Abstract
In a recent study, RNA with nucleotide sequeces specific for “Norwalk-like viruses” (NLV) was detected in 11 different brands of European mineral waters. To clarify this finding, a 1-year monitoring study was conducted. Samples of three European brands of mineral water without gas were monitored weekly by reverse transcriptase PCR using generic and genogroup-specific oligonucleotides. Additional analyses were performed to investigate a possible correlation between NLV sequence contamination and mineral water lot numbers, the long-term stability (persistence) of NLV sequences in mineral water, and the level of contamination. NLV sequences were detected in 53 of 159 samples analyzed (33%) and belonged entirely to genogroup II. Although all NLV strains identified were closely related, three mineral water brand-specific clusters could be identified for both primer systems by sequencing. Analyses of second samples from lots previously shown to be positive for NLV sequences gave corresponding results in 45 of 53 cases (85%) (within a six-pack). NLV persistence was tested by analyzing 10 positive samples after 6 and 12 months of storage in darkness at room temperature. After 6 months, all samples remained positive; after 12 months, 9 of 10 samples were still positive for NLV sequences. No NLV sequences could be detected by analysis of 0.1-liter aliquots of 53 samples shown to be positive by testing of 1-liter volumes. Based on this fact and a test sensitivity of approximately 10 viral units, levels of contamination in positive mineral water samples were estimated to be in the range of 10 to 100 genomic equivalents per liter.
“Norwalk-like viruses” (NLVs) are single-stranded RNA viruses which form a genus within the family Caliciviridae (25, 32). The “Norwalk agent” was first discovered in fecal specimens collected from an outbreak of gastroenteritis at an elementary school in Norwalk, Ohio, in 1968 (1, 19). Because these agents cannot be propagated in cell culture, the taxonomic status of NLVs remained uncertain until Norwalk virus (NV) was first cloned and sequenced in 1990 (18). The subsequent development of diagnostic tools based on reverse-transcriptase PCR (RT-PCR) and genotyping by hybridization or sequencing techniques enabled the recognition of NLVs as members of the Caliciviridae. The genus Norwalk-like virus is now divided into three distinct genogroups: genogroup I (gg I) (NV-like), gg II (Snow-Mountain-like [SMA-like]), and gg III (Sapporo-like) (6, 17, 23, 32).
The importance of NLVs has become evident with recent studies revealing that they cause more than 90% of acute viral gastroenteritis worldwide (9, 15). NLVs are highly infectious, with a minimal infectious dose of an estimated 10 to 100 virus particles (33, 38), and are frequently responsible for food-borne outbreaks due to contaminated water, ready-to-eat dishes, seafood, fruits, and vegetables (2, 4, 5, 7, 8, 10, 14, 16, 20, 21, 24, 26, 28, 31; J. N. Kuritsky, M. T. Osterholm, J. A. Korlath, K. E. White, and J. E. Kaplan, Letter, J. Infect. Dis. 151:568, 1985). In the United States, the food-borne mode of transmission seems to account for 21% (13) to 40% (27) of all recorded NLV outbreaks, and estimates made by Mead et al. (27) presume a ratio of NLV cases in the community to cases reaching national surveillance of 460:1 (for the United States), for an estimated 23 million NLV infections annually. This ratio emphasizes the importance of NLV infections, especially compared with corresponding ratios for bacterial infections such as Campylobacter (7.6:1) or Salmonella (3.2:1) infections (40). Although this viral illness usually causes no complications, it has been estimated that approximately 6.9% (27) to 11% (40) of all food-related deaths each year are caused by NLVs.
There is epidemiological evidence that the water-borne exposure route is also important in the dissemination of NLVs to human hosts. Various outbreaks have been associated with ingestion of contaminated drinking or recreational surface water (2, 14, 20, 24, 26). An example is the infection of as many as a thousand persons with NLVs and other fecal contaminants in a Swiss village in 1998 (26). To date, bottled mineral water has never been clearly identified as a source of NLV infections. On the other hand, bacterial contamination of bottled mineral waters has been reported repeatedly (22, 34, 35, 37, 39, 41), which suggests that viral contamination is also possible. This hypothesis is supported by an epidemiological evaluation of repeated NLV outbreaks in a German recovery home, where the consumption of a particular mineral water was suspected to be the vehicle of all gastrointestinal illnesses (A. Ammon [Robert Koch Institute, Berlin, Germany], personal communication). After 1 year of repeated NLV infections (confirmed by RT-PCR for 33 stool specimens), no further outbreaks occurred once the recovery home stopped distributing the suspected brand of mineral water among its guests. Subsequent analyses of the suspected mineral water for NLV revealed none. However, in a further study without an epidemiological link, we could confirm the presence of NLV sequence contamination in different mineral waters. In a screening of 63 1-liter mineral water samples of 11 different brands, 21 (33%) were found to be NLV contaminated (3).
The aim of the present project was to investigate questions such as seasonal and lot number dependence of NLV contamination, levels of NLV contamination, and the appearance of particular NLV strains. Fur this purpose, 1-liter samples of three commercially available European mineral water brands were analyzed weekly over a 1-year period by filtration, concentration of viral RNA, and subsequent RT-PCR (3). Two RT-PCR systems were used in order to detect as many NLV strains as possible. Both strands of all NLV-positive amplicons were sequenced for molecular comparisons.
MATERIALS AND METHODS
Mineral waters.
Three European mineral water (“groundwater”) brands without gas, bottled in polyethylene bottles, were chosen. Once a week, six-packs of 1.5-liter bottles of each brand were bought at a local grocery store. Mineral water bottles were stored in the dark at room temperature (20°C) until all analyses were performed.
Analytical scheme.
We performed weekly analyses on 1-liter samples of three mineral water brands for 1 year, from April 2000 to April 2001, resulting in 159 samples (3 samples × 53 weeks), numbered BAG 1 to BAG 159 (“BAG” stands for Bundesamt für Gesundheit, the Swiss Federal Office of Public Health). If NLV-positive results were obtained in the first run from the 1-liter sample of mineral water, a second analysis of 0.1 liter (from the remaining 0.5 liter in the same bottle) was performed. We also analyzed a second bottle from the same six-pack in order to investigate a possible correlation between NLV positivity and the lot number. The last analyses were performed on a third and a fourth bottle from the same six-pack after 6 and 12 months in order to compare the stability of NLV sequences in mineral waters. Every week we analyzed two positive (see below) and two negative control samples in order to evaluate the methodology. Bacteriological analyses for Enterococcus faecalis, Escherichia coli, and Pseudomonas aeruginosa were performed on each mineral water sample according to the Swiss Food Manual, chapter 56.
Positive-control samples.
Two positive-control samples were used. We included an RT-PCR (described by Gilgen et al. [11]) for poliovirus strains of a known concentration to estimate the efficiency of the detection method. The poliomyelitis vaccine used, Poloral Berna (Serum- und Impfinstitut, Berne, Switzerland), consisted of three poliovirus strains: Sabin I (2 × 106 50% tissue culture infective doses [TCID50]/ml) (1 TCID50 corresponds to 10 to 100 viral particles), Sabin II (2 × 105 TCID50/ml), and Sabin III (6 × 106 TCID50/ml). We established a detection limit of a 10−9-fold dilution, which corresponds to 8.2 × 10−3 TCID50, for our RT-PCR. The detection of less than 1 copy number is possible in this case, since there is an unknown number of inactive viruses within the vaccine, detectable by RT-PCR but not by cell culture.
The second positive-control sample consisted of NLV strains of gg I and gg II isolated from different stool specimens. The NLV gg I-positive control sample contained Southampton-related viruses (GenBank accession number L07418), and the NLV gg II-positive control sample contained Camberwell-related viruses (GenBank accession number AF145896). Detection limits for seeded NLVs in 1-liter mineral water samples were established repeatedly, and dilutions of 10−5 for NLV gg I and 10−4 for NLV gg II remained positive (data not shown).
Negative-control samples.
We included two negative-control samples, consisting of 1 liter of distilled water each, one immediately before and one immediately after the mineral water samples; these were used in order to detect possible cross-contamination.
Virus concentration and isolation from 1 liter of mineral water.
One liter of each mineral water and of each positive- and negative-control sample was analyzed by a three-step isolation method modified from the work of Gilgen et al. (11, 12) as presented in a recent publication (3). As external positive controls, three 1-liter samples of distilled water were each seeded with 100 μl of diluted NLV gg I- or gg II-infected stool samples or with 10 μl of a 106-fold dilution of poliomyelitis vaccine. The modified three-step isolation procedure used for all analyses includes two subsequent concentrations, followed by virus lysis and RNA isolation. For the first concentration step, samples were vigorously shaken and filtered through a positively charged 0.45-μm-pore-size, 47-mm-diameter membrane (Zetapor filter membrane; CUNO Inc., Meriden, Conn.). After filtration, the Zetapor membrane was transferred to a 50-ml centrifuge tube containing 4 ml of 50 mM sodium-glycine at pH 9.5 with 1% beef extract (Sigma Chemical Corporation, St. Louis, Mo.). After the tube was shaken at 500 rpm at room temperature for 20 min in order to elute the viruses, the virus-containing buffer was adjusted to pH 8 with 20 μl of 1 M HCl. The 4-ml virus-containing buffer was concentrated to 100 μl by use of a microconcentrator (Ultrafree-Biomax; Millipore Corporation, Bedford, Mass.) at 1,000 × g and room temperature according to the manufacturer's protocol. The retained concentrate was adjusted to 140 μl with 1× phosphate-buffered saline, which was used for RNA isolation by use of the QIAmp HCV minikit (Qiagen GmbH, Hilden, Germany) according to the manufacturer's protocol. Ten microliters of the 60-μl “RNA solution” obtained was used for each reverse transcription.
Oligonucleotides.
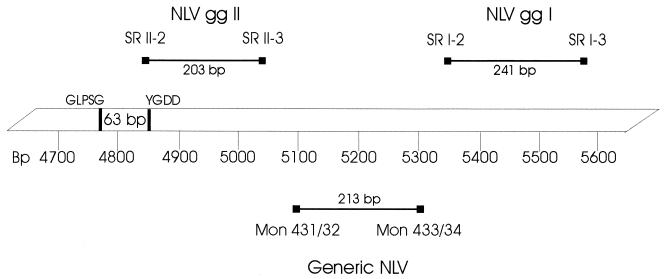
Two NLV RT-PCR systems and one enterovirus RT-PCR system were used to perform all analyses. Table 1 shows sequences and localizations of all oligonucleotides used. All primers had been synthesized by Microsynth GmbH (Balgach, Switzerland) and stored freeze-dried at −40°C until the first use. See Fig. 1 for a diagrammatic representation.
TABLE 1.
NLV and enterovirus primers used in this study
| Virus and oligonucleotides | Region | Sequence (5′→3′)a | Polarity | Localizationb |
|---|---|---|---|---|
| Genogroup-specific NLV and enterovirus oligonucleotidesc | ||||
| NLV gg I | ||||
| SRI-2 (RT-PCR) | Capsid | AAA TGA TGA TGG CGT CTA AG | Sense | 5344-5361 |
| SRI-3 (PCR) | AAA AYR TCA CCG GGK GTA T | Antisense | 5584-5566 | |
| NLV gg II | ||||
| SRII-2 (RT-PCR) | RNA Pole | TWC TCY TTY TAT GGT GAT GAT GA | Sense | 4844-4866 |
| SRII-3 (PCR) | TTW CCA AAC CAA CCW GCT G | Antisense | 5046-5028 | |
| Generic NLV oligonucleotidesd | ||||
| Mon431 (RT-PCR) | RNA Pol | TGG ACI AGR GGI CCY AAY CA | Sense | 5093-5112 |
| Mon432 (RT-PCR) | RNA Pol | TGG ACI CGY GGI CCY AAY CA | Sense | 5093-5112 |
| Mon433 (PCR) | RNA Pol | GAA YCT CAT CCA YCT GAA CAT | Antisense | 5285-5305 |
| Mon434 (PCR) | RNA Pol | GAA SCG CAT CCA RCG GAA CAT | Antisense | 5285-5305 |
| Enterovirus | ||||
| EV05 (PCR) | 5′ UTR | CAC GGA CAC CCA AAG TAG T | Antisense | 563-184 |
| EV06 (RT-PCR) | CAA GCA CTT CTG TTT CCC | Sense | 448-467 |
Mixed bases in degenerate primers are as follows: W stands for A or T, Y stands for C or T, K stands for G or T, R stands for A or G, S stands for C or G, and I stands for inosine.
Source: Gilgen et al. (12).
Source: S. S. Monroe, personal communication.
Pol, polymerase.
FIG. 1.
Diagrammatic representation of amplified RT-PCR segments within the putative RNA polymerase and the beginning of the capsid gene of NLV gg I and gg II. The central bar represents the NV (NV/F8Iia/68/US) genome (GenBank accession number M87661) beginning at nucleotide 4700. Block sequences and primer names indicate the relative locations of primers used; the 63-bp region shown within the GLPSG and YGDD motifs is commonly used by most diagnostic laboratories to detect NLV strains by RT-PCR.
The genogroup-specific NLV RT-PCR system for the separate detection of NLVs of gg I and gg II is based on degenerate primers described by Gilgen et al. (12) and located in highly conserved regions of the capsid gene for NLV gg I and of the RNA polymerase for NLV gg II. The predicted product sizes are 241 bp for NLV gg I and 203 bp for NLV gg II.
The generic NLV RT-PCR system is based on primers of region B within open reading frame (ORF) 1 (3′ end) (S. S. Monroe [Centers for Disease Control and Prevention {CDC}, Atlanta, Ga.], personal communication). The predicted product size is 213 bp.
The enterovirus-RT-PCR system was used as a second external control sample and is based on primers within the 5′ untranslated region (5′ UTR) described by Gilgen et al. (12). The predicted product size is 400 bp.
Reverse transcription.
Ten microliters of RNA was reverse transcribed for each of the three RT-PCR systems by using the Sensiscript RT Kit (Qiagen GmbH) according to the manufacturer's protocol. RNAs were reverse transcribed by incubation for 60 min at 37°C, followed by 5 min at 95°C to inactivate the reverse transcriptase. In a final volume of 20 μl, the reaction conditions were 1× RT-Sensiscript buffer, 0.5 mM concentrations of each deoxynucleoside triphosphate, 1 μM primer (SRI-2, SRII-2, Mon431-32, or EV06), 10 U of RNase inhibitor (RNasin; Promega, Madison, Wis.), 1 μl of RT-Sensiscript, and 10 μl of template-RNA.
PCR.
All 20 μl of the reverse transcription product was mixed with 60 μl of PCR mixture (final concentrations, 1× PCR buffer for recombinant Taq DNA polymerase [GIBCO BRL, Life Technologies Inc., Paisley, Scotland], 2 μg of bovine serum albumin [Fluka, Buchs, Switzerland]/ml, 0.2 mM deoxynucleoside triphosphates, 0.25 μM forward primers, MgCl2 at 1.5 mM [for all three NLV RT-PCRs] or 3 mM [for the enterovirus RT-PCR], and 1 U of recombinant Taq polymerase [Life Technologies]). Cycling was performed in a UNO II thermocycler (Biometra, Göttingen, Germany). The PCR program was as follows: denaturation for 180 s at 95°C; 40 cycles of 40 s at 95°C, 90 s at 50°C, and 60 s at 72°C; and a final extension for 420 s at 72°C. For both NLV RT-PCR systems and the enterovirus RT-PCR system, a two-tube RT-PCR was performed by using the same protocol (except for the MgCl2 concentration).
Analyses of PCR products.
Ten microliters of each amplicon was mixed with 10 μl of loading buffer and analyzed on 2.6% agarose gels. Products were visualized by ethidium bromide staining and UV transillumination. Fragment sizes were compared with a commercially available size standard (100-bp DNA ladder; Promega). To determine the viral origins of amplicons, both strands of each positive RT-PCR product, including all positive controls, were sequenced.
DNA sequencing and sequence analyses.
Both strands of RT-PCR products were directly sequenced by cycle sequencing (ABI PRISM 377 DNA sequencer; Perkin-Elmer) carried out by Microsynth GmbH. Amplicons (132 bp) of the viral RNA polymerase region from both primer systems, specific and generic, were compared with GenBank data bank entries by using the Fasta program of the Genetics Computer Group (Madison, Wis.) Wisconsin Package, version 9.1. Phylogenetic relationships were calculated by using Clustal W (36), and dendrograms were displayed by using TreeView drawing software (30).
Nucleotide sequence accession numbers.
NLV sequences isolated from mineral waters have been submitted to GenBank under accession numbers AF405523 through AF405528.
RESULTS
Weekly analyses of three mineral water brands.
In order to clarify a phenomenon observed in our recent study, where different mineral waters were determined to be contaminated by NLV sequences, we performed weekly analyses of three different brands of mineral waters over a 1-year period (April 2000 to April 2001). Of these 159 samples (three weekly analyses for 53 weeks), 53 (33%) were found to be NLV sequence positive. Each mineral water brand had the following percentage of positive results: brand 1, 30% (16 of 53); brand 2, 36% (19 of 53); brand 3, 34% (18 of 53). All RT-PCR products were further analyzed by double-strand sequencing and shown to belong to NLV gg II. During the first 8 months (from April to December 2000), 52 of 108 (48.1%) mineral water samples were positive for NLV sequences. These data correspond to the following bottling dates: brand 1, February to June 2000; brand 2, December to August 2000; brand 3, October 1999 to June 2000. However, in the next 5 months (analyses from December 2000 to April 2001), only 1 sample out of 51 (2%) was positive for NLV sequences. These data correspond to the following bottling dates: brands 1 and 3, July 2000 to January 2001; brand 2, September 2000 to January 2001.
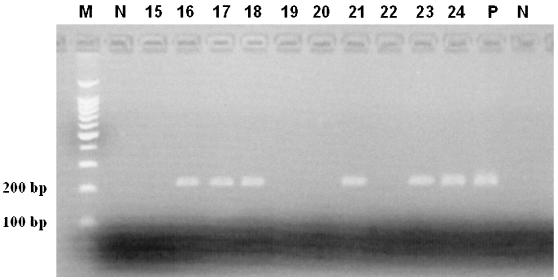
In addition, the two primer systems used had different detection peaks. From April to July 2000 (analysis data), we obtained mainly positive results with generic primers; from August 2000 to April 2001, we obtained mainly positive results with the NLV gg II-specific primers. Figure 2 shows ethidium bromide-stained NLV gg II sequences from positive mineral water samples on a 2.6% agarose gel. Six of 10 samples tested on this agarose gel show 213-bp amplicons of NLV gg II genomes.
FIG. 2.
Agarose gel (2.6%) and ethidium bromide staining of RT-PCR products obtained with generic primers for NLV sequences (213-bp amplicon) in mineral water samples BAG 15 to BAG 24. Electrophoresis was carried out at 70 mV/cm. Samples 16 to 18, 21, 23, and 24 are NLV gg II sequence positive. P, positive control; N, negative controls; M, marker.
Additional analyses.
Second samples (same lot numbers and six-packs) of each mineral water previously determined to be NLV positive were analyzed in order to investigate possible correlations within lots. Out of 53 second samples, 45 (85%) were positive for NLV gg II sequences: 87.5% (14 of 16) for brand 1, 84.2% (16 of 19) for brand 2, and 83.4% (15 of 18) for brand 3. Amplicon sequences were all identical to those previously determined within the same lot and the same six-pack. Results of analyses of 0.1-liter aliquots taken from the remaining 0.5 liter for all 53 positive samples remained entirely NLV negative. Further investigations of the stability of NLV sequences in mineral water revealed that 10 of 10 samples were still NLV sequence positive after 6 months of storage in the dark at room temperature. After 12 months, 9 of 10 samples were still positive.
Sequence analyses.
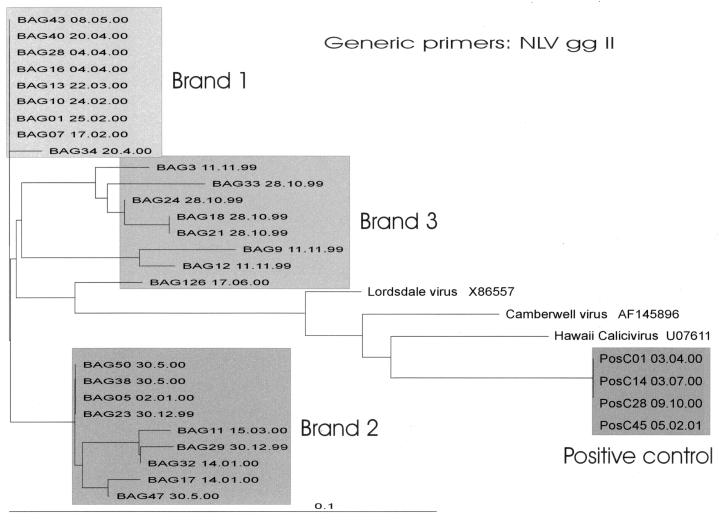
All amplicons were related to NLVs of gg II. Positive samples shared 91 to 93% sequence identity with Lordsdale virus (Hu/NLV/Lordsdale/93/United Kingdom; accession number X86557), 87 to 90% with Camberwell virus (Hu/NLV/Camberwell/101922/94/Aus; accession number AF145896), and 86 to 88% with Hawaii virus (Hawaii/71/US [serotype 2]; accession number UO7611). There were similarities between different sequences within each brand. Brand 1 revealed, with one exception, 100% similarity among all amplicon sequences with both primer systems. Both of the other brands had some sequences with similarities while others differed, without correlation to bottling dates. No amplicon sequences were identical to the NLV gg II-positive control samples. Figures 3 and 4 show dendrograms of 132-bp fragments of the viral RNA polymerase region of NLV gg II sequences amplified with genogroup-specific primers (27 samples) and generic primers (26 samples), respectively. Included in Fig. 4 is an NLV sequence detected in brand 3 during a previous study (3) with a sequence similarity of 85% with strains isolated in this study. Two NLV sequences of each brand, one isolated with the genogroup-specific primer system and one isolated with the generic primer system, were submitted to GenBank (Table 2).
FIG. 3.
Phylogenetic analyses performed on a 132-bp region of the RNA polymerase of NLV gg II sequences detected with genogroup-specific primers (12) by weekly analyses of 27 mineral water samples. An NLV gg II sequence of brand 3 isolated in the previous study (original designation, V2) (3), 4 of 53 identical positive-control sequences, and published sequences (accessed from GenBank) of Hawaii virus (U07611), Lordsdale virus (X86557), and Camberwell virus (AF145896) are also included. Three clusters of NLV sequences corresponding to each brand are shown.
FIG. 4.
Phylogenetic analyses performed on a 132-bp region of the RNA polymerase of NLV gg II sequences detected with generic primers (S. S. Monroe, personal communication) by weekly analyses of 26 mineral water samples. Four of 53 identical positive-control sequences and published sequences (accessed from GenBank) of Hawaii virus (U07611), Lordsdale virus (X86557), and Camberwell virus (AF145896) are also included. Three clusters of NLV sequences corresponding to each brand are shown.
TABLE 2.
NLV sequences isolated in mineral waters and submitted to GenBank
| Identification (primer) | Bottling date (day. mo. yr) | GenBank accession no. |
|---|---|---|
| Brand 1 | ||
| BAG 85 (SRII-3) | 27.07.00 | AF405524 |
| BAG 16 (Mon431) | 04.04.00 | AF405526 |
| Brand 2 | ||
| BAG 86 (SRII-3) | 28.08.00 | AF405525 |
| BAG 17 (Mon431) | 14.01.00 | AF405527 |
| Brand 3 | ||
| BAG 84 (SRII-3) | 06.01.00 | AF405523 |
| BAG 18 (Mon431) | 28.10.99 | AF405528 |
DISCUSSION
Samples of three different European mineral water brands (without gas) from purchased six-packs were monitored weekly for a 1-year period by RT-PCR using generic and genogroup-specific oligonucleotides. Additional analyses were performed to investigate a possible correlation between NLV sequence contamination and lot numbers of mineral water, the long-term stability of NLV sequences in mineral water, and the level of contamination. NLV sequences, all belonging to gg II, were detected in 53 of 159 samples analyzed (33%). In a previous study (3), where 63 samples of 29 different mineral water brands were analyzed, the same percentage was positive for NLV sequences, but both genogroups were present (76% of the NLV sequences detected belonged to gg I, and 24% belonged to II). This time, all three mineral water brands had approximately the same NLV sequence contamination level: brand 1, 30%, brand 2, 36%; brand 3, 34%. A sequence alignment (data not shown) with NLV gg II sequences of brand 3 of both studies shows 85% sequence similarity, suggesting recontamination of the mineral water, since NLVs cannot replicate, and thereby mutate, outside of cells.
Confirming our expectations, sequence analyses of amplified NLV gg II strains revealed the presence of strains that were closely related among all mineral water samples. Dendrograms (Fig. 3 and 4) of both RT-PCR systems present three closely related clusters, each corresponding to a specific mineral water brand. These results are in accordance with data published by Noel et al. (29), where a distinct strain of NLVs with a global distribution was identified. The “95-96 U.S. subset” described by these investigators, as well as the NLV gg II-positive strands we sequenced, is closely related to the Lordsdale virus (NC_001674) and the Camberwell virus (AF145896). Because the NLV sequences amplified in this study are different from that amplified by Noel et al. (29), direct comparisons by alignment could not be performed with this “95-96 U.S. subset.” Additional analyses of stool specimens from different Swiss NLV outbreaks during the same period revealed strains closely related to those in this study, to Lordsdale virus, and finally to the “95-96 U.S. subset.” This global distribution supports the presence of NLV sequences closely related to this “95-96 U.S. subset” in and around Switzerland.
RT-PCR is one of the most effective methods for detection of pathogens such as NLVs, which cannot be grown in vitro. A critical step of this methodology is the selection of optimized primer oligonucleotides. In order to detect the widest range of NLV strains, we decided to use a second RT-PCR system in addition to the one used in the previous study (3). It would be desirable for further monitoring studies to use a single RT-PCR system, but as our results confirm, this seems not to be achievable yet, since we obtained different results with two adjacent primer locations in highly conserved regions. This detection shift for both primer sets used could be explained by recontamination of mineral waters by closely related NLV gg II strains, since their replication in source water is impossible. Brands 1 and 2, which are bottled geographically close together (90 km), and brand 3, which is bottled more than 300 km away from the other brands, show quite similar distributions of positive samples when the bottling dates are compared. Samples of all three brands remained positive until August 2000. The reason for the sudden change to mainly negative results from September 2000 to January 2001 remains unclear. The fact that results of analyses of brand 3 samples with bottling dates of October 1999 to January 2000 were positive, compared with negative results for samples bottled from October 2000 to January 2001, led us to conclude that there is no seasonal pattern of contamination for these mineral waters. We also found no correlations between positivity and bottling date with brands 1 and 2.
Additional analyses.
Positive results obtained for the correlation of lot numbers and NLV sequence contamination support previous findings showing a significant correlation between mineral water bottles of the same lot within a six-pack (3). However, there was no correlation between mineral water bottles of the same lot among different six-packs. These data also suggest sporadic contamination within a lot, without any seasonal correlations. In order to obtain information about another crucial question, the concentration of NLV sequences, we investigated the presence of NLV sequences in 0.1 liter of all 53 previously tested NLV sequence-positive samples. This method had to be chosen in the absence of a reliable quantification method for NLVs. The fact that all samples were negative for NLV sequences and that the sensitivity of our method is estimated to be approximately 10 viral particles suggests a low concentration of NLV sequences per liter in the mineral water brands tested. Determining the stability of NLV sequences in mineral water (in the dark at room temperature) was a further step in understanding the contamination phenomenon. Our findings of NLV sequence-positive mineral waters after 6 and 12 months of storage suggest a high stability of NLV sequences in mineral water, even over a year at room temperature. Another crucial question about the possible infectivity of the NLV sequences detected cannot be answered yet, since the detection method used (RT-PCR) is not able to differentiate between infective and inactive NLVs. However, we presume that the survival of naked viral RNA in mineral water over several months is improbable. We also do not believe that concentration of viruses by adsorption to filters and elution with beef extract could possibly have succeeded unless the virions were intact. Since viral RNA sequences have to be enveloped in order to survive for 12 months in mineral water, it is possible that the NLV sequences detected are infective.
The origin of NLV sequence contamination in bottled mineral waters also remains unclear. We still consider three main contamination routes: contamination of the water sources, of the packing material, or during the bottling procedure. All three hypotheses can be supported by the results of this study. Since we were unable to monitor processing within a mineral water plant, no conclusions can be drawn. The hypothesis of a single contamination in the past seems not to be probable. Both the 15% difference in NLV amplicons of brand 3 compared with a previous study (3) and the presence of different NLV sequences within mineral water samples bottled on the same date support the possibility of several contaminations. Our results for NLV sequence distributions among different lots confirm that there is repeated contamination of some parts of a lot, since samples of the same lot but of different six-packs can be contaminated or not. These repeated contaminations may also be the reason for the distribution of a single NLV cluster among different mineral water brands. If we consider the results of our analyses in stool specimens over the last few months (data not shown), an NLV strain related to the global epidemic strain described by Noel et al. (29) is also predominant in our region. Since an NLV cluster is predominant over a region, the contamination of mineral waters by this cluster is possible. Now that we know that recontamination of mineral waters by NLV sequences is possible, we should find out where any contamination takes place. In addition, because disinfection procedures to avoid microbiological contamination in mineral waters are forbidden in some countries and therefore cannot be applied, it is necessary to carry out process checks in mineral water plants to determine the origin of contamination. As the Center for Food Safety and Applied Nutrition of the U.S. Food and Drug Administration states in its HACCP (Hazard Analyses of Critical Control Points) guidelines (http://vm.cfsan.fda.gov/∼dms/fc99-a5.html), the “Norwalk virus group” has to be considered a hazard with possible complications and therefore has to be addressed in every prevention-based food safety system in order to avoid contamination.
Future research will focus on the elaboration of a reliable quantitative real-time RT-PCR method for NLV sequences in food. As long as NLVs cannot be grown in cell culture, quantification by real-time PCR by using an appropriate primer-specific internal standard is the only option. In order to characterize risk factors related to NLVs, the Swiss Tropical Institute and the official food control authorities of the cantons of Basel-Land and Solothurn have launched a 2-year case-control study. One of the aims of this case-control study will be to evaluate whether NLV sequence-contaminated mineral water presents a risk to consumers or not.
Acknowledgments
We gratefully acknowledge the contributions of Marc Solioz (University of Berne), Stephan S. Monroe (CDC), Andrea Ammon (Robert-Koch Institute), and Peter Kohler (Official Food Control Authority of the Canton of Solothurn, Switzerland).
This study was supported by a grant from the Swiss Federal Office of Public Health (SFOPH).
REFERENCES
- 1.Adler, J. L., and R. Zickl. 1969. Winter vomiting disease. J. Infect. Dis. 199:668-673. [DOI] [PubMed] [Google Scholar]
- 2.Beller, M., A. Ellis, S. H. Lee, M. A. Drebot, S. A. Jenkerson, E. Funk, M. D. Sobsey, O. D. Simmons, S. S. Monroe, T. Ando, J. Noel, M. Petric, J. P. Middaugh, and J. S. Spika. 1997. Outbreak of viral gastroenteritis due to a contaminated well. JAMA 278:563-568. [PubMed] [Google Scholar]
- 3.Beuret, C., D. Kohler, and T. Luthi. 2000. Norwalk-like virus sequences detected by reverse transcription-polymerase chain reaction in mineral waters imported into or bottled in Switzerland. J. Food Prot. 63:1576-1582. [DOI] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1997. Viral gastroenteritis associated with eating oysters—Louisiana, December 1996-January 1997. Morb. Mortal. Wkly. Rep. 46:1109-1112. [PubMed] [Google Scholar]
- 5.Cliver, D. O. 1997. Virus transmission via food. World Health Stat. Q. 50:90-101. [PubMed] [Google Scholar]
- 6.Cubitt, D., D. W. Bradley, and M. J. Carter. 1995. Caliciviridae, p. 359-163. In F. A. Murphy, C. M. Fauquet, D. H. L. Bishop, et al. (ed.), Virus taxonomy: the classification and nomenclature of viruses. Sixth report of the International Committee on Taxonomy of Viruses. Springer-Verlag, Vienna, Austria.
- 7.Daniels, N. A., D. A. Bergmire-Sweat, K. J. Schwab, K. A. Hendricks, S. Reddy, S. M. Rowe, R. L. Fankhauser, S. S. Monroe, R. L. Atmar, R. I. Glass, and P. Mead. 2000. A foodborne outbreak of gastroenteritis associated with “Norwalk-like viruses”: first molecular traceback to deli sandwiches contaminated during preparation. J. Infect. Dis. 181:1467-1470. [DOI] [PubMed] [Google Scholar]
- 8.Denee, V. C., J. M. Hunt, C. R. Paule, R. I. James, R. G. Johnson, M. Y. Raymond, and C. W. Hedberg. 2000. The importance of foodborne calicivirus disease: the Minnesota experience. J. Infect. Dis. 181:281-283. [DOI] [PubMed] [Google Scholar]
- 9.Fankhauser, R. L., J. S. Noel, S. S. Monroe, T. Ando, and R. I. Glass. 1998. Molecular epidemiology of small round structured viruses in outbreaks of gastroenteritis in the United States. J. Infect. Dis. 178:1571-1578. [DOI] [PubMed] [Google Scholar]
- 10.Gaulin, C. D., D. Ramsay, P. Cardinal, and M. A. D'Halevyn. 1999. Epidemic gastroenteritis of viral origin associated with eating imported raspberries. Can. J. Public Health 90:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilgen, M., B. Wegmuller, P. Burkhalter, H. P. Buhler, U. Muller, J. Luthy, and U. Candrian. 1995. Reverse transcription-PCR to detect enteroviruses in surface water. Appl. Environ. Microbiol. 61:1226-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilgen, M., D. Germann, J. Luthy, and P. Hubner. 1997. Three-step isolation method for sensitive detection of enterovirus, rotavirus, hepatitis A virus, and small round structured viruses in water samples. Int. J. Food Microbiol. 37:189-199. [DOI] [PubMed] [Google Scholar]
- 13.Glass, R. I., J. Noel, T. Ando, R. Fankhauser, G. Belliot, A. Mounts, D. U. Prashar, J. S. Bresee, and S. S. Monroe. 2000. The epidemiology of enteric caliciviruses from humans: a reassessment using new diagnostics. J. Infect. Dis. 181(Suppl. 2):254-261. [DOI] [PubMed] [Google Scholar]
- 14.Gray, J. J., J. Green, C. Cunliffe, C. Gallimore, J. V. Lee, K. Neal, and D. W. Brown. 1997. Mixed genogroup SRSV infections among a party of canoeists exposed to contaminated recreational water. J. Med. Virol. 52:425-429. [PubMed] [Google Scholar]
- 15.Green, K. Y. 1997. The role of human caliciviruses in epidemic gastroenteritis. Arch. Virol. Suppl. 13:153-165. [DOI] [PubMed] [Google Scholar]
- 16.Hafliger, D., M. Gilgen, J. Luthy, and P. Hubner. 1997. Seminested RT-PCR systems for small round structured viruses and detection of enteric viruses in seafood. Int. J. Food Microbiol. 37:27-36. [DOI] [PubMed] [Google Scholar]
- 17.Jiang, X., M. Wang, K. Wang, and M. K. Estes. 1993. Sequence and genome organization of Norwalk virus. Virology 195:51-61. [DOI] [PubMed] [Google Scholar]
- 18.Jiang, X., D. Y. Graham, K. Wang, and M. K. Estes. 1990. Norwalk virus genome: cloning and characterization. Science 250:1580-1583. [DOI] [PubMed] [Google Scholar]
- 19.Kapikian, A. Z., R. G. Wyatt, R. Dolin, T. S. Thornhill, A. R. Kalica, and R. M. Chanock. 1972. Visualization by immune electron microscopy of a 27-nm particle associated with acute infectious nonbacterial gastroenteritis. J. Virol. 10:1075-1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaplan, J. E., R. A. Goodman, L. B. Schonberger, E. C. Lippy, and G. W. Gary. 1982. Gastroenteritis due to Norwalk virus: an outbreak associated with a municipal water system. J. Infect. Dis. 146:190-197. [DOI] [PubMed] [Google Scholar]
- 21.Kuritsky, J. N., M. T. Osterholm, H. B. Greenberg, J. A. Korlath, J. R. Godes, C. W. Hedberg, J. C. Forfang, A. Z. Kapikian, J. C. McCullough, and K. E. White. 1984. Norwalk gastroenteritis: a community outbreak associated with bakery product consumption. Ann. Intern. Med. 100:519-521. [DOI] [PubMed] [Google Scholar]
- 22.Lalumandier, J. A., and L. W. Ayers. 2000. Fluoride and bacterial content of bottled water vs tap water. Arch. Fam. Med. 9:246-250. [DOI] [PubMed] [Google Scholar]
- 23.Lambden, P. R., E. O. Caul, C. R. Ashley, and I. N. Clarke. 1993. Sequence and genome organization of a human small round-structured (Norwalk-like) virus. Science 259:516-519. [DOI] [PubMed] [Google Scholar]
- 24.Lawson, H. W., M. M. Braun, R. I. Glass, S. E. Stine, S. S. Monroe, H. K. Atrash, L. E. Lee, and S. J. Englender. 1991. Waterborne outbreaks of Norwalk virus gastroenteritis at a southwest US resort: role of geological formations in contamination of well water. Lancet 337:1200-1204. [DOI] [PubMed] [Google Scholar]
- 25.Matthews, R. E. 1979. Classification and nomenclature of viruses. Intervirology 12:129-296. [DOI] [PubMed] [Google Scholar]
- 26.Maurer, A. M., and D. A. Sturchler. 2000. A waterborne outbreak of small round structured virus, campylobacter and shigella co-infections in La Neuveville, Switzerland, 1998. Epidemiol. Infect. 125:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murphy, A. M., G. S. Grohmann, P. J. Christopher, W. A. Lopez, G. R. Davey, and R. H. Millsom. 1979. An Australia-wide outbreak of gastroenteritis from oysters caused by Norwalk virus. Med. J. Aust. 2:329-333. [DOI] [PubMed] [Google Scholar]
- 29.Noel, J. S., R. L. Fankhauser, T. Ando, S. S. Monroe, and R. I. Glass. 1999. Identification of a distinct common strain of “Norwalk-like viruses” having a global distribution. J. Infect. Dis. 179:1334-1344. [DOI] [PubMed] [Google Scholar]
- 30.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 31.Parashar, U. D., L. Dow, R. L. Fankhauser, C. D. Humphrey, J. Miller, T. Ando, K. S. Williams, C. R. Eddy, J. N. Noel, T. Ingram, J. S. Bresee, S. S. Monroe, and R. I. Glass. 1998. An outbreak of viral gastroenteritis associated with consumption of sandwiches: implications for the control of transmission by food handlers. Epidemiol. Infect. 121:615-621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pringle, C. R. 1999. Virus taxonomy. Arch. Virol. 144:421-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schaub, S. A., and R. K. Oshiro. 2000. Public health concerns about caliciviruses as waterborne contaminants. J. Infect. Dis. 181(Suppl. 2):374-380. [DOI] [PubMed] [Google Scholar]
- 34.Schindler, P. R. 1994. Enterobacteria in mineral, spring and table water. Gesundheitswesen 56:690-693. [PubMed] [Google Scholar]
- 35.Sefcova, H. 1998. Survey of the microbiological quality of bottled water. Cent. Eur. J. Public Health 6:42-44. [PubMed] [Google Scholar]
- 36.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tsai, G. J., and S. C. Yu. 1997. Microbiological evaluation of bottled uncarbonated mineral water in Taiwan. Int. J. Food Microbiol. 37:137-143. [DOI] [PubMed] [Google Scholar]
- 38.Vinje, J., and M. P. Koopmans. 1996. Molecular detection and epidemiology of small round structured viruses in outbreaks of gastroenteritis in The Netherlands. J. Infect. Dis. 174:610-615. [DOI] [PubMed] [Google Scholar]
- 39.Warburton, D., B. Harrison, C. Crawford, R. Foster, C. Fox, L. Gour, and P. Krol. 1998. A further review of the microbiological quality of bottled water sold in Canada: 1992-1997 survey results. Int. J. Food Microbiol. 39:221-226. [DOI] [PubMed] [Google Scholar]
- 40.Wheeler, J. G., D. Sethi, J. M. Cowden, P. G. Wall, L. C. Rodrigues, D. S. Tompkins, M. J. Hudson, and P. J. Roderick. 1999. Study of infectious intestinal disease in England: rates in the community, presenting to general practice, and reported to national surveillance. BMJ 318:1046-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wilkinson, F. H., and K. G. Kerr. 1998. Bottled water as a source of multi-resistant Stenotrophomonas and Pseudomonas species for neutropenic patients. Eur. J. Cancer Care 7:12-14. [DOI] [PubMed] [Google Scholar]