Abstract
Four strains of Pseudomonas aeruginosa (wild type, ΔpilHIJK mutant, lasI mutant, and rpoS mutant) were genetically tagged with the green fluorescent protein, and the development of flow chamber-grown biofilms by each of them was investigated by confocal laser scanning microscopy. The structural developments of the biofilms were quantified by the computer program COMSTAT (A. Heydorn, A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbøll, and S. Molin, Microbiology 146:2395-2407, 2000). Two structural key variables, average thickness and roughness, formed the basis for an analysis of variance model comprising the four P. aeruginosa strains, five time points (55, 98, 146, 242, and 314 h), and three independent rounds of biofilm experiments. The results showed that the wild type, the ΔpilHIJK mutant, and the rpoS mutant display conspicuously different types of temporal biofilm development, whereas the lasI mutant was indistinguishable from the wild type at all time points. The wild type and the lasI mutant formed uniform, densely packed biofilms. The rpoS mutant formed densely packed biofilms that were significantly thicker than those of the wild type, whereas the ΔpilHIJK mutant formed distinct microcolonies that were regularly spaced and almost uniform in size. The results are discussed in relation to the current model of P. aeruginosa biofilm development.
Adhesion of bacteria to surfaces and subsequent proliferation and exopolymer production lead to formation of surface-associated bacterial communities called biofilms. Over the past few years, genetic components required for the formation of single-species bacterial biofilms have become subjects of investigation. In Pseudomonas aeruginosa, flagella have been shown to be essential for the initial attachment to abiotic surfaces, and twitching motility has been suggested to be necessary for microcolony formation and thus necessary for normal biofilm development (23). Twitching motility depends on polar type IV pili, which are also known to play a crucial role in mediating adherence to and colonization of mucosal surfaces (10). The mechanical basis for this flagellum-independent type of surface motility is believed to be extension and retraction of the type IV pili, which propel the cells along the surface (19, 35). Cell-to-cell communication has also been shown to be important for the development of P. aeruginosa biofilms. A lasI mutant unable to synthesize the autoinducer N-(3-oxododecanyl)-l-homoserine lactone formed a flat, undifferentiated biofilm. However, when synthetic N-(3-oxododecanyl)-l-homoserine lactone was added to the medium, the lasI mutant formed a differentiated biofilm similar to that of the wild type, consisting of microcolonies separated by water channels (6). Finally, there are indications that the stationary-phase sigma factor RpoS is involved in the control of biofilm growth in P. aeruginosa (11). When grown in flow chambers, an rpoS mutant developed significantly thicker biofilms than the wild-type strain. Taken together, these reports suggest that biofilm formation is a complex process involving several different cellular functions.
Biofilm experiments are performed in many different ways, and no standardized setup for biofilm experiments exists (15, 24, 25). Many different physical factors are known to influence biofilm formation (for a review, see reference 32), and it is therefore difficult to present general models of biofilm development based on reports from different laboratories. This may be a major cause of some of the contradicting reports on biofilm research. Furthermore, the development of biofilm structures is, to a certain extent, a stochastic process and independent rounds of biofilm experiments never result in exact structural copies, even if the experimental conditions are kept constant. We found it of interest to initiate a quantitative characterization of some of the key cellular functions (i.e., twitching motility, cell-to-cell signaling, and RpoS) reported to be involved in P. aeruginosa biofilm formation under well-defined, reproducible conditions. For this purpose, we tagged four isogenic strains of P. aeruginosa (the wild type and the ΔpilHIJK, rpoS, and lasI mutants) with the green fluorescent protein (GFP) and investigated the development of flow chamber-grown biofilms by confocal laser scanning microscopy (CLSM). Biofilm development was quantified by the computer program COMSTAT (12) and analyzed in a bivariate analysis of variance model including the four P. aeruginosa strains, five time points (55, 98, 146, 242, and 314 h), and three independent rounds of biofilm experiments.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
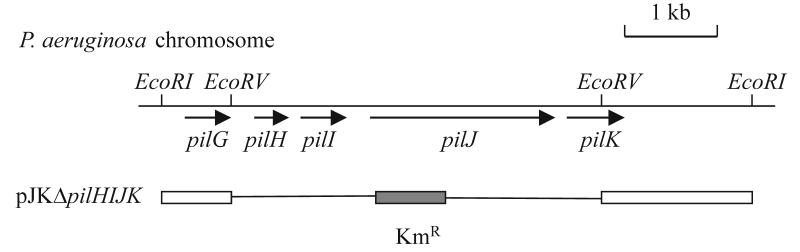
The strains used were P. aeruginosa PAO1 (13), P. aeruginosa lasI (26), and P. aeruginosa rpoS (36). P. aeruginosa ΔpilHIJK was constructed by amplifying the pilHIJ genes from genomic DNA of P. aeruginosa PAO1 by using the forward primer 5′-CCGTTGATGCTTATTTCGTGATGGGG-3′ and the reverse primer 5′-AGTTTGAAGCCGGATACCGAGTTGC-3′. A 6.4-kb EcoRI fragment containing the P. aeruginosa pilGHIJK genes was cloned into pUC118 by using the amplified pilHIJ genes as a probe. The resulting plasmid was digested with EcoRV and ligated with a HindII-flanked kanamycin resistance gene cassette from pUC4K (Pharmacia) to construct pJKΔpilHIJK (Fig. 1). pJKΔpilHIJK was introduced into P. aeruginosa PAO1 by electroporation (17), and kanamycin-resistant transformants were selected on LB medium (27) containing 1.5 g of kanamycin per liter. Disruption of the chromosomal pilHIJK genes was confirmed by Southern hybridization. The ability of ΔpilHIJK mutant and wild-type P. aeruginosa to perform twitching motility was assessed by stabbing cells into a 2-mm-thick LB agar plate (1.5% agar) with a toothpick and incubating the plate for 48 h at 37°C. Strains proficient in twitching motility are characterized by a light haze of growth at the agar-plate interface surrounding the colony (18).
FIG. 1.
Construction of a P. aeruginosa ΔpilHIJK mutant strain. A schematic diagram showing the pilGHIJK gene cluster on the P. aeruginosa PAO1 chromosome and relevant restriction sites for EcoRI and EcoRV is presented. Also shown is the region of pJKΔpilHIJK that contains P. aeruginosa DNA with a Kmr cartridge replacing part of the pilGHIJK gene cluster (described in Materials and Methods).
A PA1/04/03-gfp-T0-T1 transposon cassette was inserted into the chromosome of the four strains by using pUTtc as the delivery plasmid for the wild type, the rpoS mutant, and the ΔpilHIJK mutant and pUTtl as the delivery plasmid for the lasI mutant (7, 28). The cassette was cloned into the NotI sites of the pUT plasmids and mobilized from Escherichia coli CC118 λpir to the recipients by using the helper strain E. coli HB101(RK600). Exconjugants with mini-Tn5 cassettes inserted into the chromosome were selected on a 1:1 mixture of Pseudomonas isolation agar (Difco) and L agar supplemented with tetracycline (80 μg/ml) or tellurite (150 μg/ml). The selected GFP-tagged strains showed no sign of phenotypic changes compared to the parental strains when tested in liquid medium or flow chamber biofilms.
Cultivation of biofilms.
Biofilms were grown at 30°C in flow chambers with individual channel dimensions of 1 by 4 by 40 mm supplied at a flow of 3 ml h−1 with modified FAB medium supplemented with 0.1 mM sodium citrate (12). The flow system was assembled and prepared as described previously (3). Cultures for inoculation of the flow channels were prepared by inoculating a single colony from a plate into test tubes containing modified FAB medium supplemented with 20 mM Na-citrate and growing them at 30°C for 16 h. Cultures were diluted to an optical density at 600 nm of 0.1 in 0.9% NaCl and used for inoculation. A 350-μl volume of diluted culture was injected into each flow channel with a small syringe. After inoculation, flow channels were left at 30°C for 1 h. Medium flow was then started at a constant rate of 3 ml h−1 with a Watson Marlow 205S peristaltic pump. The mean flow velocity in the flow cells was 0.2 mm s−1, which corresponds to laminar flow with a Reynolds number of 0.02. Bacterial growth upstream of the flow channels was removed daily in order to avoid the influence of upstream biomass on the flow chamber biofilms. Cultivation of the biofilms was done carefully, and measures were taken to promote experimental reproducibility (for details, see reference 11).
Image acquisition.
Three independent rounds of biofilm experiments were performed. In each round, each of the four P. aeruginosa strains was grown in two separate channels. In each of these eight channels, nine image stacks were acquired at 55, 98, 146, 242, and 314 h after inoculation. Accidentally, no images were acquired for the P. aeruginosa rpoS mutant at the 98-h time point in the second round. All microscopic observations and image acquisitions were performed by CLSM (TCS4D; Leica Lasertechnik, GmbH, Heidelberg, Germany). Images were obtained with a 40×/0.75 air objective. Image scanning was carried out with the 488-nm laser line from an Ar-Kr laser. Simulated fluorescence projections and sections through the biofilms were generated with the IMARIS software package (Bitplane AG, Zürich, Switzerland) running on a Silicon Graphics Indigo2 workstation (Silicon Graphics, Mountain View, Calif.).
Image processing by COMSTAT.
Images from each experiment were analyzed by the computer program COMSTAT (12). A fixed threshold value and connected volume filtration were used for all image stacks.
Statistical model for analysis of data.
There are several reasons why an analysis of variance model should be used to analyze the present experiments. Essentially, by taking into account the inherent variability in biofilm experiments, an analysis of variance model allows us to compare biofilm structures in an objective manner and to make valid statements about the temporal development of each biofilm (for details, see reference 11).
Using lowercase and uppercase letters for fixed or random factors, respectively, the experiment factors are as follows: bacterial strain, b (fixed factor at four levels: 1, wild type; 2, rpoS mutant; 3, ΔpilHIJK mutant; 4, lasI mutant); experimental round, R (random factor at three levels: 1, 2, and 3); time, t (fixed factor at five levels: 55, 98, 146, 242, and 314 h). For each combination of b, R, and t, there are two repetitions, i.e., two flow channels. Each repetition comprises an average of nine pairs of values of average thickness and roughness, calculated by COMSTAT from nine image stacks acquired in each flow channel. In order to account for the correlation between consecutive measurements, time was included by analyzing the experiment as a repeated-measures analysis of variance (9). Here we used the simplest possible dependency between consecutive time points, which is a first-order autoregressive [AR(1)] structure. An AR(1) structure has a fixed correlation between neighboring time points (9). In order to fully profit from the multivariate nature of the measured variables, we subjected average thickness and roughness collectively to a multivariate analysis of variance. Thus, the response variable in the analysis of variance becomes bivariate and can be represented by an observation vector. In order to stabilize the variances, the data were transformed by taking the logarithm (loge) prior to analysis (for details, see reference 11 ). The model thus becomes
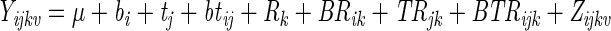 |
where Yijkv is the observed multivariate value (average thickness, roughness) for bacterial strain i, time j, experiment round k, and repetition ν. μ is the overall mean value of the experiment, bi is the additional effect of bacterial strain i, tj is the additional effect of time j, btij is the interaction between the bacterial strain and the time, Rk is the random effect of experiment round k, BRik is the random effect of a possible interaction between bacterial strain i and experiment round k, TRjk is the random effect of a possible interaction between time j and experiment round k, and BTRijk is the random effect of a possible interaction among bacterial strain i, time j, and experiment round k. Zijkν is the random effect of replication, i.e., the residual. The assumptions on the parameters are as follows:
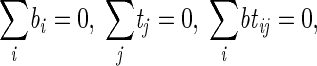 |
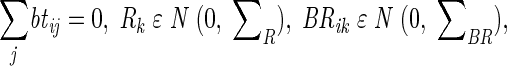 |
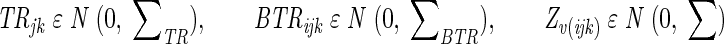 |
Furthermore, all random effects are assumed to be independent of each other, both within and between effects. The covariance matrices ΣR, ΣBR, ΣTR, and ΣBTR are structured with nonzero elements on the main diagonal and zero elements elsewhere. However, the covariance matrix for the residual Σ is unstructured and can contain nonzero elements everywhere. Finally, the residuals for each experimental unit (flow channel) are assumed to be correlated in time according to an AR(1) structure. This means that the residuals for the same flow channel between two consecutive time points (e.g., from 55 to 98 h or from 142 to 242 h) have correlation ρ. Between 55 and 142 h (or 98 and 242 h), which are nonconsecutive as there is a time point between them, the correlation is assumed to have decreased to ρ2, etc. This implies that a flow channel over the five time points has a covariance structure as follows:
 |
where ⊗ denotes the Kroenecker product of two matrices (16).
A significant effect b means the bacterial strains show overall differences. A significant effect t means that the averages of the strains show overall differences over time. Perhaps most interestingly, a significant effect bt means that the strains develop differently. If an effect is significant, pairwise tests can be used to determine which levels are significantly different. In the univariate situation, this corresponds to pairwise t tests, i.e., performance of pairwise t tests for each variable in turn. In order to perform simultaneous pairwise tests, the multivariate equivalent of the pairwise t test, i.e., Hotelling's T2, must be used. To our knowledge, no commercially available standard statistical software package is able to perform this part of the analysis in a truly multivariate fashion. However, an equivalent and very intuitive way of locating the simultaneous differences is by plotting, e.g., 90% confidence ellipses around the mean values. Nonoverlapping ellipses would indicate significant differences. Note that, in general, it is not possible to replace a multivariate test with multiple univariate tests; this would correspond to forcing the confidence ellipses into a rectangular shape. The random effects (i.e., R, BR, TR, and BTR) can all be interpreted as aspects of reproducibility. If the experiment is truly reproducible, none of these effects should be significant.
The analyses were performed with PROC MIXED from version 8 of the SAS statistical package (29). The default estimation technique in PROC MIXED, restricted maximum likelihood, was used.
RESULTS
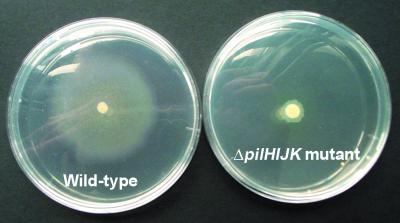
Construction of the lasI mutant (26) and that of the rpoS mutant (36) have been described elsewhere. In order to construct a type IV pilus mutant, the ΔpilHIJK gene cluster was deleted from the chromosome of P. aeruginosa (see Materials and Methods for details). The resulting mutant lacks the ability to produce intact type IV pili and is unable to translocate across surfaces by twitching motility (5). In order to verify the phenotype of the ΔpilHIJK mutant, twitching motility assays were performed (see Materials and Methods for details). The hazy zone of growth around the wild-type colony in Fig. 2 is the twitching zone, i.e., the area containing cells that have used type IV pilus-mediated twitching motility to move away from the point of inoculation. The average size of the twitching zone for the wild type was 30 mm, whereas the twitching zone for the ΔpilHIJK mutant was only 4 mm. This was measured as the distance from the edge of the colony to the edge of the twitching zone. This confirms that the ΔpilHIJK mutant is unable to move by twitching motility.
FIG. 2.
Twitching motility assays of wild-type and ΔpilHIJK mutant P. aeruginosa. Thin agar plates (1.5%) were stab inoculated with a toothpick to the bottom of the plate and incubated for 48 h at 37°C. The light haze of growth, the twitch zone, at the agar-plate interface is a measure of twitching motility. The smaller, denser zone represents surface colony growth.
Planktonic growth curves of the four strains showed that the doubling times at 37°C in citrate minimal medium were virtually identical for all four strains (53 min for the wild type, 56 min for the rpoS mutant, 54 min for the ΔpilHIJK mutant, and 56 min for the lasI mutant). Furthermore, the doubling times in glucose minimal medium at 37°C (46 min for the wild type, 46 min for the rpoS mutant, 45 min for the ΔpilHIJK mutant, and 46 min for the lasI mutant) and LB medium at 37°C (30 min for the wild type, 30 min for the rpoS mutant, 31 min for the ΔpilHIJK mutant, and 34 min for the lasI mutant) were also virtually identical. Thus, the differences in biofilm formation among the four strains do not simply reflect differences in growth rates.
In order to study the biofilm development of the four strains in flow chambers by CLSM, we tagged the strains with the GFP by chromosomal insertion of mini-Tn5 cassettes containing the gfp gene fused to a strong promoter (see Materials and Methods for details). Planktonic growth curves in citrate minimal medium and LB medium of the resulting GFP-tagged strains showed no changes in growth rate due to insertion of the mini-Tn5 cassette. Furthermore, flow chamber-grown biofilms of the GFP-tagged strains showed no observable changes compared to biofilms of the untagged parental strains.
Biofilm development of the four P. aeruginosa strains.
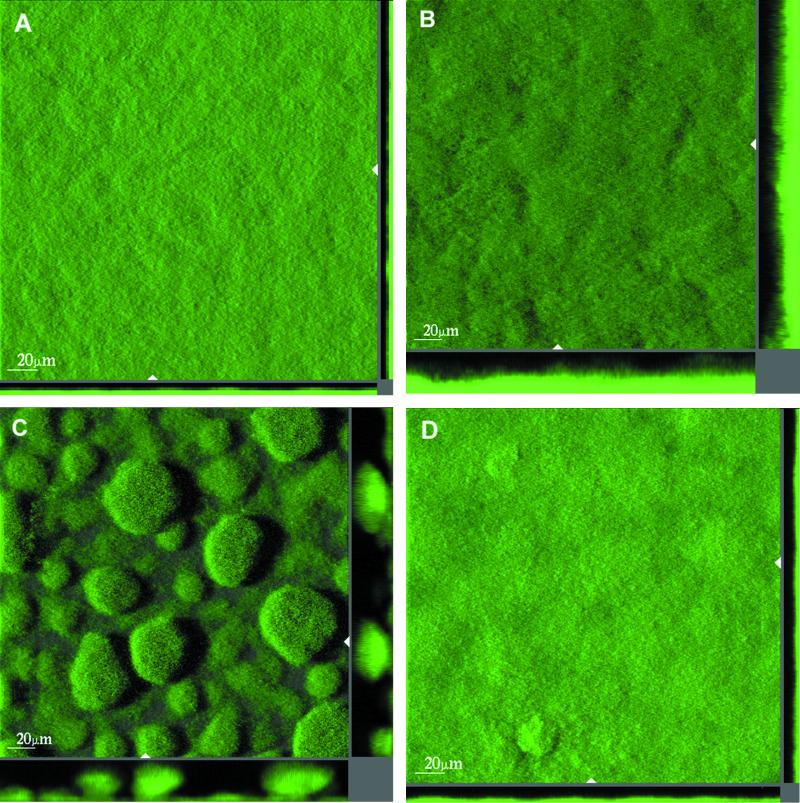
Flow chambers irrigated with citrate minimal medium were inoculated with each of the four strains, and biofilm formation was monitored by CLSM. In order to quantify the initial attachment of each of the four strains to the glass substrate, the flow chambers were observed microscopically 1 h after the flow was started. Images were acquired at 10 random spots in two flow channels for each strain. In all of the flow chambers, the bacteria attached as individual cells. A script written in MATLAB (The MathWorks Inc., Natick, Mass.) was used to count the number of cells that had attached. The number of attached cells were 3,501 cells/mm2 (σ = 426 cells/mm2) for the wild type, 3,564 cells/mm2 (σ = 437 cells/mm2) for the rpoS mutant, 18,863 cells/mm2 (σ = 547 cells/mm2) for the ΔpilHIJK mutant, and 4,089 cells/mm2 (σ = 292 cells/mm2) for the lasI mutant. The ΔpilHIJK mutant thus attached approximately five times better than the other three strains. The numbers of CFU in all of the cultures used for inoculation were determined and found to be similar (data not shown). Differences in initial attachment are therefore not due to differences in inoculum size. During the next 13 days, the flow chambers were observed microscopically daily and sets of images were acquired on days 2 (55 h), 4 (98 h), 6 (146 h), 10 (242 h), and 13 (314 h). After 55 h, the wild type, the rpoS mutant, and the lasI mutant covered the entire substrate with cells whereas the ΔpilHIJK mutant had left small areas (<1%) of the substrate uncolonized. At 98 and 146 h after inoculation, the ΔpilHIJK mutant biofilm consisted of a large number of distinct microcolonies that were regularly spaced and almost uniform in size (Fig. 3). At these time points, the wild-type and lasI mutant biofilms were rather flat and undifferentiated. Also, the rpoS mutant biofilm was relatively uniform but significantly thicker than the wild-type and lasI mutant biofilms. As the biofilms continued to grow, the microcolonies of the ΔpilHIJK mutant coalesced, forming larger and larger microcolonies, whereas the rpoS mutant biofilm became slightly more heterogeneous. The wild type and the lasI mutant remained relatively flat and uniform.
FIG. 3.
Spatial structures of 98-h-old biofilms of wild-type P. aeruginosa PAO1 (A), P. aeruginosa rpoS (B), P. aeruginosa ΔpilHIJK (C), and P. aeruginosa lasI (D) growing in citrate minimal medium. The bacteria are expressing the GFP. The larger central plots are simulated fluorescent projections in which a long shadow indicates a large, high microcolony. Shown in the right and lower frames are vertical sections through the biofilms collected at the positions indicated by the white triangles. The scale bars shown in the central plots are also valid for the right and lower frames.
Quantification and comparison of biofilm structures.
Biofilms are often described in subjective, nonquantitative terms as in the section above. In order to quantify and compare the biofilm structures formed by the four P. aeruginosa strains in the present study, we used a recently developed computer program, COMSTAT (12). COMSTAT comprises 10 different features for quantitative characterization of three-dimensional biofilm images. With COMSTAT, we analyzed all of the images from each of the three independent rounds of biofilm experiments (see Materials and Methods for details). This totaled approximately 10,800 individual images or approximately 2.8 gigabytes of data. COMSTAT calculates a range of variables, such as average thickness, roughness, surface-to-volume ratio, substratum coverage, biomass volume, etc. Theoretically, it is possible to use all of the variables calculated by COMSTAT. However, in order to make the multivariate analysis of variance robust, it is important to use only a few variables. By visual inspection of data plots, average thickness and roughness were included as the only two variables in the analysis. These two variables have straightforward biological and physical interpretations and are widely used in the biofilm literature. Furthermore, they did not seem to be correlated to any significant extent, which means that they describe different aspects of the data (for detailed descriptions of COMSTAT variables, see reference 12).
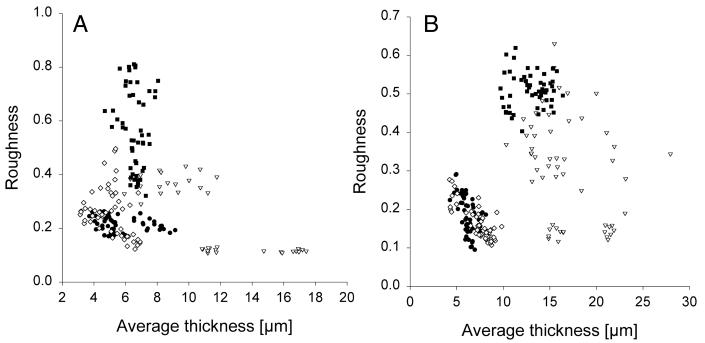
Figure 4 shows the results of the quantification at the 98-and 146-h time points (the results from the 55-, 242-, and 314-h time points are not shown). Each spot represents a single stack of images. There is clearly a natural variation in the biofilm architecture of each strain at a given time point. This variation is considerable for the rpoS and ΔpilHIJK mutant biofilms and less pronounced for the wild-type and lasI mutant biofilms. Furthermore, the plots may suggest that, at these time points, the wild type and the lasI mutant form similar biofilm structures whereas the rpoS mutant and the ΔpilHIJK mutant develop biofilm structures that are different from the wild-type biofilm structure. In the following sections, these putative differences are examined at all five time points in the framework of the analysis of variance model described above.
FIG. 4.
Roughness versus average thickness of 98-h-old (A) and 146-h-old (B) biofilms of wild-type P. aeruginosa (•), P. aeruginosa rpoS (▿), P. aeruginosa ΔpilHIJK (▪), and P. aeruginosa lasI (⋄). Images were acquired by CLSM in three independent biofilm experiments and quantified by the computer program COMSTAT. In each of the three rounds, 18 image stacks were acquired from two flow channels totaling 54 image stacks for each strain at each time point. Each spot represents a single stack of images. Note that there are only 36 spots for the P. aeruginosa rpoS mutant at the 98-h time point because, inadvertently, no images were acquired for this strain at the 98-h time point in the second round.
The statistical analyses showed the following (see Materials and Methods for details). The fixed effect of the bacterial strain, b, was highly significant (P = 0.006); i.e., there was an overall difference among the four strains. Not surprisingly, the fixed effect, time (t), was very significant (P < 0.0001). This means that there was an overall development over time. Most importantly, the interaction between the bacterial strain and time, bt, was highly significant (P = 0.002), showing that there were clear differences in the development of the biofilms of the four strains over time.
The random effects R, BR, and TR were all nonsignificant. However, the three-way interaction among the bacterial strain, the time, and the experimental round (BTR) was significant (P = 0.0012). In other words, the development over time of the four strains was not completely the same from experimental round to experimental round. This does not contradict the finding that the fixed effect, bt, describing the average development of the four strains over time was significant. It indicates, however, that conclusions about the average development of the strains over time should be based on a number of experimental rounds.
Interestingly, the covariance of the variables average thickness and roughness was not significant, so the two selected variables really describe two different (orthogonal) aspects of the data. The autoregression parameter, ρ, was highly significant (P < 0.0001) and was estimated at 0.51. This means that there was a dependence between data points over time, as should be expected when taking repeated measurements of the same experimental unit (a flow channel).
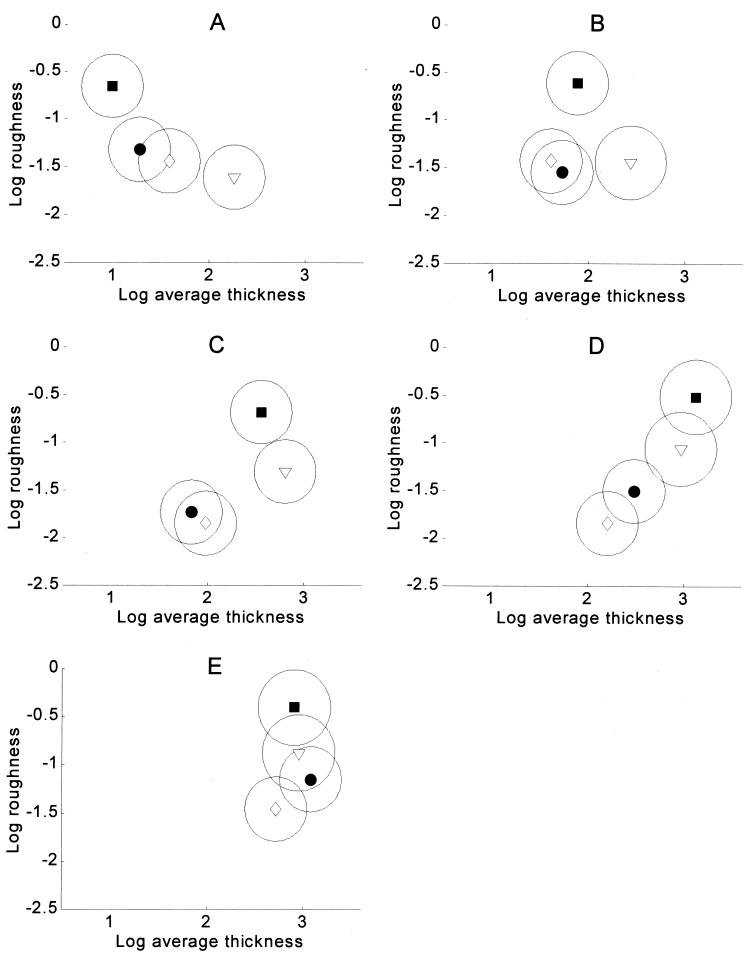
Which of the biofilms are different and at which time points are indicated in Fig. 5, which shows the mean values of the logarithmically transformed data and 90% confidence ellipses of the means. Two nonoverlapping ellipses indicate that a particular pair of biofilms were different at a specific time point. Two overlapping ellipses indicate that the corresponding biofilms were not significantly different at that particular time point. This implies not that the biofilms were identical but simply that they were not significantly different within the uncertainty of the present experiments. Note that the confidence ellipses are quite similar in size, which may seem surprising in view of the data in Fig. 4. This is because the data have been transformed by taking the logarithm, which stabilizes the variances.
FIG. 5.
Comparison of P. aeruginosa biofilm architectures. Mean values for log average thickness and log roughness together with 90% confidence ellipses is shown for wild-type P. aeruginosa (•), P. aeruginosa rpoS (▿), P. aeruginosa ΔpilHIJK (▪), and P. aeruginosa lasI (◊). Two nonoverlapping ellipses indicate that a particular pair of biofilms were different at a specific time point. Two overlapping ellipses mean that a pair of biofilms were not significantly different at a particular time point. This type of comparison corresponds to a multivariate t test on approximately a 5% level. Panels: A, 55 h; B, 98 h; C, 146 h; D, 242 h; E, 314 h.
The results in Fig. 5 are summarized in Table 1. Tables 2 and 3 show the same comparisons but univariately for average thickness and roughness individually, based on pairwise t tests. As already noted, the results of the multivariate test in Table 1 cannot be replaced by the results of the two univariate tests in Tables 2 and 3. As would be expected, however, there is good agreement between them.
TABLE 1.
Bivariate comparison of biofilms of wild-type P. aeruginosa, P. aeruginosa rpoS, P. aeruginosa ΔpilHIJK, and P. aeruginosa lasIa
| PAO1 strains compared | Significant difference at:
|
||||
|---|---|---|---|---|---|
| 55 h | 98 h | 146 h | 242 h | 314 h | |
| Wild type vs rpoS mutant | + | + | + | − | − |
| Wild type vs ΔpilHIJK mutant | + | + | + | + | + |
| Wild type vs lasI mutant | − | − | − | − | − |
| rpoS mutant vs ΔpilHIJK mutant | + | + | + | − | − |
| rpoS mutant vs lasI mutant | + | + | + | + | − |
| ΔpilHIJK mutant vs lasI mutant | + | + | + | + | + |
The variables average thickness and roughness were collectively subjected to a multivariate analysis of variance. Two nonoverlapping ellipses in Fig. 5 indicate that a particular pair of biofilms were different at that specific time point. This corresponds to a multivariate t test on approximately a 5% level. A plus sign denotes a significant difference, i.e., where the 90% confidence ellipses do not overlap, and a minus sign indicates no difference.
TABLE 2.
Univariate comparison of biofilms of wild-type P. aeruginosa, P. aeruginosa rpoS, P. aeruginosa ΔpilHIJK, and P. aeruginosa lasIa
| PAO1 strains compared | Significant difference at:
|
||||
|---|---|---|---|---|---|
| 55 h | 98 h | 146 h | 242 h | 314 h | |
| Wild type vs rpoS mutant | + | + | + | + | − |
| Wild type vs ΔpilHIJK mutant | − | − | + | + | − |
| Wild type vs lasI mutant | − | − | − | − | − |
| rpoS mutant vs ΔpilHIJK mutant | + | + | − | − | − |
| rpoS mutant vs lasI mutant | + | + | + | + | − |
| ΔpilHIJK mutant vs lasI mutant | + | − | + | + | − |
Average thickness was subjected to a univariate analysis of variance, and pairwise t tests at the 5% level were used to determine significant differences. A plus sign denotes a significant difference, and a minus sign indicates no difference.
TABLE 3.
Univariate comparison of biofilms of wild-type P. aeruginosa, P. aeruginosa rpoS, P. aeruginosa ΔpilHIJK, and P. aeruginosa lasIa
| PAO1 strains compared | Significant difference at:
|
||||
|---|---|---|---|---|---|
| 55 h | 98 h | 146 h | 242 h | 314 h | |
| Wild type vs rpoS mutant | − | − | − | − | − |
| Wild type vs ΔpilHIJK mutant | − | + | + | + | + |
| Wild type vs lasI mutant | − | − | − | − | − |
| rpoS mutant vs ΔpilHIJK mutant | + | + | + | + | − |
| rpoS mutant vs lasI mutant | − | + | + | + | + |
| ΔpilHIJK mutant vs lasI mutant | + | + | + | + | + |
Roughness was subjected to a univariate analysis of variance, and pairwise t tests at the 5% level were used to determine significant differences. A plus sign denotes a significant difference, and a minus sign indicates no difference.
Biofilm development of the four P. aeruginosa strains in different media.
The complete set of three biofilm experiments, five time points, eight channels, etc., were done in citrate minimal medium as described above. In order to investigate the biofilm structures formed by the four strains in other media, similar experiments were performed with glucose minimal medium and 50-fold-diluted LB medium. The biofilm structures formed by each of the four strains in 0.1 mM glucose minimal medium were similar to those formed in 0.1 mM citrate minimal medium (data not shown). When the flow chambers were irrigated with 50-fold-diluted LB medium (20 ml of LB medium diluted in 980 ml of Milli-Q water), the biofilms formed much faster and all four strains formed heterogeneous biofilm structures within 48 h after inoculation (data not shown). Compared to minimal glucose or citrate medium, diluted LB medium is much richer in nutrients. It is therefore not surprising that biofilm development is more rapid and very different in diluted LB medium compared to minimal medium.
DISCUSSION
Current model for P. aeruginosa biofilm development.
The genus Pseudomonas is a physiologically diverse and ecologically significant group of bacteria (30). One prominent member of this group, P. aeruginosa, has become a major model organism in biofilm research. Key results of P. aeruginosa biofilm research include the observations that cell-to-cell signaling is necessary for the development of mature biofilms containing microcolonies separated by water channels (6) and that type IV pilus-mediated twitching motility is necessary for the formation of microcolonies (23). Other observations show that flagella are necessary for the initial adherence of P. aeruginosa to plastic surfaces (23). These findings and others have led to a general model of P. aeruginosa biofilm formation (for details, see references 4 and 22) in which flagella are involved in the initial attachment to a surface. After attachment, cells are able to move along the surface by using twitching motility instead of flagellum-mediated swimming. The cells multiply and form a continuous layer covering the substrate, and later, groups of bacteria begin to form microcolonies. Type IV pilus-mediated twitching motility is essential for the formation of the microcolonies, and it has been suggested that the microcolonies are formed partly through twitching motility-mediated cell aggregation (23). In the model, the microcolonies then differentiate into a mature biofilm consisting of tower- and mushroom-shaped microcolonies interspersed with water channels. Cell-to-cell communication through the lasI-lasR quorum-sensing system is believed to be essential for this differentiation (6).
Biofilm heterogeneity must be addressed.
It is well known that biofilms are often structurally heterogeneous. This heterogeneity can be very troublesome because it makes comparisons between different types of biofilms (for example, between biofilms formed by two different strains) very complicated. This is illustrated in Fig. 4, which shows the average thickness and roughness of 98- and 146-h-old biofilms of each of the four strains. Each spot represents a single stack of images. The structural variation within each strain is significant, and it is apparent that a single image or a few images are not sufficient for a comparison of different types of biofilms. In fact, acquisition of too few images may lead to biased or even incorrect conclusions. In contrast, when a larger number of images are acquired, plots similar to the ones in Fig. 4 facilitate visualization of the results and of the variation within each biofilm. Figure 4 also emphasizes why a statistical analysis is necessary for a comparison of the strains and an investigation of their development over time. A statistical analysis is the only way to answer questions such as the following. (i) At which time points are the wild-type biofilm and each of the mutant biofilms significantly different? (ii) Is there consistent development of the biofilms over time? The experiments required to perform a statistical analysis as described above must be based on high-resolution images acquired by confocal microscopy, and such images are obtained only from fluorescently labeled specimens. This is the major reason for the GFP tagging of the strains investigated in our studies. The preferred method of tagging has been to construct chromosomal insertions to ensure a stable gene dosage of the tag sequence, and mini-Tn5 has so far been the carrier of choice. The major advantage of the Tn5 carrier is the available library of genetic tools derived from this transposon, which permit easy and efficient strain constructions whenever needed. The major drawback, however, is that this transposon inserts itself randomly into the chromosome, resulting in nonisogenic constructs. We therefore routinely compare the biofilm behavior and characteristics of the tagged strains we construct with those of the corresponding untagged strains. Although the resolution of the latter in a confocal microscope is not optimal, these control images provide sufficient details to distinguish structural differences in the different strains, and in no case were significant effects of the transposon insertions observed. The differences observed here between the wild type and the ΔpilHIJK mutant have been reproduced by employing an alternative transposon carrier known to insert itself in a genetically neutral position (unpublished data), and the identical performance of the wild type and the lasI mutant has been observed also when testing biofilm characteristics in strains carrying autonomous plasmids as GFP tag carriers (unpublished). There has not been any indication in any of the many phenotypic tests we have performed that the transposon insertions described here have had any impact on biofilm development other than those determined by the specific mutations in the different strains.
Biofilm formation of P. aeruginosa PAO1.
In the present study, P. aeruginosa PAO1 formed uniform, densely packed biofilms with an average thickness of 4 μm after 55 h, 5 μm after 98 h, 6 μm after 146 h, 14 μm after 242 h, and 23 μm after 314 h. Stewart et al. (31) quantified biofilms of P. aeruginosa grown in a continuous-flow annular biofilm reactor on a glucose minimal medium and described the biofilms as rough with a thickness between 13 and 60 μm. However, in a similar experimental setup, Murga et al. (20) described P. aeruginosa biofilms as thin and nearly uniformly thick with a roughness coefficient of 0.15, results that are similar to ours (Fig. 4). Davies et al. (6) reported that flow chamber biofilms of P. aeruginosa growing on glucose minimal medium were characterized by microcolonies separated by water channels and that the size of the microcolonies was approximately 100 μm. Nivens et al. (21) found that P. aeruginosa growing in flow chambers on minimal glutamate-glycerol medium formed densely packed biofilms with an average thickness of 6 μm. These different descriptions of P. aeruginosa biofilms, to a large extent, reflect the different experimental setups used by different laboratories, but they also illustrate the capacity of P. aeruginosa to adapt to a wide variety of different conditions by changing the structure of its biofilms. For a recent review of the factors that influence biofilm structure, see reference 32.
Biofilm formation of P. aeruginosa rpoS.
As shown in Table 2, the rpoS mutant formed significantly thicker biofilms than the wild type at all of the time points studied except the 314-h time point. Table 3 shows that the roughness of the wild-type biofilm and that of the rpoS biofilm were not significantly different at any time points. These results confirm the visual observations exemplified in Fig. 3 and indicate that RpoS is, in some way, involved in the control of biofilm growth in P. aeruginosa. Adams and McLean (1) reported that the biofilm cell density of E. coli grown in a modified Robbins device was reduced by 50% in an rpoS mutant compared to that of the parental strain. There are several differences between the roles of RpoS in E. coli and P. aeruginosa (14, 34), and it is therefore not surprising that RpoS plays different roles in biofilm development in E. coli and P. aeruginosa.
Biofilm formation of P. aeruginosa ΔpilHIJK.
The well-defined microcolonies formed by the ΔpilHIJK mutant are the reason for the significant differences in roughness between the wild-type and ΔpilHIJK mutant biofilms (Table 3). At the 55-h time point, the microcolonies are not yet formed, but from the 98-h time point on, the ΔpilHIJK mutant biofilm is significantly rougher than the wild-type biofilm. Table 1 shows that when average thickness and roughness were analyzed collectively, the two strains were different at all of the time points studied. These results seem to be in conflict with observations made by O'Toole and Kolter (23). They studied the formation of microcolonies of P. aeruginosa PA14 on polyvinyl chloride plastic tabs in glucose minimal medium by phase-contrast microscopy and monitored the formation of microcolonies during the first 8 h of biofilm formation. After 8 h, the wild type had formed a dense monolayer punctuated by microcolonies, which were approximately three to five layers of cells thick. Like the wild type, a type IV pilus mutant also formed a dense monolayer of cells but did not develop the small characteristic microcolonies. Through time lapse microscopy of 7- to 8-h-old biofilms of the wild type, they observed that microcolonies were formed by motility-driven aggregation of cells on the substrate and not solely by growth of the cells. Based on these observations, they suggested that type IV pili are necessary for microcolony formation by P. aeruginosa and that the wild-type microcolonies are formed by twitching motility-mediated cell aggregation. The biofilm setup used by O'Toole and Kolter (23) is very different from the setup used in the present study. Most importantly, they used a static system with no flow and only the first 8 h of biofilm formation was studied. In the present study, biofilms were grown in flow chambers under laminar-flow conditions and investigated over a 13-day period. The microcolonies described by O'Toole and Kolter (23) were three to five layers of cells thick (approximately 3 to 5 μm), whereas the microcolonies formed by the ΔpilHIJK mutant in the present study were approximately 20 to 50 μm thick. Thus, the results from the two different experimental approaches are, in fact, not opposed to each other but complementary. While type IV pilus-mediated twitching motility may mediate aggregation of cells in young biofilms, twitching motility is not required for the formation of microcolonies in mature biofilms. In fact, our study may suggest that twitching motility is used as a spreading mechanism in order for P. aeruginosa to colonize the entire substrate. The microcolonies formed by the ΔpilHIJK mutant could then be explained by clonal growth of individual bacteria in the biofilm caused by the inability of these bacteria to move by twitching motility. However, further information about the formation of ΔpilHIJK mutant microcolonies is necessary to address this hypothesis.
Initial attachment of P. aeruginosa ΔpilHIJK mutant.
The ΔpilHIJK mutant attached approximately five times better to the glass substrate than did the wild-type strain (see Results for details). We have no direct explanation for this observation. Interestingly, however, Déziel et al. (8) recently isolated small-, rough-colony variants of P. aeruginosa 57RP from static liquid cultures and biofilms growing on hexadecane as the substrate. These variants were defective in swimming, swarming, and twitching motility and reverted to the wild-type phenotype outside a selective environment, suggesting that the shift is regulated by a phase variation mechanism. In contrast to the parental strain, these small-colony variants rapidly initiated the formation of strongly adherent biofilms.
Biofilm formation of P. aeruginosa lasI.
The lasI mutant biofilms were indistinguishable from the wild-type biofilms at all time points with respect to both average thickness and roughness (Tables 1 and 2). Even when average thickness and roughness were analyzed collectively, there were no differences between the two strains at any time point (Table 3). Both strains formed uniform, densely packed biofilms. When the biofilms were grown in minimal glucose medium or in 50-times-diluted LB medium instead of citrate minimal medium, still no differences between the biofilms of the two strains could be observed (see Results for details). In diluted LB medium, however, both the wild type and the lasI mutant formed heterogeneous biofilm structures within 48 h after inoculation (data not shown). These results were surprising, because the lasI mutant had previously been reported to form much thinner biofilms than the wild type. Davies et al. (6) studied flow chamber biofilms of P. aeruginosa PAO1 and an isogenic lasI mutant grown on a minimal glucose medium and found that the lasI mutant formed flat, undifferentiated biofilms that were sensitive to the biocide sodium dodecyl sulfate (SDS). The wild-type strain, on the other hand, formed characteristic microcolonies separated by water channels and was resistant to SDS. The results obtained by Davies et al. (6) were enticing because they provided a link between quorum sensing, a system that controls gene expression in groups of bacteria, and biofilms, which are, in fact, organized groups of bacteria. In a recent work, Stoodley et al. (33) compared the biofilm structures of wild-type P. aeruginosa and an isogenic lasR lasI mutant in glass flow cells under laminar- and turbulent-flow conditions in minimal glucose medium. They found no marked difference in the structural complexity of the lasR lasI mutant biofilm compared to that of the wild-type biofilm under either laminar- or turbulent-flow conditions. Brooun et al. (2) compared biofilms of wild-type P. aeruginosa and an isogenic lasI mutant growing on polystyrene surfaces in microtiter plates and found no differences in susceptibility to the antibiotics ofloxacin and tobramycin or SDS. Taken together, these results suggest that cell-to-cell communication may not be necessary for P. aeruginosa biofilm formation under all conditions.
Conclusion.
Our results indicate that the current model for biofilm formation by P. aeruginosa needs modification. Under the experimental conditions of the present study, twitching motility was not required for microcolony formation in P. aeruginosa biofilms and microcolonies were not formed through twitching motility-mediated cell aggregation. Furthermore, cell-to-cell signaling through the lasI-lasR quorum-sensing system was not required for the formation of mature P. aeruginosa biofilms. The present study also suggested an important role for RpoS in the control of the growth of P. aeruginosa biofilms. As is true for all biofilm experiments, observations are influenced by the experimental conditions used and the conclusions must therefore be limited to the context of the present experiments. However, this work provides useful information that will assist in further characterization of the process of P. aeruginosa biofilm formation.
Acknowledgments
We thank Barbara Iglewski for providing P. aeruginosa lasI and Marvin Whiteley for providing P. aeruginosa rpoS. We thank Kalai Mathee for providing us with flow channels.
REFERENCES
- 1.Adams, J. L., and R. J. McLean. 1999. Impact of rpoS deletion on Escherichia coli biofilms. Appl. Environ. Microbiol. 65:4285-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooun, A., S. Liu, and K. Lewis. 2000. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 44:640-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christensen, B. B., C. Sternberg, J. B. Andersen, R. J. Palmer, A. T. Nielsen, M. Givskov, and S. Molin. 1999. Molecular tools for study of biofilm physiology. Methods Enzymol. 310:20-42. [DOI] [PubMed] [Google Scholar]
- 4.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 5.Darzins, A., and M. A. Russell. 1997. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosa as a model system—a review. Gene 192:109-115. [DOI] [PubMed] [Google Scholar]
- 6.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 7.de Lorenzo, V., M. Herrero, U. Jakubzik, and K. N. Timmis. 1990. Mini-Tn 5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J. Bacteriol. 172:6568-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diggle, P. J., K. Y. Liang, and S. L. Zeger. 1994. Analysis of longitudinal data. Oxford University Press, Oxford, United Kingdom.
- 10.Hahn, H. P. 1997. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—a review. Gene 192:99-108. [DOI] [PubMed] [Google Scholar]
- 11.Heydorn, A., B. K. Ersbøll, M. Hentzer, M. R. Parsek, M. Givskov, and S. Molin. 2000. Experimental reproducibility in flow chamber biofilms. Microbiology 146:2409-2415. [DOI] [PubMed] [Google Scholar]
- 12.Heydorn, A., A. T. Nielsen, M. Hentzer, C. Sternberg, M. Givskov, B. K. Ersbøll, and S. Molin. 2000. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 146:2395-2407. [DOI] [PubMed] [Google Scholar]
- 13.Holloway, B. W., and A. F. Morgan. 1986. Genome organization in Pseudomonas. Annu. Rev. Microbiol. 40:79-105. [DOI] [PubMed] [Google Scholar]
- 14.Jorgensen, F., M. Bally, V. Chapon-Herve, G. Michel, A. Lazdunski, P. Williams, and G. S. Stewart. 1999. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology 145:835-844. [DOI] [PubMed] [Google Scholar]
- 15.Kharazmi, A., B. Giwercman, and N. Høiby. 1999. Robbins device in biofilm research. Methods Enzymol. 310:207-215. [DOI] [PubMed] [Google Scholar]
- 16.Mardia, K. V., J. T. Kent, and J. M. Bibby. 1979. Multivariate analysis. Academic Press, London, United Kingdom.
- 17.Masduki, A., J. Nakamura, T. Ohga, R. Umezaki, J. Kato, and H. Ohtake. 1995. Isolation and characterization of chemotaxis mutants and genes of Pseudomonas aeruginosa. J. Bacteriol. 177:948-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mattick, J. S., C. B. Whitchurch, and R. A. Alm. 1996. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—a review. Gene 179:147-155. [DOI] [PubMed] [Google Scholar]
- 19.Merz, A. J., M. So, and M. P. Sheetz. 2000. Pilus retraction powers bacterial twitching motility. Nature 407:98-102. [DOI] [PubMed] [Google Scholar]
- 20.Murga, R., P. S. Stewart, and D. Daly. 1995. Quantitative analysis of biofilm thickness variability. Biotechnol. Bioeng. 45:503-510. [DOI] [PubMed] [Google Scholar]
- 21.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 23.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 24.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 25.Palmer, R. J., Jr. 1999. Microscopy flow cells: perfusion chambers for real-time study of biofilms. Methods Enzymol. 310:160-166. [DOI] [PubMed] [Google Scholar]
- 26.Pearson, J. P., E. C. Pesci, and B. H. Iglewski. 1997. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of elastase and rhamnolipid biosynthesis genes. J. Bacteriol. 179:5756-5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 28.Sanchez-Romero, J. M., R. Diaz-Orejas, and V. de Lorenzo. 1998. Resistance to tellurite as a selection marker for genetic manipulations of Pseudomonas strains. Appl. Environ. Microbiol. 64:4040-4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.SAS Institute, Inc. 1997. SAS/STAT software: changes and enhancements through release 6.12. SAS Institute, Inc., Cary, N.C.
- 30.Spiers, A. J., A. Buckling, and P. B. Rainey. 2000. The causes of Pseudomonas diversity. Microbiology 146:2345-2350. [DOI] [PubMed] [Google Scholar]
- 31.Stewart, P. S., B. M. Peyton, W. J. Drury, and R. Murga. 1993. Quantitative observations of heterogeneities in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 59:327-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stoodley, P., J. D. Boyle, D. DeBeer, and H. M. Lappin-Scott. 1999. Evolving perspectives of biofilm structure. Biofouling 14:75-90. [Google Scholar]
- 33.Stoodley, P., F. Jørgensen, P. Williams, and H. M. Lappin-Scott. 1999. The role of hydrodynamics and AHL signalling molecules as determinants of the structure of Pseudomonas aeruginosa biofilms, p. 323-330. In R. Bayston, M. Brading, P. Gilbert, J. Walker, and J. W. T. Wimpenny (ed.), Biofilms: the good, the bad, and the ugly. J. W. T. BioLine, Cardiff, United Kingdom.
- 34.Suh, S. J., L. Silo-Suh, D. E. Woods, D. J. Hassett, S. E. West, and D. E. Ohman. 1999. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J. Bacteriol. 181:3890-3897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wall, D., and D. Kaiser. 1999. Type IV pili and cell motility. Mol. Microbiol. 32:1-10. [DOI] [PubMed] [Google Scholar]
- 36.Whiteley, M., M. R. Parsek, and E. P. Greenberg. 2000. Regulation of quorum sensing by RpoS in Pseudomonas aeruginosa. J. Bacteriol. 182:4356-4360. [DOI] [PMC free article] [PubMed] [Google Scholar]