the Internet (http://www.ncbi.nlm.nih.gov) to determine the closest relatives in the database. The new sequences were added to an alignment of full prokaryotic 16S rRNA sequences by using the automated aligning tool of the ARB program package (Technical University of Munich, Munich, Germany [http://www.arb-home.de]). Analysis of sequence similarity was further done by constructing a similarity matrix with the ARB facilities.
Oligonucleotide probes and in situ hybridization.
Thiocystis- and Cytophaga-related populations were counted by FISH. An 18-nucleotide specific sequence was chosen for each of the two Thiocystis populations as a target site by using the appropriate tools of the ARB software package. Probe CHR-452 (5′-GTA TTC GCC ACG CGC TTT-3′) targeted the Thiocystis sp. that was represented by sequence DGGE-6. Optimum hybridization was found at 35% formamide. Probe CHR-626 (5′-GTA TCC ACT GCC GTT CCC-3′) targeted the Thiocystis sp. that was represented by sequence DGGE-8. Optimum hybridization was found at 20% formamide. For Cytophaga-like bacteria, we used probes described in the literature (27, 53). Some of the cytophagas did not give good hybridization signals with the specific probes designed and were targeted with the phylum probe CF319a. Oligonucleotide probes were synthesized with Cy3 fluorochrome at the 5′ end (Interactiva Biotechnologie GmbH, Ulm, Germany). Hybridization and microscopic counts were performed as previously described (21) with an Axiophot II microscope (Zeiss, Jena, Germany).
Flow cytometry and sorting.
Cell sorting for the Synechococcus-like cells was performed as described previously (50). Sorted cells were PCR amplified by using cyanobacterium-specific DGGE primers (37). PCR products were separated in a DGGE gel and sequenced as described above.
The sequences reported for Lake Vilar have been deposited in the EMBL database under the accession numbers AJ240007 to AJ240013 and AJ422234 to AJ422239.
RESULTS
Characterization of the environment and distribution of microbial populations by microscopy.
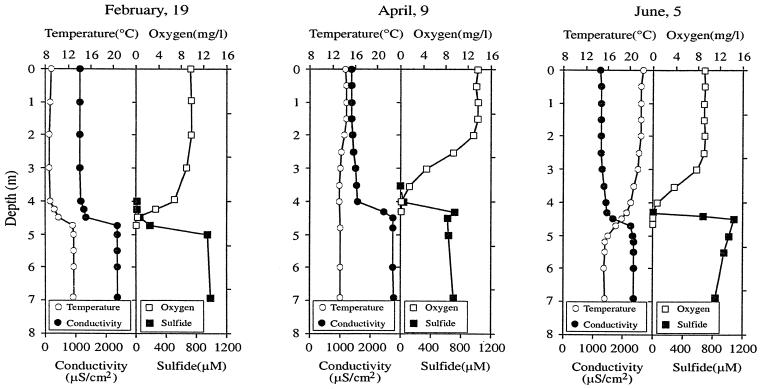
Time-depth changes in temperature, conductivity, oxygen, and sulfide are shown in Fig. 1 for three representative dates in Lake Vilar (in winter, spring, and summer). Additional data are presented in Table 1. During winter the lake had colder water (9°C at surface) overlying warmer, higher-conductivity, bottom water (13°C). Light did not penetrate further than 4.5 m. Oxygen and sulfide showed opposite gradients with coexistence at ca. 4.5 m. In spring, the surface temperature increased and the inverse thermal stratification disappeared. This favored the coexistence of sulfide and light at 4.0 m. High oxygen concentrations were measured during this period in the first 2 m of the lake (14 mg/liter), produced by the photosynthetic activity of algae and cyanobacteria. In June, however, the oxygen concentration decreased (9 mg/liter), the surface water was more transparent, and the sulfide concentration increased up to 1.2 mM in the anoxic compartment.
FIG. 1.
Depth profiles of temperature, conductivity, oxygen, and sulfide from Lake Vilar for three selected dates in 1996.
TABLE 1.
Temperature, conductivity, irradiance, oxygen, sulfide, and DAPI counts from the depths studied in Lake Vilar
| Date (day.mo) | Depth (m) | Temp (°C) | Conductivity (μS cm−2) | Light (μE m−2 s−1) | Concn of:
|
DAPI counts (105 cells/ml)
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| O2 (mg/liter) | H2S (μM) | Synechococcus sp. | Chlorobiaceae | Chromatiaceae | Other prokaryotes | |||||
| 19.02 | 0.20 | 9 | 1,121 | 800 | 9.7 | 0 | 6.20 | BD | BD | 250 |
| 4.75 | 13 | 2,380 | 10 | 0.2 | 190 | 0.32 | 11.3 | BD | 69.3 | |
| 7.00 | 13 | 2,370 | 0 | 0.0 | 986 | 0.28 | 7.0 | BD | 98.2 | |
| 09.04 | 2.00 | 14 | 1,397 | 65 | 13.1 | 0 | 13.2 | BD | BD | 31.4 |
| 4.25 | 13 | 2,430 | <5 | 0.0 | 720 | 4.00 | 41.0 | BD | 130 | |
| 7.00 | 13 | 2,770 | 0 | 0.0 | 707 | 0.61 | 14.2 | BD | 140 | |
| 23.04 | 3.75 | 15 | 1,180 | 16 | 1.4 | 4 | ND | 0.4 | 0.85 | 82.1 |
| 4.25 | 14 | 1,660 | <5 | 0.0 | 709 | ND | 260 | BD | 160 | |
| 7.00 | 14 | 2,040 | 0 | 0.0 | 837 | ND | 43.1 | BD | 122 | |
| 08.05 | 4.15 | 16 | 1,240 | <5 | 1.0 | 140 | ND | 96.3 | 0.96 | 131 |
| 4.40 | 15 | 1,600 | 0 | 0.0 | 550 | ND | 121 | 0.17 | 161 | |
| 21.05 | 4.20 | 18 | 1,480 | ND | 0.2 | 104 | ND | 2.3 | 13.3 | 64.2 |
| 4.40 | 17 | 1,910 | ND | 0.0 | 844 | ND | 180 | 0.17 | 131 | |
| 05.06 | 2.00 | 22 | 1,283 | ND | 9.4 | 0 | 8.61 | BD | BD | 51.3 |
| 4.30 | 19 | 1,467 | ND | 0.1 | 13 | ND | 0.9 | 16.1 | 97.4 | |
ND, not determined; BD, below detection limits.
Conspicuous photosynthetic populations were identified with the microscope (Table 2). In the oxic zone they mostly corresponded to cyanobacteria of the Synechococcus sp. type (autofluorescent, nonmotile, small cocci; concentration up to 106 cells/ml) and algae (Cryptomonas and Crucigenia spp. among others; concentrations up to 103 cells/ml each). In the anoxic zone conspicuous phototrophic organisms were brown-pigmented green sulfur bacteria (Chlorobium phaeobacteroides-like cells; concentration up to 107 cells/ml) and okenone-containing purple sulfur bacteria of the family Chromatiaceae (mainly Thiocystis minor-like cells, concentration up to 106 cells/ml, but also Chromatium weissei- and Chromatium okenii-like cells, concentration 103 cells/ml). These anaerobic photosynthetic populations were vertically segregated along gradients of light and sulfide, i.e., purple sulfur bacteria bloomed above green sulfur bacteria (Table 1). In addition, blooms of each photosynthetic populations were separated in time. Thus, the bloom of Synechococcus sp. occurred in March, followed by the bloom of Chlorobium sp. in April and, finally, followed by the bloom of Thiocystis sp. in May. Therefore, microscopy revealed successive changes in the photosynthetic populations of Lake Vilar involving three distantly related phylogenetic groups: cyanobacteria, green sulfur bacteria, and purple sulfur bacteria.
TABLE 2.
Microbial morphotypes observed microscopically and sequences retrieved which might correspond to them
| Population (genus) | Morphology | Cell size (μm) | DGGE sequence | Closest relative in database | % Similaritya |
|---|---|---|---|---|---|
| Synechococcus | Coccus | 1.5-2.0 | DGGE-2, CYA-1 | Synechococcus sp. strain PCC6307 | 97.1 |
| Synechococcus | Coccus | 1.5-2.0 | DGGE-7, CYA-2 | Synechococcus sp. strain PCC6307 | 98.1 |
| Chlorobium | Rod | 0.8 by 2.0-2.5 | DGGE-5 | Chlorobium phaeobacteroides | 95.2 |
| Thiocystis | Rod | 2.0-4.0 by 4.0-6.0 | DGGE-6 | Thiocystis minor | 93.7 |
| Thiocystis | Rod | 2.0-4.0 by 4.0-6.0 | DGGE-8 | Thiocystis minor | 93.0 |
| Cryptomonas | Chloroplast | 15 by 30 | CYA-3 | Cryptomonas sp. | 95.1 |
| Cryptomonas | Chloroplast | 15 by 30 | CYA-4 | Cryptomonas sp. | 95.2 |
| Pseudanabaena | Filaments | 2.5 by 35 | CYA-5 | Pseudanabaena limnetica | 97.0 |
| Cytophaga | Unknown | Unknown | DGGE-3 | Cytophaga WCHB | 93.3 |
| Cytophaga | Small rod, coccus | 0.5-0.8 | DGGE-1 | Cytophaga fermentans | 88.2 |
| Cytophaga | Unknown | Unknown | DGGE-4 | Cytophaga fermentans | 88.6 |
The percent similarity of the 16S rRNA sequence to the closest relative in the database.
DGGE fingerprints and 16S rDNA sequences.
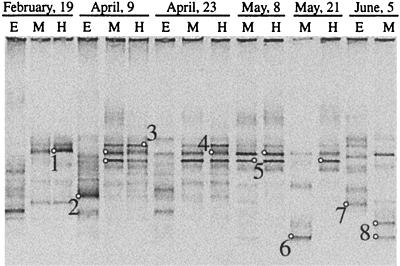
As expected from microscopic observations, the fingerprints showed different banding patterns when the aerobic and the anaerobic assemblages were compared (Fig. 2). Up to 17 bands were observed in the aerobic epilimnion, whereas in the sulfide-rich depths no more than 8 bands were detected. In the hypolimnion, only a few bands (one to three) were very strong. Finally, changes with time in the number and intensity of the bands were more marked in the aerobic than in the anaerobic depths.
FIG. 2.
Negative image of ethidium bromide-stained DGGE gel containing PCR-amplified segments of 16S rRNA genes obtained by using universal bacterial primers. Lanes E, M, and H correspond to the aerobic epilimnion, the oxic-anoxic metalimnion, and the anaerobic rich-sulfide hypolimnion, respectively. Small circles correspond to the bands excised from the gel and then sequenced, and numbers are given for these sequences.
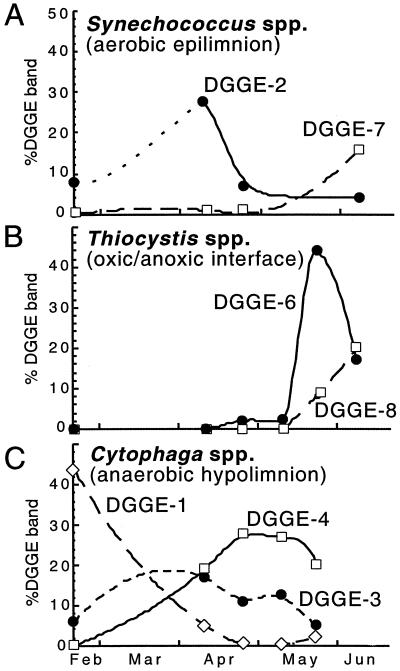
Predominant bands from the gel were excised and sequenced. Bands that were in the same position in the DGGE gel were excised from different lanes and sequenced to confirm that the same position corresponded to the same sequence all along the temporal survey (bands marked with a small round circle in Fig. 2). The 16S rRNA fingerprinting and sequencing approach recovered the predominant microorganisms identified under the microscope (Synechococcus, Chlorobium, and Thiocystis spp.), as well as three sequences belonging to the Cytophaga-Flavobacterium-Bacteroides phylum, as the most abundant sequences (Table 2). In most cases, however, more than one sequence was recovered that was affiliated with the same morphotype. Thus, two different sequences (DGGE-2 and DGGE-7, 97.3% similarity to each other) were related to Synechococcus sp., and two sequences (DGGE-6 and DGGE-8, 92.3% similarity to each other) were related to Thiocystis minor. In addition, two sequences (DGGE-1 and DGGE-4, 96.2% similarity to each other) were related to Cytophaga fermentans and a third sequence (DGGE-3, 91.1% similarity to the former) was related to Cytophaga sp. Only one sequence related to Chlorobium phaeobacteroides (DGGE-5) was recovered. Changes in the relative abundance of such sequences were monitored in time by changes in the intensity of the DGGE bands. Thus, temporal changes for the different sequences of Synechococcus, Thiocystis, and Cytophaga spp. were observed in the aerobic epilimnion, the oxic-anoxic interface, and the anaerobic hipolimnion, respectively (Fig. 3). These data indicated a shift with time in the relative abundance of the different sequences of each morphotype. In order to reject PCR artifacts or the presence of 16S rRNA multioperons in these populations, PCR-independent techniques (i.e., FISH with specific probes and flow cytometry coupled to sorting and further sequencing) were subsequently used.
FIG. 3.
Seasonal changes in the relative contributions of selected DGGE bands to the total intensity for each sample in Fig. 2. (A) Bands from Synechococcus spp. in the aerobic epilimnion; (B) bands from Thiocystis spp. in the oxic-anoxic interface; (C) bands of Cytophaga spp. in the anaerobic rich-sulfide hipolimnion.
FISH counts and flow cytometry sorting.
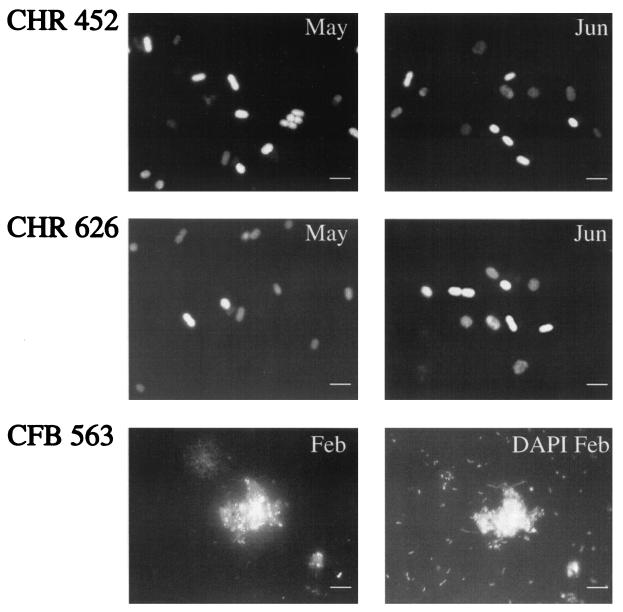
Two different approaches were carried out depending on the population targeted. For the Cytophaga- and Thiocystis-related populations, FISH with specific probes was used. Between 55 and 75% of all prokaryote-shaped particles that were stained with DAPI hybridized with the general bacterial probe EUB338. Fewer than 0.1% of DAPI-stained nontargeted cells (i.e., nontypical Chromatiaceae-like cells) were found to cross-react with the specific probes designed for Thiocystis, and these cells were not considered. FISH demonstrated that the different Thiocystis-related sequences corresponded to different populations and were not a result of methodological problems (Fig. 4). The two Thiocystis populations were rod shaped with similar ranges of cell sizes (Table 2). No shifts in pigment composition (bacteriochlorophyll a and the carotenoid okenone, both found in Thiocystis minor) were detected in spring or in summer after high-pressure liquid chromatography analyses (L. Bañeras, unpublished data). Therefore, both Thiocystis spp. did not differ either in morphology or in pigment composition. The Cytophaga population present in the winter (band DGGE-1) was specifically targeted by probe CFB563, and most of the cells were coccoid and mostly attached to particles (Fig. 4) whereas the cytophagas, which hybridized with the phylum probe CF319a, showed more variable morphology. For instance, filamentous cytophagas were observed to hybridize with probe CF319a but not with probe CFB563 (data not shown).
FIG. 4.
FISH analysis with specific probes for both Thiocystis spp. (CHR-452 and CHR-626; upper and middle panels, respectively) and one Cytophaga sp. (CFB563, lower panels). Bars, 5 μm. Positive Thiocystis cells appear white; autofluorescent negative cells of Thiocystis appear gray. Cytophagas are shown to correspond to the DAPI-stained cells attached to particles.
Temporal shifts detected by DGGE were confirmed after FISH counts. Thus, in the bloom of May ca. 70% of the total Chromatiaceae-shaped cells (i.e., 9 × 105 cells/ml) hybridized with probe CHR-452 (specific for DGGE-6) and ca. 20% (i.e., 2 × 105 cells/ml) hybridized with probe CHR-626 (specific for DGGE-8). In June, however, both Thiocystis spp. contributed to the Chromatiaceae-like assemblage in similar percentages (Table 3). In contrast, the winter Cytophaga population detected in sulfide-rich depths by probe CFB563 (DGGE-1) accounted for 23% (i.e., 2 × 106 cells/ml) of the total cell counts in February but decreased to below detection limits in May (Table 4), in agreement with the decrease in DGGE band intensity (Fig. 3).
TABLE 3.
Thiocystis sp.-related FISH counts in the oxic-anoxic interface of Lake Vilar as determined with the specific probes CHR-452 (DGGE-6) and CHR-626 (DGGE-8)
| Date (day.mo) | Depth (m) | Mean FISH count (104 cells/ml) ± SD (% DAPI)a with:
|
|
|---|---|---|---|
| CHR-452 | CHR-626 | ||
| 19.02 | 4.75 | BD | BD |
| 09.04 | 4.25 | BD | BD |
| 23.04 | 3.75 | 6.0 ± 0.3 (0.7) | BD |
| 08.05 | 4.15 | 8.3 ± 0.2 (0.4) | 0.77 ± 0.03 (0.03) |
| 21.05 | 4.20 | 91.1 ± 3.5 (11.5) | 26.3 ± 4.2 (3.3) |
| 05.06 | 4.30 | 74.2 ± 3.8 (6.5) | 61.2 ± 3.3 (5.4) |
BD, below detection limits. The % DAPI concentration is given in parentheses.
TABLE 4.
Cytophaga sp.-related FISH counts in the sulfide-rich waters of Lake Vilara
| Date (day.mo) | Depth (m) | Mean FISH count (106 cells/ml) ± SD (% DAPI) with:
|
|
|---|---|---|---|
| CF319a | CFB563 | ||
| 19.02 | 7.00 | 3.2 ± 0.7 (30.2) | 2.4 ± 0.8 (22.7) |
| 09.04 | 7.00 | 4.9 ± 0.6 (32.0) | 3.1 ± 0.7 (2.0) |
| 23.04 | 7.00 | 4.7 ± 0.4 (28.7) | 1.6 ± 0.7 (0.9) |
| 08.05 | 4.40 | 3.1 ± 0.3 (11.2) | BD |
CF319a refers to total Cytophaga-like bacteria. CFB563 is a probe specific for the Cytophaga population represented by the DGGE-1 sequence. BD, below detection limits. The % DAPI concentration is given in parentheses.
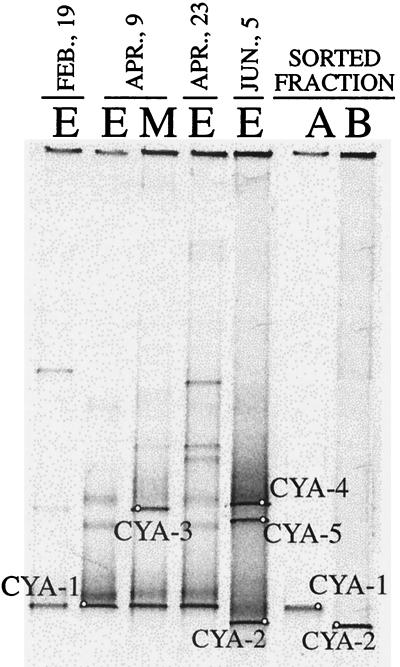
Characterization of Synechococcus spp. was carried out by using flow cytometry and cell sorting based on autofluorescence. Both Synechococcus-like cells showed autofluorescence in the red channel (presence of chlorophyll a), but the summer population (DGGE-7) lacked autofluorescence in the orange channel (phycoerythrin either absent or present in a low concentration). The sorted populations were PCR amplified with specific chloroplast-cyanobacterium primers and were compared by DGGE to the natural oxygenic photosynthetic assemblage of Lake Vilar over time (Fig. 5). The sorted Synechococcus populations of spring and summer yielded a single band each in the DGGE (CYA-1 and CYA-2, respectively) that matched DGGE bands from the natural assemblage. CYA-1 (positive in both the red and the orange channels) was present in winter and spring, and CYA-2 (positive in the red channel and negative in the orange channel) was present mostly in the summer. In addition, the sequence CYA-1 matched the sequence of band DGGE-2, whereas the sequence CYA-2 matched the sequence of band DGGE-7. Thus, the primer combinations 341f-907r for bacteria and 359f-781r for cyanobacteria recovered the same populations. Interestingly, different but closely related 16S rRNA chloroplast sequences were also observed for the alga Cryptomonas sp. in spring (CYA-3) and in summer (CYA-4) (Fig. 5 and Table 2), and the cyanobacterium Pseudanabaena sp. appeared in summer as an important DGGE band (CYA-5) but not in winter or early spring. Therefore, the temporal shift in Synechococcus populations was accompanied also by changes in other members of the oxygenic photosynthetic assemblage.
FIG. 5.
Negative image of ethidium bromide-stained DGGE gel containing PCR-amplified segments of 16S rRNA genes obtained by using cyanobacterium-chloroplast primers. Lanes A and B correspond to the sorted populations from spring and summer, respectively. Identification of the bands excised from the gel is presented in Table 2.
DISCUSSION
The meaning of the small-scale variability in 16S rDNA sequences commonly observed in molecular surveys (such as in hot springs (5, 15), soils (6, 25), oral cavities (10), wastewater treatment plants (2), and ocean waters (16, 19, 29, 33), among others, is a controversial matter. Early evidence of widespread environmental 16S rRNA microheterogeneity was obtained from the first PCR-generated genetic libraries in marine samples. Paradigmatic examples are the SAR 11 cluster (20) and the marine archaea (11, 19). Later on, a combination of molecular and physiological studies on cultured strains and on flow cytometrically sorted field samples of Prochlorococcus sp. demonstrated the physiological and genetic diversity of cyanobacterial populations with a high similarity in their 16S rRNA sequences (13, 32, 43, 44, 48). Further research revealed a specific vertical distribution for such Prochlorococcus populations in natural samples (14, 46, 54, 55), giving some clues on the ecology of these microorganisms. In the present work, we provide new evidence for the ecological significance of environmental 16S rDNA microdiversity. We have shown that the 16S rDNA-defined populations with limited sequence diversity changed with time. Therefore, these data support the hypothesis that clusters of closely related 16S rRNA environmental sequences may actually represent numerous closely related, yet ecologically distinct, populations (18, 51).
In Lake Vilar we first assigned by traditional methods photosynthetic cells to the genus or even to the species level (i.e., Synechococcus sp., Chlorobium phaeobacteroides, and Thiocystis minor), and we studied their temporal dynamics. Populations with no morphologic traits, however, could not be studied in this way and required a molecular approach based on the 16S rRNA gene. DGGE identified such unknown populations and, in addition, revealed that more than one population was hidden behind some single morphotypes that we had distinguished with the microscope. Quantitative analysis of the DGGE band patterns indicated temporal changes in the relative abundance of these populations. However, limitations described for PCR-based techniques might prevent an accurate quantitative estimation of cell abundance (47). For that reason, we were cautious in the number of PCR cycles run, we used the same amount of template in each reaction, and the samples that we compared were all amplified in the same PCR run and were analyzed in the same DGGE gel. Thus, any biases should have been the same for all samples, and the comparison should still be valid. When these populations were counted with specifically designed FISH probes, we found good correlation between microscopic counts and DGGE fingerprints, and both methods showed the same temporal change in the populations. Because Lake Vilar is a very small lake, and one sample at the center of the basin is representative of the pelagic zone of the lake, we can rule out spatial variability due to our sampling strategy. To characterize the vertical profile, our sampling scheme first detected the depths of the chemo- or thermocline, and then we sampled these layers every few centimeters with a special laminar sampler (42). Thus, we could select the depths representative of the epilimnion (which is mixed) the oxic-anoxic interface (where oxygen and sulfide coexist at low concentrations) and the hypolimnion (which is uniform) with precision.
On the other hand, the use of 16S rDNAs for the identification of microorganisms calls for caution because such sequences evolve so slowly that more than one population may have the same sequence (17, 40) and, therefore, closely related 16S rDNA populations could have very different phenotypic attributes. In Lake Vilar we found differences in closely related populations ranging between 8% (Thiocystis spp.) and 3% (Synechococcus spp.). We have to consider, however, that the partial 16S rDNA sequence analyzed (positions 341 to 907, Escherichia coli numbering) included some of the most highly variable regions of this ribosomal gene. Thus, lower differences (close in the best case to 1 and 3%, respectively) are expected when complete 16S rDNAs are compared. In the case of Thiocystis sp., we found that the two genotypes had very similar physiological properties and a very characteristic morphotype (rod-shaped cells up to 6 μm in length, with internal sulfur globules and with bacteriochorophyll a and okenone as carotenoid). Considering the temporal patterns of occurrence for these microorganisms, they were certainly distinct populations. In the case of the two Synechococcus sp. and the two Cytophaga sp., the differences in their 16S rRNA sequences were lower but both had populations with different phenotypes and different temporal distributions. Therefore, the winter-to-summer transition study in Lake Vilar revealed that closely related 16S rDNA populations of Synechococcus, Thiocystis, and Cytophaga distributed differently with time and for some of them we could detect differences in the phenotype. In the case of the green sulfur bacteria, however, we detected only one population of Chlorobium. Nevertheless, we cannot rule out that other related populations of Chlorobium present in the lake were not recovered from the gel (e.g., those that corresponded to weaker bands) or that the organisms might have been present but at lower values than the detection limit of DGGE (9) (i.e., ca. 0.5% of the total DAPI counts).
Temporal population dynamics raises interesting questions about the ecology and physiology of these uncultured microdiverse microbes, and by using a combination of several methodological tools, we could answer them to a certain extent. Thus, both Synechococcus spp. showed different pigment compositions when they were analyzed by flow cytometry (phycoerythrin was absent or present at very low concentration in the summer population), and this might be an indication of a different light-dependent physiology. Changes in nutrient concentrations, however, could also play an important role in selecting one or the other population (43, 55). It is interesting to consider the shift detected also in other members of the oxygenic photosynthetic assemblage. In marine cyanobacteria of the genus Prochlorococcus, small-scale variability in 16S rRNA sequences has been clearly correlated with niche specialization of the different ecotypes (32, 54, 55), and this can also be the case for freshwater cyanobacteria. For Cytophaga populations, the temporal shift was also linked to changes in the habitat colonized (particle-attached versus free-living cells). The case of the purple photosynthetic Thiocystis bacterium is, however, more intriguing. Differences were not found either in morphology or in pigment composition. Thus, the coexistence of both ecotypes might be related to different substrate affinities (49). A better ability to grow under light-limited conditions by increasing the specific pigment content or a better response to fluctuating conditions by increasing the concentration of storage products is also a possible explanation (28). It is extremely difficult to extract further conclusions without having the appropriate pure cultures to examine. Previous molecular studies, however, have revealed that the photosynthetic sulfur bacteria isolated thus far from these environments do not match those that bloom in situ (8, 9). In addition, highly abundant Cytophaga-related organisms found in the anaerobic, sulfide-rich, freshwaters of Lake Vilar indicated that the organisms available in culture collections do not account for the full range of metabolic capabilities present in this group. Therefore, the phenotypic characteristics of the organisms that successfully grow in the field will remain unknown as long as the isolation process is not successful with the naturally abundant strains. Time-depth distribution patterns provided a good idea of the habitat of these microorganisms and under what physicochemical conditions they grow in situ, and this can help in the isolation process.
Overall, we can imagine a natural scenario in which an ecologically unique microbial population becomes predominant for a short period of time but that can easily be outcompeted by a close relative better adapted to newly established environmental conditions. These bacteria could represent a tremendous potential for rapid changes in community composition, since they could grow quickly and multiply over several orders of magnitude under appropriate conditions. Thus, the function carried out by the microbial guild in the environment persists, but the identity of the microbial populations may change with time as an adaptative response to environmental pressures and to fluctuating conditions. This would provide an explanation as to why so many related isolates can be obtained from a natural sample (see, for example, reference 45) or why these isolates do not match sequences of predominant organisms at a certain time (see, for example, reference 9). The successful members of the SAR 11 cluster (20) that comprises the most ubiquitous bacterial gene type recovered from seawater, clusters of marine archaea of cosmopolitan distribution (29), and the multiple ecotypes of Prochlorococcus that colonize a remarkable range of depths in the water column of the world's oceans (54, 55) might owe their ecological success to the versatility and plasticity of the closely related populations that form these groups.
We have shown here that the diversity recovered in Lake Vilar fits this scenario. The small-scale 16S rRNA diversity corresponded to ecologically significant diversity that occurred in several phylogenetically nonrelated groups. This may be a general strategy in the microbial world. Certainly, we need to bring into culture microorganisms that show 16S rRNA small-scale variability but we also have to combine molecular methods and in situ chemical measurements for a better understanding of the rules that govern temporal changes in such closely related populations.
Acknowledgments
This work was funded by the Max-Planck Society and by DGICyT grant PB95-0222 from the Spanish Ministerio de Educación y Cultura. Parts of this work were funded by the MIDAS project (Microbial Diversity in Aquatic Systems; grant MAS3-CT97-0154) from the European Union. E.O.C. benefited also from the Max-Planck Society-CSIC exchange program.
We thank Ramon Rosselló, Enric Llobet, and Hendrik Schäfer for help. Lluis Bañeras and Carles Abellà from the University of Girona provided sampling facilities and shared field data. We thank Bernard Fuchs and Mike Zubkov for help with flow cytometry and sorting analyses and Nyree West for grammar corrections and comments.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Amann, R., J. Snaidr, M. Wagner, W. Ludwig, and K. H. Schleifer. 1996. In situ visualization of high genetic diversity in a natural microbial community. J. Bacteriol. 178:3496-3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bañeras, L., J. Rodriguez-Gonzalez, and L. J. Garcia-Gil. 1999. Contribution of photosynthetic sulfur bacteria to the alkaline phosphatase activity in anoxic aquatic ecosystems. Aquat. Microb. Ecol. 18:15-22. [Google Scholar]
- 5.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borneman, J., P. W. Skroch, K. M. O'Sullivan, J. A. Palus, N. G. Rumjanek, J. L. Jansen, J. Nienhuis, and E. W. Triplett. 1996. Molecular microbial diversity of an agricultural soil in Wisconsin. Appl. Environ. Microbiol. 62:1935-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casamayor, E. O., G. Muyzer, and C. Pedrós-Alió. 2001. Composition and temporal dynamics of planktonic archaeal assemblages from anaerobic sulfurous environments studied by 16S rDNA denaturing gradient gel electrophoresis and sequencing. Aquat. Microb. Ecol. 25:237-246. [Google Scholar]
- 8.Casamayor, E. O., M. T. Núñez-Cardona, J. I. Calderón-Paz, J. Mas, and C. Pedrós-Alió. 2000. Comparison of pure cultures and natural assemblages of planktonic photosynthetic sulfur bacteria by low molecular mass RNA fingerprinting. FEMS Microbiol. Ecol. 32:25-34. [DOI] [PubMed] [Google Scholar]
- 9.Casamayor, E. O., H. Schäfer, L. Bañeras, C. Pedrós-Alió, and G. Muyzer. 2000. Identification of and spatio-temporal differences between microbial assemblages from two neighboring sulfurous lakes: comparison by microscopy and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 66:499-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi, B. K., B. J. Paster, F. E. Dewhirst, and U. B. Goebel. 1994. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect. Immun. 62:1889-1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Díez, B., C. Pedrós-Alió, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ernst, A., P. Marschall, and C. Postius. 1995. Genetic diversity among Synechococcus spp. (cyanobacteria) isolated from the pelagial of Lake Constance. FEMS Microbiol. Ecol. 17:197-203. [Google Scholar]
- 14.Ferris, M. J., and B. Palenik. 1998. Niche adaptation in ocean cyanobacteria. Nature 396:226-228. [Google Scholar]
- 15.Ferris, M. J., G. Muyzer, and D. M. Ward. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl. Environ. Microbiol. 62:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field, K. G., D. Gordon, T. Wright, M. Rappe, E. Urbach, K. Vergin, and S. J. Giovannoni. 1997. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl. Environ. Microbiol. 63:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fox, G. E., J. D. Wisotzkey, and P. Jurtshuk, Jr. 1992. How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity. Int. J. Syst. Bacteriol. 42:166-170. [DOI] [PubMed] [Google Scholar]
- 18.Fuhrman, J. A., and L. Campbell. 1998. Microbial microdiversity. Nature 393:410-411. [Google Scholar]
- 19.Fuhrman, J. A., K. McCallum, and A. A. Davis. 1992. Novel major archaebacterial group from marine plankton. Nature 356:148-149. [DOI] [PubMed] [Google Scholar]
- 20.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 21.Glöckner, F. O., R. Amann, A. Alfreider, J. Pernthaler, R. Psenner, K. Trebesius, and K. H. Schleifer. 1996. An in situ hybridization protocol for detection and identification of planktonic bacteria. Syst. Appl. Microbiol. 19:403-406. [Google Scholar]
- 22.Guerrero, R., C. Pedrós-Alió, I. Esteve, and J. Mas. 1987. Communities of phototrophic sulfur bacteria in lakes of the Spanish Mediterranean region. Acta Acad. Abo. 47:125-151. [Google Scholar]
- 23.Guerrero, R., E. Montesinos, C. Pedrós-Alió, I. Esteve, J. Mas, H. van Gemerden, P. A. G. Hofman, and J. F. Bakker. 1985. Phototrophic sulfur bacteria in two Spanish lakes: vertical distribution and limiting factors. Limnol. Oceanogr. 30:919-931. [Google Scholar]
- 24.Guerrero, R., E. Montesinos, I. Esteve, and C. Abellà. 1980. Physiological adaptation and growth of purple and green sulfur bacteria in a meromictic lake (Vilà) as compared to a holomictic lake (Sisó), p. 161-171. In M. Dokulil, H. Metz, and D. Jewson (ed.), Shallow lakes. Contribution to their limnology. Developments in hydrobiology, vol. 3. Dr. W. Junk Publishers, The Hague, The Netherlands. [Google Scholar]
- 25.Liesack, W., and E. Stackebrandt. 1992. Occurrence of novel groups of the domain Bacteria as revealed by analysis of genetic material isolated from an Australian terrestrial environment. J. Bacteriol. 174:5072-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.López-García, P., F. Rodríguez-Valera, C. Pedrós-Alió, and D. Moreira. 2001. Unexpected diversity of small eukaryotes in deep-sea Antarctic plankton. Nature 409:603-607. [DOI] [PubMed] [Google Scholar]
- 27.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 28.Mas, J., and H. van Gemerden. 1995. Storage products in purple and green sulfur bacteria, p. 973-990. In R. E. Blankenship, M. T. Madigan, and C. E. Bauer (ed.), Anoxygenic photosynthetic bacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 29.Massana, R., E. F. DeLong, and C. Pedrós-Alió. 2000. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl. Environ. Microbiol. 66:1777-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montesinos, E., R. Guerrero, C. Abellà, and I. Esteve. 1983. Ecology and physiology of the competition between Chlorobium limicola and Chlorobium phaeobacteroides. Appl. Environ. Microbiol. 46:1007-1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon-van der Staay, S. Y., R. De Wachter, and D. Vaulot. 2001. Oceanic 18S rDNA sequences from picoplankton reveal unsuspected eukaryotic diversity. Nature 409:607-610. [DOI] [PubMed] [Google Scholar]
- 32.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 33.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 34.Muyzer, G. 1998. Structure, function, and dynamics of microbial communities: the molecular biological approach, p. 87-117. In G. R. Carvalho (ed.), Advances in molecular ecology. IOS Press, Amsterdam, The Netherlands.
- 35.Muyzer, G., T. Brinkhoff, U. Nübel, C. Santegoeds, H. Schäfer, and C. Wawer. 1998. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 36.Muyzer, G., and K. Smalla. 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Leeuwenhoek 73:127-141. [DOI] [PubMed] [Google Scholar]
- 37.Nübel, U., F. García-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 39.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Ecol. 9:1-55. [Google Scholar]
- 40.Palys, T., L. K. Nakamura, and F. M. Cohan. 1997. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 47:1145-1156. [DOI] [PubMed] [Google Scholar]
- 41.Pedrós-Alió, C. 1993. Diversity of bacterioplankton. Trends Ecol. Evol. 8:86-90. [DOI] [PubMed] [Google Scholar]
- 42.Pedrós-Alió, C., and R. Guerrero. 1993. Microbial ecology in Lake Cisó. Adv. Microb. Ecol. 13:155-209. [Google Scholar]
- 43.Postius, C., and A. Ernst. 1999. Mechanisms of dominance: coexistence of picocyanobacterial genotypes in a freshwater ecosystem. Arch. Microbiol. 172:69-75. [DOI] [PubMed] [Google Scholar]
- 44.Postius, C., U. Kenter, A. Wacker, A. Ernst, and P. Boger. 1998. Light causes selection among two phycoerythrin-rich Synechococcus isolates from Lake Constance. FEMS Microbiol. Ecol. 25:171-178. [Google Scholar]
- 45.Sass, H., E. Wieringa, H. Cypionka, H. D. Babenzien, and J. Overmann. 1998. High genetic and physiological diversity of sulfate-reducing bacteria isolated from an oligotrophic lake sediment. Arch. Microbiol. 170:243-251. [DOI] [PubMed] [Google Scholar]
- 46.Scanlan, D. J., W. R. Hess, F. Partensky, J. Newman, and D. Vaulot. 1996. High degree of genetic variation in Prochlorococcus (Prochlorophyta) revealed by RFLP analysis. Eur. J. Phycol. 31:1-9. [Google Scholar]
- 47.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Urbach, E., and S. W. Chisholm. 1998. Genetic diversity in Prochlorococcus populations flow cytometrically sorted from the Sargasso Sea and Gulf Stream. Limnol. Oceanogr. 43:1615-1630. [Google Scholar]
- 49.van Gemerden, H. 1974. Coexistence of organisms competing for the same substrate: an example among the purple sulfur bacteria. Microb. Ecol. 1:104-119. [DOI] [PubMed] [Google Scholar]
- 50.Wallner, G., B. Fuchs, S. Spring, W. Beisker, and R. Amann. 1997. Flow sorting of microorganisms for molecular analysis. Appl. Environ. Microbiol. 63:4223-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ward, D. M. 1998. A natural species concept for prokaryotes. Curr. Opin. Microbiol. 1:271-277. [DOI] [PubMed] [Google Scholar]
- 52.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 53.Weller, R., F. O. Glöckner, and R. Amann. 2000. 16S rRNA-targeted oligonucleotide probes for the in situ detection of members of the phylum Cytophaga-Flavobacterium-Bacteroides. Syst. Appl. Microbiol. 23:107-114. [DOI] [PubMed] [Google Scholar]
- 54.West, N. J., W. A. Schonhuber, N. J. Fuller, R. I. Amann, R. Rippka, A. F. Post, and D. J. Scanlan. 2001. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147:1731-1744. [DOI] [PubMed] [Google Scholar]
- 55.West, N. J., and D. J. Scanlan. 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65:2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Whitman, W. B., D. C. Coleman, and W. J. Wiebe. 1998. Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. USA 95:6578-6583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wintzingerode, F. V., U. B. Goebel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-229. [DOI] [PubMed] [Google Scholar]
- 58.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]