Glucose, xylose, and N-acetylglucosamine are elementary building blocks for disaccharides, oligomers, and polymers, as well as key molecules allowing fine modifications of many kinds of biological compounds. The highly diverse molecules containing these three sugars are ubiquitous components of the architecture of eukaryotic and prokaryotic cells and may also be involved in signaling between cells or organisms (all of these roles are illustrated in this minireview). Any enzymes modifying the structure of these molecules, such as glycosylhydrolases (GHs) or glycosyltransferases, are essential to modulate the biological properties of these glycosides and therefore are likely to modify their functions in vivo. Based on the comparison of their amino acid sequences, GHs are presently classified in more than eighty families (20). Among them, family-3 encompasses more than one hundred enzymes, including eubacterial and eukaryotic β-glucosidases, β-xylosidases, and β-N-acetylhexosaminidases.
Most of the cloned and sequenced genes encoding family-3 GHs, as well as the purified family-3 GHs, were investigated as encoding β-glucosidases and therefore characterized as such in cellulolytic and xylanolytic microorganisms. This bias led scientists to think, originally, that these enzymes were involved mainly in the degradation of macromolecules by microbes. Recent data on their functions in vivo (Table 1) and their distribution among sequenced genomes (Fig. 1) suggest that family-3 GHs play roles in addition to the assimilation of plant polymer products. This prompted me to select several well-studied family-3 GHs to investigate their functions in vivo. These functions range from the assimilation of exogenous saccharides by bacteria and fungi to the turnover of cell architecture components such as cell wall polymers and from the modification of biologically active molecules such as antibiotics and antifungal compounds to interactions between pathogens and the immune systems of their hosts.
TABLE 1.
Main functions of the family-3 GHs
| Functions | Enzyme(s) | Organism | Reference |
|---|---|---|---|
| Assimilation of glycosides | |||
| Cellobiose and cellodextrins | CelA | Azospirillum irakense | 13 |
| Laminaribiose and laminarins | BglB | Thermotoga neapolitana | 69 |
| Salicin (aryl-β-glucoside) | SalA, SalB | Azospirillum irakense | 12 |
| Xylooligosaccharides | XlnD | Aspergillus niger | 61 |
| Chitooligomers | NagA, | Streptomyces thermoviolaceus | 60 |
| Cht60 | Alteromonas sp. | 59 | |
| Recycling of cell components | |||
| Muropeptides from peptidoglycan | NagZ | Escherichia coli | 8 |
| β-1,3-Glucans from cell wall | Bgl2 | Coccidioides immitis | 25 |
| β-1,3- and β-1,4-glucans from cell wall | Exg1 | Zea mays | 29 |
| Xyloglucans from seedlings | TMA7501 | Tropaeolum majus | 9 |
| Modifications of free glycosides | |||
| Transglycosylation to produce cellulase inducer, sophorose | Bgl1 | Trichoderma reesei | 34 |
| Deglycosylation | |||
| To activate antibiotic, oleandomycin | OleR | Streptomyces antibioticus | 51 |
| To detoxify saponins | Avenacinase | Gaeumannomyces graminis | 3 |
| B2Tom | Septoria lycopersici | 53 | |
| Sap1 | Botrytis cinerea | 48 |
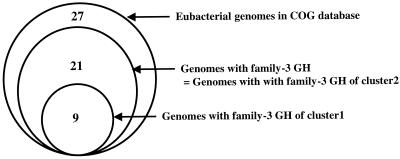
FIG. 1.
Occurrence of family-3 GHs in eubacterial genomes. Of the 27 eubacterial genomes of the COG database (57), 21 contain at least one putative open reading frame belonging to COG1472, i.e., family-3 GHs. A brief phylogenetic analysis of this COG (http://www.ncbi.nlm.nih.gov/COG) revealed two clusters: cluster 1 includes β-glucosidases and β-xylosidases, whereas N-acetylglucaminidases are in cluster 2. Surprisingly, the occurrence of cluster 1 is lower than that of cluster 2. Moreover, all genomes harboring family-3 GHs contain at least one GH of cluster 2, suggesting that a selective constraint preferentially maintained the N-acetylglucaminidase cluster among eubacterial genomes.
THE FAMILY-3 GHS
The β-glucosidases (EC 3.2.1.21), β-xylosidases (EC 3.2.1.37), and β-N-acetylhexosaminidases (EC 3.2.1.52) remove successive β-d-glucose, β-d-xylose, and β-N-acetylglucosamine residues, respectively, from the nonreducing termini. These GHs do more than cleave simple homodisaccharides (13), -oligomers (13, 61), and -polymers (23). Indeed, several enzymes can also remove glycosidic units from heterogeneous molecules such as xyloglucans (9), arylglycosides (7, 12), a glucosylated antibiotic (49), or saponins (3). The substrates of these enzymes are described in Table 2 and Fig. 2. Many family-3 GHs exhibit a combination of different activities, exemplified by the frequent association of β-glucosidase and β-xylosidase activities (4, 12, 63, 64). Such a feature complicates the identification of their natural substrates as well as their denomination since the latter is based on the hydrolytic capacities of these enzymes. However, in an attempt to facilitate the predictive analysis of the open reading frames of the sequenced genomes, a collection of clusters of orthologous genes (COGs) was constructed (57). Because the classification of GHs (http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html) and that of COGs (http://www.ncbi.nlm.nih.gov/COG) are based on sequence comparisons, it is not surprising that a COG matches a GH family, as is the case for COG1472 and family-3 GHs. Both databases provide efficient analytical and molecular tools to clearly identify a new member of this phylogenetic cluster. It is noteworthy that the three-dimensional structure of one representative member of this family, a β-glucan hydrolase from barley, was recently elucidated (62) and used as a reference to predict the three-dimensional structure of other family-3 GHs (19).
TABLE 2.
Standard substrates of family-3 GHs
| Released residue | Substratesa |
|---|---|
| β-d-Glucose | Cellobiose {[Glc (1→4)]2}, gentiobiose {[Glc (1→6)]2}, sophorose {[Glc (1→2)]2}, laminaribiose {[Glc (1→3)]2}, aryl-β-glucosides (such as salicin,∗ arbutin,∗ pNPG,∗ MUG,∗ etc.), 1,4-β-glucans {[Glc (1→4)]n}, 1,3-β-glucans {[Glc (1→3)]n} |
| β-d-Xylose | Xylobiose {[Xyl (1→4)]2}, 1,4-β-xylans {[Xyl (1→4)]n}, pNPX, MUX |
| β-N-Acetylglucosamine | N,N′-Diacetylchitobiose {[Xyl (1→4)]2}, chitosaccharides {[Xyl (1→4)]n}, pNPGNac, MUGNac |
Other substrates, as well as the chemical structure of the glycosides with an asterisk, are shown in Fig. 2. pNPG, p-nitrophenyl-β-d-glucopyranoside; pNPX, p-nitrophenyl-β-d-xylopyranoside; pNPGNac, p-nitrophenyl-N-acetyl-β-d-glucosaminide; MUG, methylumbelliferyl-β-d-glucopyranoside; MUX, methylumbelliferyl-β-d-xylopyranoside; MUGNac, methylumbelliferyl-N-acetyl-β-d-glucosaminide (these six are chromogenic substrates).
FIG. 2.
Examples of substituted glycosides as substrates of family-3 GHs. The diversity of the substituted glycosides that are hydrolyzed by family-3 GHs is illustrated. Plant-derived compounds such as salicin, arbutin, coniferin, and avenacin-1, as well as the glucosylated antibiotic oleandomycin, are cleaved by fungal or bacterial enzymes (3, 7, 12, 49). The fungal elicitors shown are putative substrates of family-3 GHs, as discussed in the text.
DEGRADATION AND ASSIMILATION OF EXOGENOUS GLYCOSIDES
Historically, the first function that was proposed for bacterial and fungal family-3 β-glucosidases was their involvement in the last steps of the cellulose degradation pathway (2), a feature further extended to the assimilation of xylan and chitin derivatives. The assimilative family-3 GHs may be extracellular, periplasmic, or cytoplasmic and take part in the assimilation of cellobiose and cellodextrins (13), β-1,4-xylosides (61), and acetylchitooligomers (59, 60), as well as that of aryl-β-glucosides (12) and β-1,3-glucosides (69). The direct contribution of family-3 GHs to the assimilation of glycosides is often difficult to demonstrate because of the presence of several enzymes exhibiting a similar activity in the same organism. The family-1 β-glucosidases, the family-39, -43, and -52 β-xylosidases, and the family-20 N-acetylglucosaminidases may also contribute to the assimilation of these glycosides by microbes. In this respect, the use of fine molecular tools, such as site-specific mutagenesis and analysis of gene expression, seems to be an obligatory approach to investigate the involvement of family-3 GHs in assimilative functions. In the particular case of the hydrolysis of heterogeneous glycosides, the fate of the aglycones, which can exhibit cell toxicity properties—as exemplified by cyanogenic glucosides (65)—remains to be clarified.
These assimilative pathways are often controlled by a catabolic repression exerted by glucose (47, 60). Such a negative control is not observed in the case of the salCAB operon of Azospirillum irakense (56). In this bacterium, the pathway of assimilation of salicin encodes enzymes defining a novel pathway for the assimilation of aryl-β-glucosides in bacteria (14). This original system lacks inner membrane transporters of aryl-β-glucosides that are usually present in the assimilative pathways containing family-1 GH (5, 11, 32, 55) but contains a salC gene encoding an outer membrane transporter exhibiting dual functions. Such a protein may act as (i) a highly sensitive receptor to detect and transport aryl-β-glucosides present at low concentrations in the environment but also as (ii) a sensor to further positively activate gene expression through a cascade of specific proteins. In addition to its assimilative function, this pathway may be implied in chemotaxis of Azospirillum through plant-derived aryl-β-glucosides.
RECYCLING AND REMODELING OF CELLULAR COMPONENTS
Cell wall recycling by family-3 GHs was recently demonstrated in the case of an N-acetylglucosaminidase of Escherichia coli. During each generation, about 40% of the cell wall murein is broken down to anhydro-muropeptides. These molecules are transported into the cytoplasm via the AmpG permease and rapidly degraded by the combined action of several enzymes, including the family-3 β-N-acetylglucosaminidase NagZ (27, 46). NagZ hydrolyzes the β-1,4 glycosidic bond between N-acetylglucosamine and anhydro-N-acetylmuramic acid (8). From amino acid sequence comparisons, proteins homologous to NagZ appear to be conserved in several gram-negative bacteria in which they would be expected to exhibit a similar housekeeping function. Such a conserved function may explain why the distribution of N-acetylglucosaminidases among bacterial genomes may be biased when compared with that of β-glucosidases and β-xylosidases (Fig. 1). In addition, NagZ can cleave p-nitrophenyl-N-acetyl-β-d-glucosaminide but does not hydrolyze N,N′-diacetylchitobiose. This feature is consistent with the presence of a specific catalytic pathway for the hydrolysis and assimilation of N,N′-diacetylchitobiose in E. coli, in which the phospho-chitobiase, ChbF, belongs to the family-4 GHs (28).
Two other remarkable works reported the stage-specific expression of family-3 β-glucosidases in the filamentous fungus Coccidioides immitis (25) and in the amoeba Dictyostelium discoideum during a cell differentiation process (6). Convergent evidence about the hydrolytic properties of Bgl2 of C. immitis, such as its cell wall localization, its stage-specific expression during the parasitic cycle, and the use of a β-glucosidase-specific inhibitor to block cell growth (25, 33), suggests that this family-3 β-glucosidase plays a crucial role in the cell wall morphogenesis and/or recycling of cell wall β-1,3-glucans. In the case of the β-glucosidase of D. discoideum, fewer data on its hydrolytic capacities are available but its lysosomal compartmentalization and its time-specific expression during the starvation-induced differentiation of D. discoideum into a multicellular aggregate suggest that this enzyme may be a putative recycling function of cell components (6). It should be emphasized that the Bgl2 protein of C. immitis exhibits highly antigenic properties. Therefore, the detection of Bgl2 antibodies appears to be a useful immunodiagnostic test for coccidioidomycosis (33). A glycosylated family-3 β-glucosidase, named antigen H, is also one of the major antigens present in the culture filtrate of the pathogenic fungus Histoplasma capsulatum (15, 16).
In plants, the implication of family-3 enzymes in cell wall turnover has also been investigated. A β-glucosidase, Exg1, was purified and immunolocalized in the shoots of maize seedlings (29). Exg1 hydrolyzes different disaccharides (β-1,3-, β-1,4-, β-1,2-, and β-1,6-), and exhibits an exo-β-d-glucanase activity towards β-1,3- and β-1,4-oligosaccharides. This developmentally regulated enzyme seems to be involved in the turnover of β-1,3- and β-1,4-glucans. Exg1 could also take part, together with endoglucanase (40), in the assembly of cellulose-hemicellulose during cell growth. Interestingly, a gene encoding a family-3 β-glucosidase was discovered downstream of the cellulose synthase operon of the cellulose-producing proteobacterium Acetobacter xylinus (58). While the role of a secreted β-1,4-endoglucanase in cellulose fiber formation was already demonstrated in this bacterium (31), the part that the family-3 β-glucosidase plays in this process is still unknown and should be regarded with attention.
Finally, in addition to their role in turnover and assembly of cell wall components, the family-3 enzymes may be involved, in concert with a set of different hydrolases, in the postgermination mobilization of the xyloglucan stored in grains of many dicotyledonous seeds. Purified from the cotyledons of germinated Tropaeolum majus seedlings, the β-glucosidase TMA7501 hydrolyzes β-1,3-, β-1,4-, β-1,2- and β-1,6-diglucosides and cellooligosaccharides and in vitro contributes to the total degradation of xyloglucan oligosaccharides, in conjunction with β-d-galactosidase and α-xylosidase (9). A similar function is also hypothesized for two family-3 exo-β-d-glucanases from barley. These two enzymes, ExoI and ExoII, were purified from 8-day-old plants and were extensively characterized (23, 24, 62), but their precise location in cell tissue remains unknown.
MODIFYING THE BIOLOGICAL ACTIVITY OF FREE GLYCOSIDES
Three well-studied models describe the role of family-3 enzymes in the interaction between the organisms and their environment via the modification of the biological activity of self-produced or exogenous glycosides. The first model is related to the production of antibiotic by bacteria of the genus Streptomyces. During the biosynthesis of the macrolide oleandomycin by Streptomyces antibioticus, an intracellar glycosyltransferase, OleI, inactivates the newly synthetized oleandomycin by the addition of a single glucose unit. Thereafter, the glycosylated oleandomycin is excreted, and an extracellular family-3 β-glucosidase, OleR, releases the active form of the antibiotic (49, 50, 51). These glycosylation and hydrolysis steps are therefore involved in the self-resistance mechanism of S. antibioticus during oleandomycin biosynthesis. A similar function has been proposed for the family-3 β-glucosidase DesR in Streptomyces venezuelae (66). Surprisingly, in Saccharopolyspora erythraea, the eryBI gene, encoding a family-3 β-glucosidase, is not involved in the biosynthesis of erythromycin A despite its position within the biosynthesis gene cluster (18). An alternative mechanism of self-resistance may therefore exist.
In the second system, the fungus Trichoderma reesei modifies the structure of cellulose-derived glucosides to generate sophorose, an inducer of the expression of cellulolytic enzymes. The cellulolytic system of T. reesei is complex. In addition to two cellobiohydrolases and four endoglucanases, a cell-associated β-glucosidase and an extracellular β-glucosidase are expressed in T. reesei. The excreted β-glucosidase Bgl1 belongs to the family-3 enzymes. A Bgl1-deficient strain is still able to grow on several carbon sources, such as cellulose and cellobiose; nevertheless, a significant lag is observed for the cellulase induction (17, 34), suggesting that the enzyme is involved in a regulatory function rather than in an assimilative function. The cellulase inducer, sophorose, is supposed to be formed from cellooligosaccharides by the transglycosylation activity of β-glucosidase. The transglycosylation activity has also been reported in other family-3 GHs (9, 64). Besides Bgl1, T. reesei excretes another family-3 enzyme, a β-d-xylosidase/α-l-arabinofuranosidase (21, 35).
In the last example, the substrates of the family-3 GHs are plant-derived saponins. Saponins are glycosylated triterpenoids, steroids, or steroidal alkaloids that are present constitutively in many plant species and have potent antifungal activity (44, 45). Several phytopathogenic fungi are resistant to saponins because they inactivate them by deglycosylation. The first gene encoding a saponin-detoxifying enzyme, termed avenacinase, was cloned from Gaeumannomyces graminis. This avenacinase is a family-3 β-glucosidase that hydrolyzes the saponin avenacin A-1 (Fig. 2). A fungal mutant lacking avenacinase is not able to infect the saponin-producing host oat (3). A tomato leaf-infecting fungus, Septoria lycopersici, also excretes a family-3 β-glucosidase, B2Tom, which specifically acts on saponin α-tomatine produced by tomato, and therefore was named tomatinase (43, 53). Tomatinase-defective mutants are more sensitive to α-tomatine than the wild-type Septoria lycopersici, but no obvious consequence of the disruption of the tomatinase-encoding gene was observed upon analysis of the macroscopic symptoms of Septoria lycopersici infection in tomato leaves (36). Nevertheless, the expression of tomatinase in Nectria haematococca resulted in its ability to detoxify α-tomatine and to parasitize green tomato fruit, an ability not shared by the wild-type N. haematococca (54). A third pathogen, Botrytis cinerea, contains at least three distinct saponin-detoxifying glycosidase activities, one of which is characterized as the family-3 β-glucosidase Sap1. A sap1 mutant, which has lost the ability to deglycosylate avenacin, is still able to hydrolyze tomatin, digitonin, and avenacosides (48). It should be emphasized that not all of the saponin-detoxifying enzymes belong to family-3. The saponin-hydrolyzing enzyme excreted by Fusarium oxysporum f. sp. lycopersici belongs to the family-10 GHs, in which are clustered many fungal xylanases (52). Another enzyme, an α-rhamnosidase that is secreted by Stagonospora avenae, suffices to inactivate the saponin 26-desglucoavenacosides. This fungus also produces a family-3 β-glucosidase that releases glucose units from the α-rhamnosidase-inactivated form of saponin (39). In this case, participation of this GH in glucose assimilation was suggested but its involvement in the hydrolysis of other, unknown, compounds should not be excluded.
In addition to these models, the role of family-3 GHs as signal-modifying enzymes has been suggested, but still not demonstrated, in the case of the phytopathogen Agrobacterium tumefaciens for the modification of virulence inducers (7, 38), such as coniferin (Fig. 2). Biotechnologically oriented research also investigates the modifying activity of β-glucosidase to produce economically relevant aglycones or to modify the characteristics of flavor molecules (22, 26, 30, 70).
EMERGING FIELDS FOR STUDY OF THE FAMILY-3 GHS IN HOST-MICROBE INTERACTIONS
The interest in the family-3 enzymes may be illustrated by recent publications in the fast-moving field of host-microbe interactions. In the case of animal models, a purified protein, STI, from Salmonella enterica serovar Typhimurium that causes systemic infection in mice has been identified as an inhibitor of T-cell responsiveness to interleukin-2 (1). The protein STI is a family-3 GH and shows high homologies to BglX from E. coli (37), the function of which is still unknown (68). The mechanism of this puzzling link between a family-3 GH and the suppression of T-cell proliferation remains to be clarified and should also be investigated in the case of BglX in E. coli. Interestingly, another recent publication reported that one of the most antigenic proteins of Histoplasma capsulatum may be used to immunize mice and protect them from intranasal infection with this pathogenic fungus (10). This protein is a family-3 β-glucosidase, the amino acid sequence of which is closely related to that of the immunoreactive β-glucosidase Bgl2 of Coccidioides immitis. This feature suggests that Bgl2 or other family-3 GHs, which are implicated in the morphogenesis of pathogenic fungi, should be assayed during immunization procedures. On the other hand, in the field of plant-microbe interactions, fungal elicitors, such as β-1,3-glucosides (67) and N-acetylchitooligomers (41), are potential substrates for family-3 enzymes (Fig. 2). These cell wall-derived compounds activate the plant defenses at a nanomolar level, but their length is crucial in retaining this biological activity. The family-3 enzymes may play a key role in the modulation of the plant elicitor response by modifying the structure and the concentration of these signals. Recently, a family-3 GH was identified as one of the six major proteins secreted from cultured tobacco cells (42), and the degradation of fungal elicitors by plant β-glucosidases was observed in suspension cultures of rice plant cells (67). The impact of these plant enzymes on the elicitor response remains to be investigated.
CONCLUSIONS
By modifying the structure of essential glycosides, the family-3 GHs take part in the housekeeping functions of eukaryotic and prokaryotic organisms, as well as in their interactions with the environment. These functions are highly diverse, and more sound information is required in some instances, especially in the case of cell wall remodeling and host-microbe interactions. Still, the functions of these GHs cannot be predicted by the simple elucidation of a substrate range in vitro or by the comparison of amino acid sequences. It is noteworthy that many organisms contain more than one family-3 GH, as exemplified by the thirteen putative family-3 GHs that were discovered in the genome of Arabidopsis thaliana. In this respect, increased knowledge of the physiological roles of representative members of the GH family-3 in model organisms, as well as of the relationships between the three-dimensional structure and catalytic properties, is still required before there can be any predictive analysis of the role of family-3 GHs emerging from genomic databases.
Acknowledgments
I thank Y. Dessaux and S. Ross for critical reading of the manuscript.
REFERENCES
- 1.Arai, T., and K. Matsui. 1997. A purified protein from Salmonella typhimurium inhibits high-affinity interleukin-2 receptor expression on CTLL-2 cells. FEMS Immunol. Med. Microbiol. 17:155-160. [DOI] [PubMed] [Google Scholar]
- 2.Béguin, P. 1990. Molecular biology of cellulose degradation. Annu. Rev. Microbiol. 44:219-248. [DOI] [PubMed] [Google Scholar]
- 3.Bowyer, P., B. R. Clarke, P. Lunness, M. J. Daniels, and A. E. Osbourn. 1995. Host range of a plant pathogenic fungus determined by a saponin detoxifying enzyme. Science 267:371-374. [DOI] [PubMed] [Google Scholar]
- 4.Breeves, R., K. Bronnenmeier, N. Wild, F. Lottspeich, W. L. Staudenbauer, and J. Hofemeister. 1997. Genes encoding two different β-glucosidases of Thermoanaerobacter brockii are clustered in a common operon. Appl. Environ. Microbiol. 63:3902-3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown, G. D., and J. A. Thomson. 1998. Isolation and characterization of an aryl-β-glucoside uptake and utilization system (abg) from gram-positive ruminal Clostridium species C. longisporum. Mol. Gen. Genet. 257:213-218. [DOI] [PubMed] [Google Scholar]
- 6.Bush, J., J. Richardson, and J. Cardelli. 1994. Molecular cloning and characterization of the full-length cDNA encoding the developmentally regulated lysosomal enzyme β-glucosidase in Dictyostelium discoideum. J. Biol. Chem. 269:1468-1476. [PubMed] [Google Scholar]
- 7.Castle, L. A., K. D. Smith, and R. O. Morris. 1992. Cloning and sequencing of an Agrobacterium tumefaciens β-glucosidase gene involved in modifying a vir-inducing plant signal molecule. J. Bacteriol. 174:1478-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng, Q., H. Li, K. Merdek, and J. T. Park. 2000. Molecular characterization of the β-N-acetylglucosaminidase of Escherichia coli and its role in cell wall recycling. J. Bacteriol. 182:4836-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crombie, H. J., S. Chengappa, A. Hellyer, and J. S. Grant Reid. 1998. A xyloglucan oligosaccharide-active, transglycosylating β-d-glucosidase from the cotyledons of nasturtium (Tropaeolum majus L.) seedlings—purification, properties and characterization of a cDNA clone. Plant J. 14:27-38. [DOI] [PubMed] [Google Scholar]
- 10.Deepe, G. S., Jr., and R. Gebbons. 2001. Protective efficacy of H antigen from Histoplasma capsulatum in a murine model of pulmonary histoplasmosis. Infect. Immun. 69:3128-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Hassouni, M., B. Henrissat, M. Chippaux, and F. Barras. 1992. Nucleotide sequence of the arb genes, which control β-glucoside utilization in Erwinia chrysanthemi: comparison with the Escherichia coli bgl operon and evidence for a new β-glycohydrolase family including enzymes from eubacteria, archeabacteria, and humans. J. Bacteriol. 174:765-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faure, D., J. Desair, V. Veijers, M. A. Bekri, P. Proost, B. Henrissat, and J. Vanderleyden. 1999. Growth of Azospirillum irakense KBCI on the aryl β-glucoside, salicin, requires either SalA or SalB. J. Bacteriol. 181:3003-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Faure, D., B. Henrissat, D. Ptacek, A. Bekri, and J. Vanderleyden. 2001. The celA gene, encoding a glycoside hydrolase family 3 β-glucosidase, is required for optimal growth of Azospirillum irakense on cellodextrins. Appl. Environ. Microbiol. 67:2380-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Faure, D., M. H. Saier, Jr., and J. Vanderleyden. 2001. An evolutionary alternative system for aryl-β-glucosides assimilation in bacteria. J. Mol. Microbiol. Biotechnol. 3:467-470. [PubMed] [Google Scholar]
- 15.Fisher, K. L., G. S. Deepe, and J. P. Woods. 1999. Histoplasma capsulatum strain variation in both H antigen production and β-glucosidase activity and overexpression of HAG1 from a telomeric linear plasmid. Infect. Immun. 67:3312-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fisher, K. L., and J. P. Woods. 2000. Determination of β-glucosidase enzymatic function of the Histoplasma capsulatum H antigen using a native expression system. Gene 247:191-197. [DOI] [PubMed] [Google Scholar]
- 17.Fowler, T., and R. D. Brown, Jr. 1992. The bgl1 gene encoding extracellular β-glucosidase from Trichoderma reesei is required for rapid induction of the cellulase complex. Mol. Microbiol. 6:3225-3235. [DOI] [PubMed] [Google Scholar]
- 18.Gaisser, S., G. A. Böhm, M. Doumith, M. C. Raynal, N. Dhillon, J. Cortés, and P. F. Leadlay. 1998. Analysis of eryBI, eryBIII and eryBVII from the erythromycin biosynthetic gene cluster in Saccharopolyspora erythaea. Mol. Gen. Genet. 258:78-88. [DOI] [PubMed] [Google Scholar]
- 19.Harvey, A. J., M. Hrmova, R. De Gori, J. N. Varghese, and G. B. Fincher. 2000. Comparative modeling of the three-dimensional structures of family 3 glycoside hydrolases. Proteins 41:257-269. [DOI] [PubMed] [Google Scholar]
- 20.Henrissat, B., and A. Bairoch. 1996. Updating the sequence-based classification of glycosyl hydrolases. Biochem. J. 316:695-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermann, M. C., M. Vrsanska, M. Jurickova, J. Hirsch, and C. P. Kubicek. 1997. The β-D-xylosidase of Trichoderma reesei is a multifunctional β-D-xylan xylohydrolase. Biochem. J. 321:375-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hessler, P. E., P. E. Larsen, A. I. Constantinou, K. H. Schram, J. M. Weber. 1997. Isolation of isoflavones from soy-based fermentations of the erythromycin-producing bacterium Saccharopolyspora erythraea. Appl. Microbiol. Biotechnol. 47:398-404. [DOI] [PubMed] [Google Scholar]
- 23.Hrmova, M., A. J. Harvey, J. Wang, N. J. Shirley, G. P. Jones, B. A. Stones, P. B. Hoj, and G. B. Fincher. 1996. Barley β-D-glucan exohydrolases with β-D-glucosidase activity. J. Biol. Chem. 271:5277-5286. [DOI] [PubMed] [Google Scholar]
- 24.Hrmova, M., and G. B. Fincher. 1998. Barley β-D-glucan exohydrolases. Substrate specificity and kinetic properties. Carbohydr. Res. 305:209-221. [Google Scholar]
- 25.Hung, C. Y., J. J. Yu, P. F. Lehmann, and G. T. Cole. 2001. Cloning and expression of the gene which encodes a tube precipitin antigen and wall-associated β-glucosidase of Coccidioides immitis. Infect. Immun. 69:2211-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwashita, K., T. Nagahara, H. Kimura, M. Takano, H. Shimoi, and K. Ito. 1999. The bglA gene of Aspergillus kawachii encodes both extracellular and cell wall-bound β-glucosidases. Appl. Environ. Microbiol. 65:5546-5553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normak, and J. Trak. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keyhani, N. O., and S. Roseman. 1997. Wild-type Escherichia coli grows on the chitin disaccharide, N,N′-diacetylchitobiose, by expressing the cel operon. Proc. Natl. Acad. Sci. USA 94:14367-14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim, J. B., A. T. Olek, and N. C. Carpita. 2000. Cell wall and membrane-associated exo-β-D-glucanases from developing maize seedlings. Plant Physiol. 123:471-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kitamoto, N., S. Yoshino, K. Ohmiya, and N. Tsukagoshi. 1999. Sequence analysis, overexpression, and antisense inhibition of a β-xylosidase gene xylA, from Aspergillus oryzae KBN616. Appl. Environ. Microbiol. 65:20-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koo, H. M., S. H. Song, Y. R. Pyun, and Y. S. Kim. 1998. Evidence that a beta-1,4-endoglucanase secreted by Acetobacter xylinum plays an essential role for the formation of cellulose fiber. Biosci. Biotechnol. Biochem. 62:2257-2259. [DOI] [PubMed] [Google Scholar]
- 32.Krüger, S., and M. Hecker. 1995. Regulation of the putative bglPH operon for aryl-β-glucoside utilization in Bacillus subtilis. J. Bacteriol. 177:5590-5597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kruse, D., and G. T. Cole. 1992. A seroreactive 120-kilodalton β-1,3-glucanase of Coccidioides immitis which may participate in spherule morphogenesis. Infect. Immun. 60:4350-4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mach, R. L., B. Seiboth, A. Myasnikov, R. Gonzalez, J. Strauss, A. M. Harkki, and C. P. Kubicek. 1995. The bgl1 gene of Trichoderma reesei QM 9414 encodes an extracellular, cellobiose-inducible β-glucosidase involved in cellulase induction by sophorose. Mol. Microbiol. 15:687-697. [DOI] [PubMed] [Google Scholar]
- 35.Margolles-Clark, E., M. Tenkanen, T. Nakari-Setälä, and M. Penttilä. 1996. Cloning of genes encoding α-L-arabinofuranosidase and β-xylosidase from Trichoderma reesei by expression in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 62:3840-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martin-Hernandez, A. M., D. Dufresne, V. Hugovieux, R. Melton, and A. Osbourn. 2000. Effects of targeted replacement of the tomatinase gene on the interaction of Septoria lycopersici with tomato plants. Mol. Plant-Microbe Interact. 12:1301-1311. [DOI] [PubMed] [Google Scholar]
- 37.Matsui, K., K. Nagano, T. Arai, I. Hirono, and T. Aoki. 1998. DNA sequencing of the gene encoding Salmonella typhimurium-derived T-cell inhibitor (STI) and characterization of the gene product, cloned STI. FEMS Immunol. Med. Microbiol. 22:341-349. [DOI] [PubMed] [Google Scholar]
- 38.Morris, J. W., and R. O. Morris. 1990. Identification of an Agrobacterium tumefaciens virulence gene inducer from the pinaceous gymnosperm Pseudotsuga menziesii. Proc. Natl. Acad. Sci. USA 87:3614-3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morrissey, J. P., J. P. Wubben, and A. E. Osbourn. 2000. Stagonospora avenae secretes multiple enzymes that hydrolyze oat leaf saponins. Mol. Plant-Microbe Interact. 13:1041-1052. [DOI] [PubMed] [Google Scholar]
- 40.Nicol, F., I. His, A. Jumeau, S. Verhettes, H. Canut, and H. Höfte. 1998. A plasma membrane-bound putative endo-1,4-β-D-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17:5563-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishizawa, Y., A. Kawakami, T. Hibi, D. Y. He, N. Shibuya, and E. Minami. 1999. Regulation of the chitinase gene expression in suspension-cultured rice cells by N-acetylchitooligosaccharides: differences in the signal transduction pathways leading to the activation of elicitor-responsive genes. Plant Mol. Biol. 39:907-914. [DOI] [PubMed] [Google Scholar]
- 42.Okushima, Y., N. Koizumi, T. Kusano, and H. Sano. 2000. Secreted proteins of tobacco cultured BY2 cells: identification of a new member of pathogenesis-related proteins. Plant Mol. Biol. 42:479-488. [DOI] [PubMed] [Google Scholar]
- 43.Osbourn, A., P. Bowyer, P. Lunness, B. Clarke, and M. Daniels. 1995. Fungal pathogens of oat roots and tomato leaves employ closely related enzymes to detoxify different host plant saponins. Mol. Plant-Microbe Interact. 8:971-978. [DOI] [PubMed] [Google Scholar]
- 44.Osbourn, A. E. 1996. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 8:1821-1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papadopoulou, K., R. E. Melton, M. Leggett, M. J. Daniels, and A. E. Osbourn. 1999. Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 96:12923-12928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, J. T. 2001. Identification of a dedicated recycling pathway for anhydro-N-acetylmuramic acid and N-acetylglucosamine derived from Escherichia coli cell wall murein. J. Bacteriol. 183:3842-3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Gonzalez, J. A., N. N. van Peij, A. Bezoen, A. P. MacCabe, D. Ramon, and L. H. de Graaff. 1998. Molecular cloning and transcriptional regulation of the Aspergillus nidulans xlnD gene encoding a β-xylosidase. Appl. Environ. Microbiol. 64:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Quidde, T., P. Büttner, and P. Tudzynski. 1999. Evidence for three different specific saponin-detoxifying activities in Botrytis cinerea and cloning and functional analysis of a gene coding for a putative avenacinase. Eur. J. Plant Pathol. 105:273-283. [Google Scholar]
- 49.Quiros, L. M., C. Hernandez, and J. A. Salas. 1994. Purification and characterization of an extracellular enzyme from Streptomyces antibioticus that converts inactive glycosylated oleandomycin into the active antibiotic. Eur. J. Biochem. 222:129-135. [DOI] [PubMed] [Google Scholar]
- 50.Quiros, L. M., and J. A. Salas. 1995. Biosynthesis of the macrolide oleandomycin by Streptomyces antibioticus. J. Biol. Chem. 270:18234-18239. [DOI] [PubMed] [Google Scholar]
- 51.Quiros, L. M., I. Aguirrezabalaga, C. Olano, C. Mendez, and J. A. Salas. 1998. Two glycosyltransferase and a glycosidase are involved in oleandomycin modification during its biosynthesis by Streptomyces antibioticus. Mol. Microbiol. 28:1177-1185. [DOI] [PubMed] [Google Scholar]
- 52.Roldan-Arjona, T., A. Pérez-Espinosa, and M. Ruiz-Rubio. Tomatinase from Fusarium oxysporum f.sp. lycopersici defines a new class of saponinases. Mol. Plant-Microbe Interact. 12:852-861. [DOI] [PubMed]
- 53.Sandrock, R. W., D. DellaPenna, and H. D. VanEtten. 1995. Purification and characterization of β2-tomatinase, an enzyme involved in the degradation of α-tomatine and isolation of the gene encoding β2-tomatinase from Septoria lycopersici. Mol. Plant-Microbe Interact. 8:960-970. [DOI] [PubMed] [Google Scholar]
- 54.Sandrock, R. W., and H. D. VanEtten. 2001. The relevance of tomatinase activity in pathogens of tomato: disruption of the β2-tomatinase gene in Colletotrichum coccodes and Septoria lycopersici and heterologous expression of the Septoria lycopersici β2-tomatinase in Nectria haematococca, a pathogen of tomato fruit. Physiol. Mol. Plant Pathol. 58:159-171. [Google Scholar]
- 55.Schnetz, K., C. Toloczyki, and B. Rak. 1987. β-Glucoside (bgl) operon of Escherichia coli: nucleotide sequence, genetic organization, and possible evolutionary relationship to regulatory components of two Bacillus subtilis genes. J. Bacteriol. 169:2579-2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Somers, E., V. Keijers, M. H. Ottoy, M. Srinivasan, J. Vanderleyden, and D. Faure. 2000. The salCAB operon of Azospirillum irakense, required for growth on salicin, is negatively regulated by SalR, a transcriptional regulator of the LacI/GalR family. Mol. Gen. Genet. 263:1038-1046. [DOI] [PubMed] [Google Scholar]
- 57.Tatusov, R. L., M. Y. Galperin, D. A. Natale, and E. V. Koonin. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 28:33-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tonouchi, N., N. Tahara, Y. Kojima, T. Nakai, F. Sakai, T. Hayashi, T. Tsuchida, and F. Yoshinaga. 1997. A β-glucosidase gene downstream of the cellulose synthase operon in cellulose-producing Acetobacter. Biosci. Biotechnol. Biochem. 61:1789-1790. [DOI] [PubMed] [Google Scholar]
- 59.Tsujibo, H. T., K. Fujimoto, H. Tanno, K. Miyamoto, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1994. Gene sequence, purification and characterization of N-acetyl-β-glucosaminidase from a marine bacterium, Alteromonas sp. strain O-7. Gene 146:111-115. [DOI] [PubMed] [Google Scholar]
- 60.Tsujibo, H. T., N. Hatano, T. Mikami, A. Hirasawa, K. Miyamoto, and Y. Inamori. 1998. A novel β-N-acetylglucosaminidase from Streptomyces thermoviolaceus OPC-520: gene cloning, expression and assigment to family 3 of the glycosyl hydrolases. Appl. Environ. Microbiol. 64:2920-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Peij, N. N., J. Brinkmann, M. Vrsanska, J. Visser, and L. H. de Graaff. 1997. β-Xylosidase activity, encoded by xlnD, is essential for complete hydrolysis of xylan by Aspergillus niger but not for induction of the xylanolytic enzyme spectrum. Eur. J. Biochem. 245:164-173. [DOI] [PubMed] [Google Scholar]
- 62.Varghese, J. N., M. Hrmova, and G. B. Fincher. 1999. Three-dimensional structure of a barley β-D-glucan exohydrolase, a family 3 glycosyl hydrolase. Structure 7:179-190. [DOI] [PubMed] [Google Scholar]
- 63.Vroemen, S., J. Heldens, C. Boyd, B. Henrissat, and N. T. Keen. 1995. Cloning and characterization of the bgxA gene from Erwinia chrysanthemi D1 which encodes a β-glucosidases/xylosidase enzyme. Mol. Gen. Genet. 246:465-477. [DOI] [PubMed] [Google Scholar]
- 64.Watt, D. K., H. Ono, and K. Hayashi. 1998. Agrobacterium tumefaciens β-glucosidase is also an effective β-xylosidase, and has a high transglycosylation activity in the presence of alcohols. Biochim. Biophys. Acta 1385:78-88. [DOI] [PubMed] [Google Scholar]
- 65.Wulff-Strobel, C. R., and D. B. Wilson. 1995. Cloning, sequencing, and characterization of a membrane-asociated Prevotella ruminicola B14 β-glucosidase with cellodextrinase and cyanoglycosidase activities. J. Bacteriol. 177:5884-5890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue, Y., L. Zhao, H. W. Liu, and D. H. Sherman. 1998. A gene cluster for macrolide biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 93:12111-12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yamaguchi, T., A. Yamada, N. Hong, T. Ogawa, T. Ishii, and N. Shibuya. 2000. Differences in the recognition of glucan elicitor signals between rice and soybean: β-glucan fragments from the rice blast disease fungus Pyricularia oryzae that elicit phytoalexin biosynthesis in suspension-cultured rice cells. Plant Cell 12:817-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yang, M., S. M. Luoh, A. Goddard, D. Reilly, W. Henzel, and S. Bass. 1996. The bglX gene located at 47.8 min on the Escherichia coli chromosome encodes a periplasmic β-glucosidase. Microbiology 143:1659-1665. [DOI] [PubMed] [Google Scholar]
- 69.Zverlov, V. V., I. Y. Volkov, T. V. Velikodvorskaya, and W. H. Schwarz. 1997. Thermotoga neapolitana bglB gene, upstream of lamA, encodes a highly thermostable β-glucosidase that is a laminaribiase. Microbiology 143:3537-3542. [DOI] [PubMed] [Google Scholar]
- 70.Zverlov, V. V., C. Hertel, K. Bronnenmeier, A. Hroch, J. Kellermann, and W. H. Schwarz. 2000. The thermostable α-L-rhamnosidase RamA of Clostridium stercorarium: biochemical characterization and primary structure of a bacterial α-L-rhamnoside hydrolase, a new type of inverting glycoside hydrolase. Mol. Microbiol. 35:173-179. [DOI] [PubMed] [Google Scholar]