Abstract
We studied the microbial diversity of benthic cyanobacterial mats inhabiting a heavily polluted site in a coastal stream (Wadi Gaza) and monitored the microbial community response induced by exposure to and degradation of four model petroleum compounds in the laboratory. Phormidium- and Oscillatoria-like cyanobacterial morphotypes were dominant in the field. Bacteria belonging to different groups, mainly the Cytophaga-Flavobacterium-Bacteriodes group, the γ and β subclasses of the class Proteobacteria, and the green nonsulfur bacteria, were also detected. In slurry experiments, these communities efficiently degraded phenanthrene and dibenzothiophene completely in 7 days both in the light and in the dark. n-Octadecane and pristane were degraded to 25 and 34% of their original levels, respectively, within 7 days, but there was no further degradation until 40 days. Both cyanobacterial and bacterial communities exhibited noticeable changes concomitant with degradation of the compounds. The populations enriched by exposure to petroleum compounds included a cyanobacterium affiliated phylogenetically with Halomicronema. Bacteria enriched both in the light and in the dark, but not bacteria enriched in any of the controls, belonged to the newly described Holophaga-Geothrix-Acidobacterium phylum. In addition, another bacterial population, found to be a member of green nonsulfur bacteria, was detected only in the bacteria treated in the light. All or some of the populations may play a significant role in metabolizing the petroleum compounds. We concluded that the microbial mats from Wadi Gaza are rich in microorganisms with high biodegradative potential.
There is general interest in studying the diversity of indigenous microorganisms capable of degrading pollutants such as crude oil, polycyclic aromatic hydrocarbons, and polychlorinated biphenyls in different environments (32, 33, 43, 48). Identification of the key organisms that play a role in pollutant degradation processes is relevant to the development of optimal in situ bioremediation strategies. Indeed, efforts have been made to characterize bacterial communities and their responses to pollutants (17, 43), to isolate potential degraders (9), and to identify functional genes involved in particular degradation processes (37, 40, 45, 57). Evidence has been presented that microbial communities dominated by phototrophic cyanobacteria can be actively involved in the degradation of petroleum and its derivatives. Observations made after oil spills in the Arabian Gulf showed that there was intensive colonization of polluted sites by cyanobacterium-dominated microbial mats that correlated with the disappearance of hydrocarbons (29, 30). Other studies have demonstrated the capacity of cyanobacterial isolates to degrade hydrocarbons (2, 10-12). While many studies to describe bacterial diversity and community changes in various pollutant-degrading communities have been performed by using recently developed culture-independent molecular techniques, little research has focused on microbial communities dominated by photosynthetic organisms in relation to oil degradation. We studied the microbial diversity of a benthic cyanobacterium-dominated community inhabiting a heavily polluted site in Wadi Gaza (Gaza Strip, Palestine), a river that receives a variety of pollutants, such as diesel oil, other petroleum products, sewage, pesticides, solid waste, agricultural discharge, and industrial discharge. We also investigated the ability of native communities to degrade model compounds (n-octadecane, pristane, phenanthrene, and dibenzothiophene) in the laboratory, and we monitored changes in the microbial community linked to exposure to these compounds. The compounds which we used represent the most important groups of petroleum constituents (straight-chain alkanes, branched alkanes, aromatic hydrocarbons, and organosulfur compounds). These pollutants were found to occur in Wadi Gaza sediments at variable levels (53). In general, alkanes and polycyclic aromatic hydrocarbons are ubiquitous environmental pollutants. Particular attention was paid to polycyclic aromatic hydrocarbons as they exhibit toxic, mutagenic, and carcinogenic properties (14, 16, 38). Combustion of sulfur-containing organic compounds, such as dibenzothiophene, emits noxious sulfur dioxide, which contributes to acid rain and air pollution (19, 36). Petroleum compounds are considered to be recalcitrant to microbial degradation and persist in ecosystems because of their hydrophobic nature (low water solubility) and low volatility, and thus they pose a significant threat to the environment.
MATERIALS AND METHODS
Environmental setting of the microbial mats.
The mat samples used in this study were obtained from Wadi Gaza, Gaza Strip, Palestine. This wadi originates at Hebron in the West Bank and flows in the east-west direction across the Gaza Strip. It is the only surface water in the Gaza Strip and is heavily used for disposal of sewage, solid waste, and agricultural and industrial wastewater. In the west, Wadi Gaza reaches the Mediterranean Sea. The connection to the sea is intermittent, depending on the rainfall. Here, where the samples were collected, most of the cyanobacterial mats develop in the presence of high levels of pollutants, mainly diesel oil and other petroleum products (53). The salinity, temperature, and water level of the wadi change seasonally, which results in marked changes in the appearance of the mats. In the western part of the wadi, the salinity ranges from 1% (wt/vol) total salts in the winter to 3.5% (wt/vol) total salts in the summer, and the average daily temperature varies between 15°C in the winter and 35°C in the summer. At the time of sampling (May 2000), the mats were submerged, the measured water temperature was 25°C, and the salinity was 2%.
Preparation of organo-clay complexes.
n-Octadecane, pristane, phenanthrene, and dibenzothiophene (Sigma-Aldrich) were used as model compounds for petroleum constituents. Hydrophobic clay was used as a carrier for the model petroleum compounds in our experiments in order to enhance the contact of the compounds with the microbial community and to increase their bioavailability. A 2% aqueous suspension of montmorillonite KSF (Aldrich) was prepared, and 0.8 mmol of benzyl-trimethylammonium (BTMA) chloride per g of clay was added slowly as a 10 mM solution (modified as described by El-Nahhal et al. [20]). The mixture was stirred for 24 h, washed three times to remove excess BTMA chloride, and then freeze-dried. To adsorb the model compounds, the hydrophobic clay (BTMA-montmorillonite) was suspended in n-hexane. The mixture of model compounds (20 mg per 100 mg of hydrophobic clay, dissolved in n-hexane) was slowly added with continuous stirring. The slurry was dried in a vacuum rotary evaporator, which removed the n-hexane. This yielded a homogeneous powder consisting of hydrophobic clay loaded with 16.67% (wt/wt) petroleum model compounds (designated the organo-clay complex). The adsorbed model compounds were reextracted with dichloromethane (DCM) and analyzed by gas chromatography.
Experimental design.
Experiments were carried out in sterile 250-ml Erlenmeyer flasks. The medium used was prepared by mixing equal volumes of natural seawater and distilled water. The salinity was adjusted to 2% to mimic the salinity of the site at the time of sampling. Nitrogen and phosphate sources were added to the medium by adding 1 mM ammonium chloride and 8 μM sodium dihydrophosphate, respectively. Each Erlenmeyer flask except the control flasks received 100 ml of autoclaved medium, 1 g of Wadi Gaza mat material, and 100 mg of organo-clay complex. The following four controls, two for chemical analysis and two for community structure comparison, were used: (i) medium and organo-clay complex without mat material (incubated in the light and in the dark); (ii) medium and organo-clay complex with autoclaved mat material; (iii) medium and mat material without organo-clay complex (control G); and (iv) medium, mat material, and hydrophobic clay without model compounds (control GC). The first two controls were used to check for photooxidation of the model compounds and to account for adsorption of the organo-clay complex particles to the mat material, respectively. The other two controls (controls G and GC) were used to monitor changes in the community in the absence of model organic compounds. All flasks were incubated at 28°C with constant shaking at 100 rpm and a light regime consisting of 12 h of light and 12 h of darkness. The light intensity was 80 μmol of photons m−2 s−1 (photosynthetically available radiation). One flask containing medium, organo-clay complex particles, and mat inoculum was incubated in complete darkness. The experiment was performed for 40 days, and samples used for chemical analysis (2 ml each) were taken every 4 days during the first 2 weeks and every 10 days until the end of the experiment. At the end of the experiment, the contents of the flasks were collected by centrifugation.
Microscopy and cultivation.
Small pieces of microbial mats from Wadi Gaza were torn apart and mounted in water on glass microscopic slides. The slides were examined by transmitted light microscopy, phase-contrast microscopy, and fluorescence microscopy. Different cyanobacterial morphotypes were identified and photographed. Different cyanobacteria were cultivated in BG11 defined medium (52) enriched with the four model compounds (1 mg of organo-clay complex per ml medium). The salinity was adjusted to 2%, and incubation was at 28°C.
Chemical analyses.
Samples (2 ml) were taken from the Erlenmeyer flasks with disposable pipettes. The flasks were shaken vigorously to suspend the solid material in order to obtain homogeneous samples. The samples were extracted ultrasonically with a mixture of methanol, DCM, and water (1:0.5:0.4, vol/vol/vol; modified as described by Bligh and Dyer [4]). After centrifugation, the supernatant was collected in a separatory funnel. This procedure was repeated four times. DCM and water were added to the combined supernatant to give a methanol/DCM/H2O ratio of 1:1:0.9 (vol/vol/vol), which resulted in phase separation. The DCM layer was collected, and the methanol-water phase was washed three times with DCM. The solvent of the combined DCM phase was removed with a rotary evaporator, and the extract was diluted to concentrations appropriate for gas chromatography analysis.
The extracts were analyzed with a Hewlett-Packard 6890 gas chromatograph equipped with a Gerstel KAS3 temperature-programmable injector, a flame ionization detector, and a fused silica column (J&W DB-5HT; 30 m by 0.25 mm; film thickness, 0.15 μm). After an isothermal phase at 60°C for 2 min, the oven was heated at a rate of 20°C/min to 150°C and then at a rate of 3°C/min to 310°C and then kept at 310°C for 15 min.
Model compounds were quantified by integration of the flame ionization detector signals and comparison with the signal of an internal standard (squalane) which was added immediately after extraction. The calculated initial amount of each model compound in a 2-ml sample was 66.67 μg. The abundance of the model compounds was determined relative to this value.
Molecular analysis.
The field material (Wadi Gaza microbial mats) and the biomass collected from controls G and GC and the two treatment preparations incubated in the light and in the dark were subjected to nucleic acid extraction, PCR, and denaturing gradient gel electrophoresis (DGGE) as previously described (1). PCR amplification of 16S rRNA was carried out by using two sets of oligonucleotide primers. CYA359F (with a 40-nucleotide clamp at the 5′ end) and CYA781R were used as specific primers for cyanobacteria (49), and GM5F with a GC clamp and universal primer 907RC were used for all bacteria (54). A hot-start program was used for the cyanobacterium-specific primers, as described by Nübel et al. (49), whereas in case of the universal bacterial primers (GM5F and 907R), a hot-start touchdown program was used to minimize nonspecific amplification (54). One thousand nanograms of the PCR-amplified material was loaded on a DGGE gel, which was subsequently electrophoresed at 60°C at a constant voltage of 200 V for 3.5 h (1). The DGGE gradient concentrations were 20 to 60% in case of the cyanobacterial primers and 20 to 80% in case of the universal primers. The DGGE bands were excised manually, the DNA was allowed to diffuse out in buffer overnight, and another PCR was performed. Products were then sequenced commercially after their purity was confirmed by using another DGGE gel.
Phylogenetic affiliation.
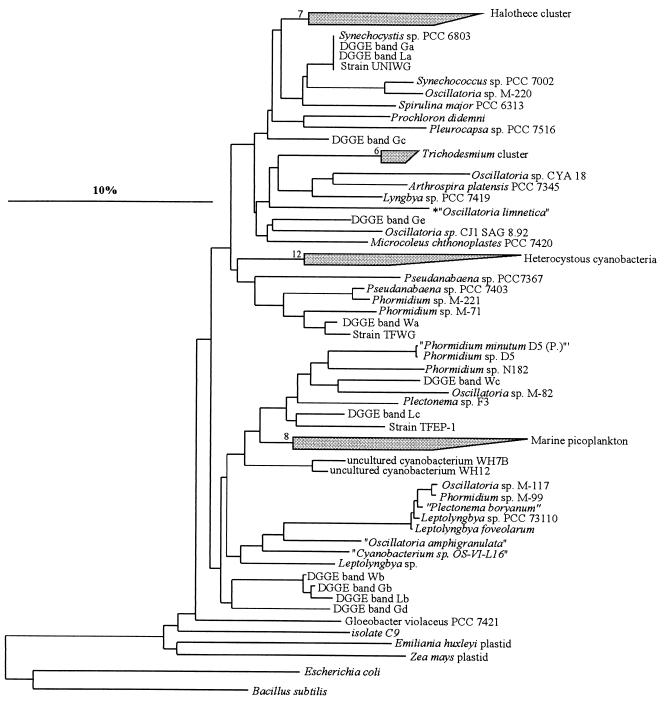
Cyanobacterial phylogenetic trees were constructed based on long (more than 1,300-bp) 16S rRNA sequences by using different methods integrated in the ARB software (42); these methods included maximum-likelihood, maximum-parsimony, and neighbor-joining procedures (a maximum-likelihood tree is shown in Fig. 2). The 16S rRNA sequences of Escherichia coli and Bacillus subtilis were included in the calculations as outgroup sequences. Partial cyanobacterial sequences (ca. 400 bp) obtained in this study were aligned with the sequences in the ARB database using the alignment tool of the ARB software package (42). These sequences were then inserted into the preestablished tree by using the ARB parsimony tool and maintaining the overall tree topology without changes. Partial bacterial sequences were also inserted into a preestablished stable tree containing all bacterial sequences in the ARB database without allowing changes in the overall tree topology. The final tree was minimized for simplicity of presentation.
FIG. 2.
Phylogenetic tree (maximum likelihood) for cyanobacteria and plastids based on publicly available, almost complete 16S rRNA genes from members of the cyanobacterial line. E. coli and B. subtilis were used as outgroups. The numbers next to the collapsed clusters indicate the number of sequences in each cluster. The 400-bp 16S rRNA sequences from our excised DGGE bands were placed phylogenetically by using parsimony criteria without changing the topology of the preestablished tree. The bar indicates 10% sequence divergence. The identities of strains correspond to the identities given in the databases, and we do not imply that they are necessarily taxonomically correct.
Nucleotide sequence accession numbers.
The sequences determined in this study have been deposited in the GenBank database under accession numbers AF423347 to AF423377.
RESULTS
Composition of the cyanobacterial community in the native microbial mats from Wadi Gaza.
Microscopic observation of the native mat samples revealed the presence of at least three morphotypes of filamentous cyanobacteria with different trichome widths. One of these morphotypes was a cyanobacterium with a thin filamentous, Phormidium-like morphology (trichome width, 1 μm). A second Phormidium-like morphotype with wider trichomes (trichome width, 2 to 4 μm) and clearly visible cross walls was also observed. The cells of this cyanobacterium were longer than they were wide (length, 3 to 6 μm) and had prominent constrictions at the cross walls. The third morphotype was an Oscillatoria-like cyanobacterium with trichomes that were 6 to 8 μm wide whose cells were discoid. In addition to these cyanobacterial morphotypes we also observed very low levels of some pennate diatoms and unicellular green algae.
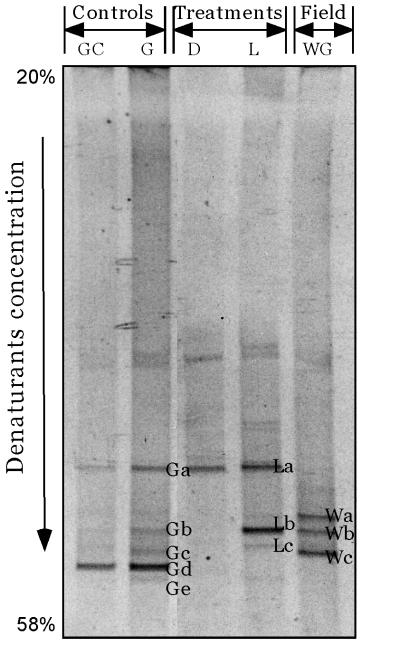
DGGE analysis of the community using cyanobacterium-specific primers revealed three distinguishable bands (Fig. 1, bands Wa, Wb, and Wc). The band patterns were very similar for triplicate extracts from the same mat sample. The sequence represented by band Wa was affiliated closely with a group of 16S rRNA gene sequences from thin filamentous cyanobacteria belonging to the genera Phormidium and Pseudanabaena (Fig. 2). Upon enrichment cultivation, a thin filamentous cyanobacterium (width, 1 μm) was isolated (strain TFWG), and its sequence was found to be closely affiliated with the sequence of this DGGE band (99% sequence similarity). This indicates that this cultivated cyanobacterium is most probably a representative of the field population represented by DGGE band Wa. The sequence of DGGE band Wb fell next to a cluster of sequences of filamentous cyanobacteria which included strains assigned to the genera Phormidium, Leptolyngbya, Plectonema, and Oscillatoria. However, the sequence of band Wb was at least 9% dissimilar when it was compared with any of the sequences in this cluster. The sequence of DGGE band Wc mapped phylogenetically close to the sequence of Oscillatoria sp. strain M-82 (31) (sequence similarity, ca. 94%).
FIG. 1.
DGGE band patterns of PCR-amplified 16S rRNA fragments obtained by using cyanobacterium-specific primers and the Wadi Gaza field microbial mats (lane WG) and mats collected at the end of the experiments from the microbiological controls (lanes G and GC) and from preparations incubated in the light (lane L) and in the dark (lane D). The labeled bands were excised, reamplified, and sequenced.
Bacterial community structure of the native microbial mats from Wadi Gaza.
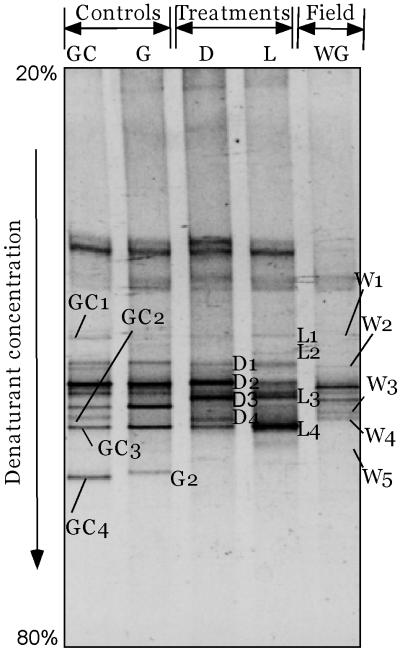
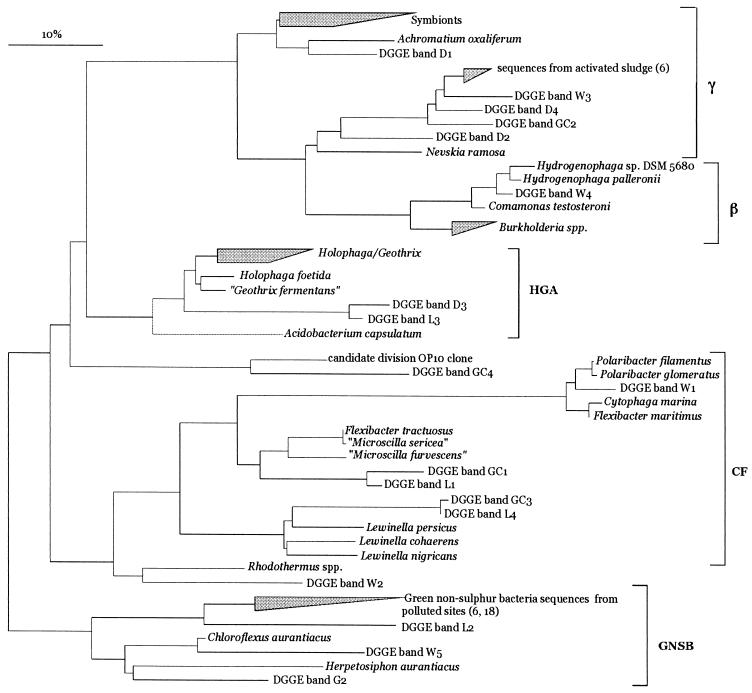
The band pattern generated with the universal eubacterial primers (GM5F and 907RC) was more complex than the pattern generated with the cyanobacterial primers (Fig. 3). This was expected, as these primers amplify not only 16S rRNAs of dominant bacteria but also 16S rRNAs of cyanobacteria. Eight clear bands were obtained, and three of them were cyanobacterial, as determined by subsequent sequencing and phylogenetic analysis. This analysis yielded results virtually identical to those obtained with the cyanobacterium-specific primers, and these results are not discussed further. The phylogenetic reconstruction obtained by using the remaining bands is shown in Fig. 4. The band W1 sequence mapped to the Cytophaga-Flavobacterium-Bacteriodes group and was affiliated with the sequence of the marine bacterium Polaribacter glomeratus (formerly Flectobacillus glomeratus) (level of sequence similarity, 93.7%) (25). The band W3 sequence fell within the sequences of the γ subclass of the class Proteobacteria next to a group of environmental bacterial sequences obtained from activated sludges (level of sequence similarity, 95.7%) (6). On the other hand, the band W4 sequence fell within the sequences of the β subclass of the Proteobacteria and was closely affiliated with the sequences of Hydrogenophaga palleroni (level of sequence similarity, 97.6%) and Hydrogenophaga sp. strain DSM 5680 (level of sequence similarity, 95.7%). These two organisms have the ability to aerobically degrade the sulfonated azo compounds 4-carboxy-4′-sulfoazobenzene (5) and 4-aminobenzenesulfonic acid (21), respectively. Sequences from bands W2 and W5 exhibited at most 82% similarity to any other sequences available in the database. The band W2 sequence fell next to sequences from Rhodothermus sp., while the band W5 sequence fell next to a group of sequences belonging to green nonsulfur bacteria (Chloroflexaceae), suggesting that this sequence is probably a member of this group. It is thought that some of the environmental sequences closely related to our sequences may be sequences of members of pollutant-degrading microbial consortia (6, 18).
FIG. 3.
DGGE profiles of PCR-amplified 16S rRNA fragments obtained by using universal bacterial primers from the Wadi Gaza field microbial mats (lane WG) and mats collected at the end of the experiments from the microbiological controls (lanes G and GC) and from preparations incubated in the light (lane L) and in the dark (lane D). The labeled fragments were excised, reamplified, and sequenced. The unlabeled bands include those containing cyanobacterial sequences and bacterial bands with sequences similar to the sequences of the labeled bands.
FIG. 4.
Unrooted phylogenetic tree showing affiliations based on partial bacterial 16S rRNA sequences and selected sequences from members of different bacterial clusters, including the γ and β subclasses of the Proteobacteria, the Cytophaga-Flavobacterium-Bacteriodes group (CF), the newly described Holophaga-Geothrix-Acidobacterium phylum (HGA), and the green nonsulfur bacteria (GNSB). The tree was simplified for clarity by omitting all sequences between clusters. The numbers in parentheses indicate the references in which the clustered sequences were described. The scale bar indicates 10% estimated sequence divergence.
Degradation of four model compounds.
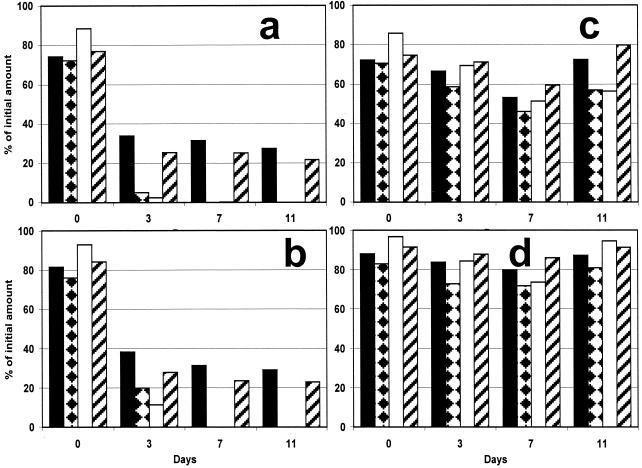
Degradation of pristane, dibenzothiophene, phenanthrene, and n-octadecane was studied by monitoring the disappearance of these compounds during experiments (Fig. 5). The amount of each model compound added to the experimental flasks was 3.33 mg (corresponding to 13.1, 13.9, 18.7, and 18.1 μmol of n-octadecane, pristane, dibenzothiophene, and phenanthrene, respectively). Extraction of the suspension at zero time yielded 76 to 84% of the theoretical amounts of the four model compounds. Such levels of recovery are commonly obtained as a result of experimental and analytical conditions. In the control samples with and without dead biomass (Fig. 5c and d), the amounts and unaltered relative compositions of the model compounds indicated that no degradation occurred. Slightly lower yields were obtained for the control containing dead biomass (Fig. 5c). A lower level of recovery and a slight decrease in the amount of n-octadecane relative to the amounts of the other model compounds were also observed for the control without biomass incubated in the light at day 11; this pattern remained unaltered until 40 days. After 3 days of treatment in the light, 3% of the phenanthrene initially added and 5% of the dibenzothiophene initially added were present. In the dark, the amounts of these two aromatic compounds were 11 and 20% of the amounts initially added (Fig. 5a and b). In the samples collected on and after day 7, these compounds were not present either in the preparations incubated in the light or in the preparations incubated in the dark. After 3 days, the aliphatic compounds, pristane and n-octadecane, were also degraded. In case of n-octadecane, ca. 25% remained after 3 days of incubation in the light or in the dark. Similarly, pristane was degraded so that 34% remained in the preparations incubated in the light and 38% remained in the preparations incubated in the dark (Fig. 5b). During the rest of the experiment until day 40, the amounts of pristane and n-octadecane remained constant at 30 and 25% of the amounts added initially, respectively.
FIG. 5.
Plots of quantities of model compounds in slurry experiments with Wadi Gaza microbial mats versus time. (a) Twelve hours of light and 12 h of darkness; (b) 24 h of darkness; (c) control with autoclaved biomass, 12 h of light and 12 h of darkness; (d) control without biomass, 24 h of darkness. The data are expressed as percentages relative to the amounts of the compounds initially introduced into the system (i.e., 3.33 mg of each model compound adsorbed on 100 mg of organo-clay complex particles). Solid bars, pristane; bars with diamonds, dibenzothiophene; open bars, phenanthrene; cross-hatched bars, n-octadecane.
Cyanobacterial community changes following exposure to pollutants.
DGGE analysis showed that there were noticeable changes in the cyanobacterial communities both in the controls and in the treated preparations (Fig. 1). The band pattern suggested that dominant species of cyanobacteria were replaced by other species. This was observed previously when mats were examined under laboratory conditions (1). Some field bands, such as bands Wa and Wc, disappeared from the treated mats as well as from the control mats; band Wb persisted in one treated preparation (preparation incubated in the light) and one control (control G). A new population represented by band La developed in all treated preparations and controls. The sequence of this band was phylogenetically affiliated with the sequence of Synechocystis sp. strain PCC 6803 (level of sequence similarity, 100%). This population was successfully isolated on a medium enriched with model compounds and, as expected, was found to be a unicellular cyanobacterium (diameter of cells, 2 to 3 μm) with the morphology of Synechocystis species. In the presence of model compounds, the mat incubated in the light produced three bands, one of which was affiliated with Syenchocystis sp. strain PCC 6803. The second band was at the same DGGE position as band Wb obtained from the native mat, and its sequence was closely related to the sequence of this band from the native mat. The third band (DGGE band Lc) was observed only in this preparation; it was not obtained with any of the controls or the native mats. This indicates that it was a direct development in response to the addition of the model compounds. The sequence of this band was similar to that of strain TFEP1, a representative of the newly proposed genus Halomicronema, which has extremely thin filaments (level of sequence similarity, 94.5%) (R. M. M. Abed, F. Garcia-Pichel, and M. Hernández-Mariné, submitted for publication). In the dark, all of the field cyanobacteria disappeared, and amplification of DNA using cyanobacterium-specific primers yielded a product whose concentration was very low. Upon DGGE of a concentrated product obtained from several PCRs, only one cyanobacterial band was present on the gel; the sequence of this band was affiliated with the sequence of Synechocystis sp. strain PCC 6803. The controls produced band patterns different from those of the treated preparations. The mat incubated in the absence of the model compounds exhibited greater diversity, represented by the development of five distinguishable bands. Two of these bands (bands Ga and Gb) were similar to bands detected in the mat incubated in the presence of the model compounds in the light. The other three bands (bands Gc, Gd, and Ge) did not show more than 93% sequence similarity to any of the cyanobacterial sequences available in the database. In the presence of clay without model compounds, only two bands were detected, and these bands were similar to those detected with control G. This implies that the only change directly attributable to the addition of the model compounds was enrichment of the population represented by band Lc in the light treatment.
Changes in bacterial community following degradation of model compounds.
Almost all DGGE bands corresponding to field bacteria were replaced by other bands during the treatments, demonstrating that there were dramatic shifts in the community structure. Some of the new bands were present in all experiments, including the controls, whereas others were specific for certain treatments (Fig. 3). The total number of bands was higher for the controls than for the treated preparations, indicating that the model compounds inhibited the growth of some bacteria. The sequences of the new common bands fell phylogenetically within different groups, such as the γ subclass of the Proteobacteria (band D1), the β subclass of the Proteobacteria (band D2), and the Cytophaga-Flavobacter cluster (bands GC1, L1, GC3, and L4). The sequences of these bands did not exhibit more than 85% similarity to any of the sequences available in the database. In the presence of the model compounds, there were four bands which were not detected in any of the controls, bands L3, D3, L2, and D4. Bands L3 and D3 were detected after the light and dark treatments, respectively. Phylogenetically, the sequences of these bands fell within a group of sequences belonging to the newly described Holophaga-Geothrix-Acidobacterium phylum, which was defined mainly on the basis of environmental sequences (41). This phylum includes only three cultivated isolates, one of which is an aerobic organism (Acidobacterium capsulatum) (34) and two of which are anaerobic organisms (Holophaga foetida and Geothrix fermentans) (15, 39). Band L2 was detected only in preparations incubated in the light, and its sequence was related to sequences from green nonsulfur bacteria. On the other hand, band D4 was detected only in preparations in the dark, and its sequence was similar to the sequence of native band W3, which exhibited ca. 95.7% sequence similarity to environmental bacterial sequences obtained from phosphate-removing and non-phosphate-removing activated sludges (6). Bands G2 and GC4 were detected only in controls G and GC, respectively. Their phylogenetic affiliations are shown in Fig. 4.
DISCUSSION
The indigenous microorganisms inhabiting the highly polluted Wadi Gaza site which we investigated seem to have a strong potential to effectively degrade petroleum compounds. This conclusion supports the results of previous studies, which indicated that chronic exposure to high levels of petroleum results in microbial communities that are adapted to hydrocarbons and have higher concentrations of hydrocarbon-degrading bacteria (7, 8).
Use of the organo-clay complex as a carrier.
The poor water solubility of most petroleum compounds (e.g., phenanthrene, <1.3 mg/liter [50]; dibenzothiophene, 1.47 mg/liter [28]) limits their biodegradation in nature. Strategies to enhance the accessibility of such compounds to microorganisms include increasing the solubility using natural or synthetic surfactants and emulsifiers (3, 26) and adsorbing them to solid substrates, such as clay or resins (22, 35). In this study we used organo-clay complexes to simulate the natural conditions in which hydrophobic contaminants are present in the adsorbed form. However, it is likely that the bacteria from Wadi Gaza are adapted to grow on solid surfaces since the inoculum was obtained from a surface biofilm on fine-grained sediment. In the dark control without biomass, more than 75% of the model compounds was recovered at the end of the experiment, and this indicated that the organo-clay complex was stable throughout the experiment and there was no biodegradation (Fig. 5c). The level of recovery was slightly lower than that obtained upon direct extraction of organo-clay complex suspensions (more than 90% [Köster, unpublished data]). This could have been due to the experimental procedures used, such as the sampling and chemical analysis procedures. A comparison of the light control without biomass (data not shown) and the dark control without biomass (Fig. 5d) showed that the model compounds were not photooxidized. The control with autoclaved biomass (Fig. 5c) indicated that adsorption was not significant (Fig 5d). Consequently, we attributed the disappearance of the model compounds in the presence of living biomass to biodegradation activity. The aromatic compounds were completely degraded, indicating that these compounds were accessible to the degrading bacteria. In contrast, the amounts of aliphatic hydrocarbons decreased after 3 days and then remained constant for up to 40 days. We assumed that alkanes were only partially accessible to bacteria due to strong binding in the BTMA-montmorillonite interlayers but were completely extractable with organic solvents. Indeed, it has been shown for phenanthrene adsorbed on unmodified montmorillonite that different modes of binding may lead to different rates of biodegradation (35, 58). There have been no detailed studies of the competitive adsorption behaviors of several compounds on organo-clay complexes. We observed that both cyanobacterial and bacterial communities in the absence of clay were different from the communities in samples containing either hydrophobic clay or organo-clay complex particles. The hydrophobic clay seemed to inhibit the growth of some cyanobacteria (Fig. 1) but enhanced the growth of other bacteria (Fig. 4). Therefore, the changes in the community induced by the hydrophobic clay could be discriminated by examining the control containing only this hydrophobic clay. Such differences in microbial communities in the presence of a sorptive phase compared to microbial communities in the absence of a sorbent have been described previously (27, 22).
Role of cyanobacteria in biodegradation.
Recent reports demonstrated that photosynthetic microorganisms, particularly cyanobacteria, may play a direct or indirect role in the metabolism and degradation of hydrocarbons. Cyanobacteria such as Anabaena cylindrica, Phormidium faveolarum, Oscillatoria sp. strain JCM, and Agmenellum quadruplicatum can degrade different aromatic compounds (10, 11, 51). Narro (47) demonstrated the ability of A. quadruplicatum to oxidize phenanthrene to trans-9,10-dihydroxy-9,10-dihydrophenanthrene and 1-methoxy phenanthrene with a monooxygenase system (47). Degradation of n-alkanes by two nonaxenic organisms, Microcoleus chthonoplastes and Phormidium corium isolated from oil-contaminated sediments, was also demonstrated (2). The n-alkanes were oxidized to fatty acids, which were subsequently incorporated into cell lipids. In mat systems, cyanobacteria are present in association with oil-degrading bacteria and prevent them from being washed out by immobilizing them in their mucilage. In addition, cyanobacteria also supply these bacteria with oxygen produced by photosynthesis and the fixed nitrogen needed for their activity in the degradation processes. This indirect role of cyanobacteria can be important to the overall success of the biodegradation process.
In our experiment, shifts in cyanobacterial community composition were dramatic, and two field species were eliminated. These species might have been sensitive to cultivation conditions in the laboratory. However, new cyanobacteria became conspicuous in the experiment, although they were not detectable in the field sample. These species were probably present in the field at very low levels. The maximum number of species was detected in the control in which the mat sample was cultivated in the absence of model compounds, lending credibility to the notion that cyanobacteria exhibit maximum diversity in the absence of petroleum compounds. Many studies have demonstrated that crude oil includes constituents that are inhibitory to cyanobacteria even at low concentrations (reference 51 and references therein). Aromatic compounds have been shown to have more drastic effects than alkanes. These compounds inhibit photosynthesis and growth, reduce enzyme activity and microbial biomass, and induce changes in cyanobacterial species composition (44). In the presence of model compounds in the light experiment, the conditions favored the growth of three cyanobacterial populations. The population represented by band Lc, which appeared only in this experiment, might play a role in detoxification of any of the model compounds. In order to demonstrate this, experiments with axenic cultures of the respective population are needed. Another population, affiliated with Synechocystis sp. strain PCC 6803, developed during this treatment and also in the controls. A search of the complete genome of Synechocystis sp. strain PCC 6803 (http://www.ncbi.nlm.nih.gov) revealed that this cyanobacterium possesses dioxygenase genes, which catalyze incorporation of both atoms of an oxygen molecule into an aromatic ring, producing dihydrodiols (23). This suggests that this cyanobacterium might also have played a role in the aerobic degradation of aromatic compounds like dibenzothiophene and phenanthrene in our experiment. In any case, these three populations certainly tolerated the concentrations of the model compounds supplied, indicating that sensitive species present in the field were replaced by more resistant species. As cyanobacteria are sensitive to pollution, any alteration of the community composition could be useful as a bioindicator of pollution (44).
Bacteria in Wadi Gaza.
DGGE analysis suggested that the bacterial community in Wadi Gaza was less diverse than communities that developed during the experiments. This was apparently due to the environmental stress caused by the high level of pollutants, which allowed only a restricted number of species that tolerated such conditions. However, the site studied seems to be very rich in novel microorganisms since in many cases phylogenetic analysis of sequences obtained from the bacterial bands did not result in close matches with any known organisms. Many sequences fell within the Cytophaga-Flavobacterium and the proteobacterial (γ and β) groups. These groups have previously been reported to be present in marine ecosystems (24), wastewaters (13), and oil-polluted sites (43, 59). The presence of some sequences (bands W3, D4, GC2, and D2) related to sequences found in activated sludge is consistent with the sewage pollution at the site, which must favor the growth of such species. The field species represented by band W4 fell within a group of organisms having degradation abilities which suggest that this organism may play a role in biodegradation of pollutants in the field. In the experiments in which the model compounds were degraded, phylogenetic characterization of the major bands indicated that they do not represent genera containing known hydrocarbon-degrading bacteria, such as Sphingomonas, Pseudomona, Burkholderia, and Mycobacterium. Instead, these bands were found to represent the newly described Holophaga-Geothrix-Acidobacterium phylum (bands D3 and L3), the γ subclass of the Proteobacteria (band D4), and the green nonsulfur bacteria (band L2). The appearance of these bands only in the presence of the model compounds and not in the controls implies that these organisms might contribute to degradation of the compounds. Bands L3 and D3 were obtained from the preparations incubated in the light and in the dark, respectively, and were found to have similar sequences and phylogenies. These sequences fell within the Holophaga-Geothrix-Acidobacterium phylum (41). One of the representative isolates of this group is the Fe(III)-reducing bacterium G. fermentans, which was isolated from a contaminated aquifer that was adapted to rapid anaerobic oxidation of toluene coupled to Fe(III) reduction (15). Another member of this group is the obligately anaerobic bacterium H. foetida, which has the ability to degrade methoxylated aromatic compounds (39). This suggests that these compounds might have also been metabolized anaerobically. Anaerobic degradation of petroleum compounds has been demonstrated previously (60). Our experiments were performed in an open, oxic system, but it is possible that there were anaerobic niches and anaerobic microorganisms. It is likely that anaerobic processes are more important in the natural environment of Wadi Gaza, in which the fine-grained sediment is oxygen depleted at depths below a few millimeters. The presence and metabolic activity of anaerobic organisms in mainly oxic zones of microbial mats have been demonstrated previously (46, 55, 56). The presence of band L2, phylogenetically shown to represent green nonsulfur bacteria, only in the presence of organo-clay complexes in the light opens the unexplored possibility that anoxygenic phototrophs, particularly those with known photoheterotrophic modes of growth, such as Chloroflexus, could be used in bioremediation studies.
Conclusions.
Our chemical results support the observation made following oil spills in the Arabian Gulf that microbial communities dominated by cyanobacteria can be involved in the degradation of petroleum. Microbial communities inhabiting Wadi Gaza effectively degraded both aliphatic compounds (pristane and n-octadecane) and aromatic compounds (phenanthrene and dibenzothiophene). This site seems to harbor phylogeneticlly diverse and novel microbial populations that are able to tolerate high levels of pollutants. Organo-clay complexes were used successfully as a carrier system for model compounds and allowed complete degradation of aromatic compounds and degradation of most of the aliphatic compounds examined. Degradation of these compounds was accompanied by changes in both the cyanobacterial and bacterial communities. The development of certain populations was attributed directly to incubation with the model compounds and thus is thought to play a role in the metabolism of these compounds. The apparent contributions of members of the Holophaga-Geothrix-Acidobacterium and green nonsulfur bacterial groups are novel findings. The data show that polluted sites contain significant hidden diversity of unknown and uncultured microorganisms that contribute to biodegradation. However, it will be necessary to isolate key microorganisms in order to fully explore their individual potentials to degrade different petroleum compounds.
Acknowledgments
We gratefully acknowledge the executive board of the Environmental Protection and Research Institute, Gaza, Palestine, and Jamal Safi for their continuous encouragement and for supplying the samples. We thank Enric Llobet-Brossa for his assistance with the ARB program.
This research was financially supported by the Deutsche Forschungsgemeinschaft (grant Ru 458/18) and by the Max Planck Society.
REFERENCES
- 1.Abed, R. M. M., and F. Garcia-Pichel. 2001. Long-term compositional changes after transplant in a microbial mat cyanobacterial community revealed using a polyphasic approach. Environ. Microbiol. 3:53-62. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hasan, R. H., D. A. Al-Bader, N. A. Sorkhoh, and S. S. Radwan. 1998. Evidence for n-alkane consumption and oxidation by filamentous cyanobacteria from oil-contaminated coasts of the Arabian Gulf. Mar. Biol. 130:521-527. [Google Scholar]
- 3.Barkay, T., S. Navon-Venezia, E. Z. Ron, and E. Rosenberg. 1999. Enhancement of solubilization and biodegradation of polyaromatic hydrocarbons by the bioemulsifier alasan. Appl. Environ. Microbiol. 65:2697-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 5.Blümel, S., M. Contzen, M. Lutz, A. Stolz, and H.-J. Knackmuss. 1998. Isolation of a bacterial strain with the ability to utilize the sulfonated azo compound 4-carboxy-4′-sulfoazobenzene as the sole source of carbon and energy. Appl. Environ. Microbiol. 64:2315-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond, P. L., P. Hugenholtz, J. Keller, and L. L. Blackall. 1995. Bacterial community structure of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl. Eviron. Microbiol. 61:1910-1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carman, K. R., J. W. Fleeger, J. C. Means, S. M. Pomarico, and D. J. McMillan. 1995. Experimental investigation of the effects of polynuclear aromatic hydrocarbons on an estuarine sediment food web. Mar. Environ. Res. 40:289-318. [Google Scholar]
- 8.Carman, K. R., J. C. Means, and S. C. Pomarico. 1996. Response of sedimentary bacteria in a Louisiana salt marsh to contamination by diesel fuel. Aquat. Microbiol. Ecol. 10:231-241. [Google Scholar]
- 9.Cerniglia, C. E. 1992. Biodegradation of polycyclic aromatic hydrocarbons, p. 351-368. In E. Rosenberg (ed.), Microorganisms to combat pollution. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 10.Cerniglia, C. E., C. V. Baalen, and D. T. Gibson. 1980. Metabolism of naphthalene by the cyanobacterium Oscillatoria sp., strain JCM. J. Gen. Microbiol. 116:485-494. [Google Scholar]
- 11.Cerniglia, C. E., C. V. Baalen, and D. T. Gibson. 1980. Oxidation of biphenyl by the cyanobacterium Oscillatoria sp., strain JCM. Arch. Microbiol. 125:203-207. [DOI] [PubMed] [Google Scholar]
- 12.Cerniglia, C. E., D. T. Gibson, and C. V. Baalen. 1980. Oxidation of naphthalene by cyanobacteria and microalgae. J. Gen. Microbiol. 116:495-500. [Google Scholar]
- 13.Cho, J.-C., and S.-J. Kim. 2000. Increase in bacterial community diversity in subsurface aquifers receiving livestock wastewater input. Appl. Environ. Microbiol. 66:956-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Churchill, S. A., J. P. Harper, and P. F. Churchill. 1999. Isolation and characterization of a Mycobacterium species capable of degrading three- and four-ring aromatic and aliphatic hydrocarbons. Appl. Environ. Microbiol. 65:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coates, J. D., D. J. Ellis, C. V. Gaw, and D. R. Lovely. 1999. Geothrix fermentans gen. nov., sp. nov., a novel Fe(III)-reducing bacterium from a hydrocarbon-contaminated aquifer. Int. J. Syst. Bacteriol. 49:1615-1622. [DOI] [PubMed] [Google Scholar]
- 16.Coates, J. D., J. Woodward, J. Allen, P. Philp, and D. R. Lovley. 1997. Anaerobic degradation of polycyclic aromatic hydrocarbons and alkanes in petroleum-contaminated marine harbor sediments. Appl. Environ. Microbiol. 63:3589-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colores, G. M., R. E. Macur, D. M. Ward, and W. P. Inskeep. 2000. Molecular analysis of surfactant-driven microbial population shifts in hydrocarbon-contaminated soil. Appl. Environ. Microbiol. 66:2959-2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duarte, G. F., A. S. Rosado, L. Seldin, W. D. Araujo, and J. D. V. Elsas. 2001. Analysis of bacterial community structure in sulfurous-oil-containing soils and detection of species carrying dibenzothiophene desulfurization (dsz) genes. Appl. Environ. Microbiol. 67:1052-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.El-Nahhal, Y., S. Nir, C. Serban, O. Rabinovitch, and B. Rubin. 2000. Montmorillonite-phenyltrimethyl ammonium yields environmentally improved formulations of hydrophobic herbicides. J. Agric. Food Chem. 48:4791-4801. [DOI] [PubMed] [Google Scholar]
- 21.Feigel, B. J., and H.-J. Knackmuss. 1993. Syntrophic interactions during degradation of 4-aminobenzenesulfonic acid by a two species bacterial culture. Arch. Microbiol. 159:124-130. [DOI] [PubMed] [Google Scholar]
- 22.Friedrich, M., R. J. Grosser, E. A. Kern, W. P. Inskeep, and D. M. Ward. 2000. Effect of model sorptive phases on phenanthrene biodegradation: molecular analysis of enrichments and isolates suggests selection based on bioavailability. Appl. Environ. Microbiol. 66:2703-2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson, D. T. 1984. Microbial degradation of aromatic hydrocarbons, p. 181-251. In D. T. Gibson (ed.), Microbial degradation of organic compounds. Marcel Dekker, New York, N.Y.
- 24.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Envrion. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gosink, J. J., C. R. Woese, and J. T. Staley. 1998. Polaribacter gen. nov., with three new species, P. irgensii sp. nov., P. franzmannii sp. nov. and P. filamentus sp. nov., gas vacuolate polar marine bacteria of the Cytophaga-Flavobacterium-Bacteriodes group and reclassification of “Flectobacillus glomeratus” as Polaribacter glomeratus comb. nov. Int. J. Syst. Bacteriol. 48:223-235. [DOI] [PubMed] [Google Scholar]
- 26.Grimberg, S. J., W. T. Stringfellow, and M. D. Aitken. 1996. Quantifying the biodegradation of phenanthrene by Pseudomonas stutzeri P16 in the presence of a nonionic surfactant. Appl. Environ. Microbiol. 62:2387-2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grosser, R. J., M. Friedrich, D. M. Ward, and W. P. Inskeep. 2000. Effect of model sorptive phases on phenanthrene biodegradation: different enrichment conditions influence bioavailability and selection of phenanthrene-degrading isolates. Appl. Environ. Microbiol. 66:2695-2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hassett, J. J., J. C. Means, and W. L. Banwart. 1980. Sorption of benzidine by sediments and soils. J. Environ. Qual. 9:184-186. [Google Scholar]
- 29.Hoffmann, L. 1996. Recolonization of the intertidal flats by microbial mats after the Gulf War oil spill, p. 96-115. In F. Krupp, A. H. Abuzinada, and I. A. Nader (ed.), A marine wildlife sanctuary for the Arabian Gulf: environmental research and conservation following the 1991 Gulf War oil spill. NCWCD, Riyad, Saudi Arabia, and Senckenberg Research Institute, Frankfurt, Germany.
- 30.Höpner, T., M. Yousef, L. Berthe-Corti, H. Felzmann, H. Struck, and A. Al-Thukair. 1996. Cyanobacterial mats on oil-polluted sediments--start of a promising self-remediation process?, p. 85-95. In F. Krupp, A. H. Abuzinada, and I. A. Nader (ed.), A marine wildlife sanctuary for the Arabian Gulf: environmental research and conservation following the 1991 Gulf War oil spill. NCWCD, Riyad, Saudi Arabia, and Senckenberg Research Institute, Frankfurt, Germany.
- 31.Ishida, T., A. Yokota, and J. Sugiyama. 1997. Phylogenetic relationships of filamentous cyanobacterial taxa inferred from 16S rRNA sequence divergence. J. Gen. Appl. Microbiol. 43:237-241. [DOI] [PubMed] [Google Scholar]
- 32.Kanaly, R. A., R. Bartha, K. Watanabe, and S. Harayama. 2000. Rapid mineralization of benzo[a] pyrene by a microbial consortium growing on diesel fuel. Appl. Environ. Microbiol. 66:4205-4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kasai, Y., H. Kishira, K. Syutsubo, and S. Harayama. 2001. Molecular detection of marine bacterial populations on beaches contaminated by the Nakhodka tanker oil-spill accident. Environ. Microbiol. 3:246-255. [DOI] [PubMed] [Google Scholar]
- 34.Kishimoto, N., Y. Kosako, and T. Tano. 1991. Acidobacterium capsulatum gen. nov., sp. nov.: an acidophilic chemoorganotrophic bacterium containing menaquinone from acidic mineral environment. Curr. Microbiol. 22:1-7. [DOI] [PubMed] [Google Scholar]
- 35.Lahlou, M., and J. J. Ortega-Calvo. 1999. Bioavailability of labile and desorption-resistant phenanthrene sorbed to montmorillonite clay containing humic fractions. Environ. Toxicol. Chem. 18:2729-2735. [Google Scholar]
- 36.Larose, C. D., D. Labbe, H. Bergeron, A. M. Jones, C. W. Greer, J. Al-Hawari, M. J. Grossman, B. M. Sankey, and P. C. K. Lau. 1997. Conservation of plastid-encoded dibenzothiophene desulfurization genes in several rhodococci. Appl. Environ. Microbiol. 63:2915-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laurie, A. D., and G. Lloyd-Jones. 2000. Quantification of phnAc and nahAc in contaminated New Zealand soils by competitive PCR. Appl. Environ. Microbiol. 66:1814-1817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Law, A. T., and K. S. Teo. 1997. Oil biodegradation in the Straits of Malacca: phenanthrene degradation by AR-3. J. Mar. Biotechnol. 5:162-167. [Google Scholar]
- 39.Liesack, W., F. Bak, J.-U. Kreft, and E. Stackebrandt. 1994. Holophaga foetida gen. nov., sp. nov., a new, homoacetogenic bacterium degrading methoxylated aromatic compounds. Arch. Microbiol. 162:85-90. [DOI] [PubMed] [Google Scholar]
- 40.LIoyd-Jones, G., A. D. Laurie, D. W. F. Hunter, and R. Fraser. 1999. Analysis of catabolic genes for naphthalene and phenanthrene degradation in contaminated New Zealand soils. FEMS Microbiol. Ecol. 29:69-79. [Google Scholar]
- 41.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, S. Spring, A. Hartmann, and K. H. Schleifer. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 42.Ludwig, W., O. Strunk, S. Klugbauer, N. Klugbauer, M. Weizenegger, J. Neumaier, M. Bachleitner, and K. H. Schleifer. 1998. Bacterial phylogeny based on comparative sequence analysis. Electrophoresis 19:554-568. [DOI] [PubMed] [Google Scholar]
- 43.Macnaughton, S. J., J. R. Stephen, A. D. Venosa, G. A. Davis, Y.-J. Chang, and D. C. White. 1999. Microbial population changes during bioremediation of an experimental oil spill. Appl. Environ. Microbiol. 65:3566-3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Megharaj, M., I. Singleton, N. C. McClure, and R. Naidu. 2000. Influence of petroleum hydrocarbon contamination on microalgae and microbial activities in a long-term contaminated soil. Arch. Environ. Contam. Toxicol. 38:439-445. [DOI] [PubMed] [Google Scholar]
- 45.Mesarch, M. B., C. H. Nakatsu, and L. Nies. 2000. Development of catechol 2,3-dioxygenase-specific primers for monitoring bioremediation by competitive quantitative PCR. Appl. Environ. Microbiol. 66:678-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minz, D., J. L. Flax, S. J. Green, G. Muyzer, Y. Cohen, M. Wagner, B. E. Rittmann, and D. A. Stahl. 1999. Diversity of sulfate-reducing bacteria in oxic and anoxic regions of a microbial mat characterized by comparative analysis of dissimilatory sulfite reductase genes. Appl. Environ. Microbiol. 65:4666-4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narro, M. L. 1985. Oxidation of aromatic hydrocarbons by marine cyanobacteria. Ph.D. thesis. University of Texas at Austin, Austin.
- 48.Nogales, B., E. R. B. Moore, E. Llobet-Brossa, R. Rossello-Mora, R. Amann, and K. N. Timmis. 2001. Combined use of 16S rRNA ribosomal DNA and 16S rRNA to study the bacterial community of polychlorinated biphenyl-polluted soil. Appl. Environ. Microbiol. 67:1874-1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pearlman, R. S., S. H. Yalkowsky, and S. Banerjee. 1984. Water solubility of polynuclear aromatic and heteroaromatic compounds. J. Phys. Chem. Ref. Data 13:555-562. [Google Scholar]
- 51.Radwan, S. S., and R. H. Al-Hasan. 2000. Oil pollution and cyanobacteria, p. 307-319. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 52.Rippka, R., and M. Herdman. 1992. Culture media and growth, p. 61. In Pasteur Culture Collection of Cyanobacterial Strains in Axenic Culture catalogue & taxonomic handbook, vol. I. Institute Pasteur, Paris, France.
- 53.Safi, N. M. D., J. Köster, and J. Rullkötter. 2001. Fossil fuel pollution in Wadi Gaza and biodegradation of petroleum model compounds by cyanobacterial mats, p. 209. In J. Mascle, A. Lascaratos, S. Fowler, D. Gutnick, C. Papaconstantinou, and F. Boero (ed.), Rapport du 36eme Congrés de la Commission Internationale pour l’Exploration Scientifique de la Mer Méditerranée (CIESM), 36. CIESM, Monte Carlo, Monaco, France.
- 54.Santegoeds, C. M., T. G. Ferdelman, G. Muyzer, and D. de Beer. 1998. Structural and functional dynamics of sulfate-reducing populations in bacterial biofilms. Appl. Environ. Microbiol. 64:3731-3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sigalevich, P., M. V. Baev, A. Teske, and Y. Cohen. 2000. Sulfate reduction and possible aerobic metabolism of the sulfate-reducing bacterium Desulfovibrio oxyclinae in a chemostat coculture with Marinobacter sp. strain MB under exposure to increasing oxygen concentrations. Appl. Environ. Microbiol. 66:5013-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sigalevich, P., E. Meshorer, Y. Helman, and Y. Cohen. 2000. Transition from anaerobic to aerobic growth conditions for the sulfate-reducing bacterium Desulfovibrio oxyclinae results in flocculation. Appl. Environ. Microbiol. 66:5005-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stapleton, R. D., and G. S. Sayler. 1998. Assessment of the microbiological potential for the natural attenuation of petroleum hydrocarbons in a shallow aquifer system. Microb. Ecol. 36:349-361. [DOI] [PubMed] [Google Scholar]
- 58.Theng, B. K. G., R. H. Newman, and J. S. Whitton. 1998. Characterization of an alkylammonium-montmorillonite-phenanthrene intercalation complex by carbon-13 nuclear magnetic resonance spectroscopy. Clay Min. 33:221-230. [Google Scholar]
- 59.Whiteley, A. S., and M. J. Bailey. 2000. Bacterial community structure and physiological state within an industrial phenol bioremediation system. Appl. Environ. Microbiol. 66:2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Widdel, F., and R. Rabus. 2001. Anaerobic biodegradation of saturated and aromatic hydrocarbons. Curr. Opin. Biotechnol. 12:259-276. [DOI] [PubMed] [Google Scholar]