Abstract
Mercury-contaminated chemical wastewater of a mercury cell chloralkali plant was cleaned on site by a technical-scale bioremediation system. Microbial mercury reduction of soluble Hg(II) to precipitating Hg(0) decreased the mercury load of the wastewater during its flow through the bioremediation system by up to 99%. The system consisted of a packed-bed bioreactor, where most of the wastewater's mercury load was retained, and an activated carbon filter, where residual mercury was removed from the bioreactor effluent by both physical adsorption and biological reduction. In response to the oscillation of the mercury concentration in the bioreactor inflow, the zone of maximum mercury reduction oscillated regularly between the lower and the upper bioreactor horizons or the carbon filter. At low mercury concentrations, maximum mercury reduction occurred near the inflow at the bottom of the bioreactor. At high concentrations, the zone of maximum activity moved to the upper horizons. The composition of the bioreactor and carbon filter biofilms was investigated by 16S-23S ribosomal DNA intergenic spacer polymorphism analysis. Analysis of spatial biofilm variation showed an increasing microbial diversity along a gradient of decreasing mercury concentrations. Temporal analysis of the bioreactor community revealed a stable abundance of two prevalent strains and a succession of several invading mercury-resistant strains which was driven by the selection pressure of high mercury concentrations. In the activated carbon filter, a lower selection pressure permitted a steady increase in diversity during 240 days of operation and the establishment of one mercury-sensitive invader.
Increasing exposure to toxic mercury compounds is a considerable threat for human health (13, 14). Global natural emissions of mercury have been estimated at 3,000 tons per year, and anthropogenic sources account for approximately 3,600 tons per year (15, 17). Man-made sources of mercury include the combustion of fossil fuels, mining, and mercury cell chloralkali plants. If released into the environment, inorganic mercury can be methylated abiotically or biotically (1, 20). Methylmercury (CH3Hg+) is more toxic and mobile than its precursor Hg(II) and biomagnifies in the food chain, endangering ecosystems and public health. In freshwater ecosystems, methylmercury accumulation is more common than in saline environments (5, 9). Today, seafood is a major source of mercury to which humans are exposed (2). In the United States, some 60,000 babies per year may be born with neurological damage caused by mercury poisoning of their mothers from consumption of large amounts of fish from polluted locations during pregnancy (23). It is of great environmental and public health importance to stop mercury dumping into river and marine ecosystems. An end-of-pipe technology for mercury-emitting industries that suits both industrialized and developing countries has to be efficient as well as cost effective.
In previous experiments the efficient and cost-effective use of Hg(II)-resistant bacteria in packed-bed bioreactors was demonstrated for the retention of mercury from wastewater of three European chloralkali plants, both in a laboratory-scale plant (27, 29) and in a technical-scale pilot plant (31). This study elucidates the active part of the bioremediation process, the mercury-reducing biofilms, during the operation of the pilot plant as reported previously (31).
The basic principle of this process is the enzymatic reduction of water-soluble, ionic mercury Hg(II) to insoluble, metallic mercury Hg(0) by Hg(II)-resistant bacteria. The metallic mercury is retained in the matrix of the bioreactor in the form of mercury droplets (3, 30). This energy-dependent reaction is an ancient and ubiquitous mechanism of bacteria to detoxify their surrounding environment (11, 16, 18, 24).
Mercury-reducing bioreactors operated with nonsterile chloralkali wastewater have been shown to be colonized by invading mercury-resistant bacteria, and they exhibited increased microbial diversity and stable performance (7, 28, 30). To better understand the structure and activity of microbial communities in response to an invasion of Hg(II)-resistant or Hg(II)-sensitive bacteria and in response to the selection pressure of the pollutant, the culture-independent 16S-23S ribosomal DNA intergenic spacer polymorphism analysis (RISA) was applied to examine the bacterial community on the strain level.
In a previous experiment, biofilm diversity and activity during operation were monitored by analyzing bioreactor effluent samples (28, 29). In this study, the biofilm community was monitored directly by sampling both the solid and liquid phases of four bioreactor horizons during operation. Biofilm composition and mercury and oxygen concentrations were simultaneously determined. In such a way, it was possible to identify spatial and temporal variations in the structure (in terms of community composition) and activity (in terms of oxygen consumption and mercury reduction) of the bioreactor biofilms during 8 months of operation on the site of a chloralkali plant and their dependence on daily oscillating mercury concentrations.
MATERIALS AND METHODS
Design and operation of a technical-scale mercury bioremediation system.
As described in detail previously (31), the pH of the incoming wastewater was adjusted to pH 7.0 ± 0.5 by the addition of NaOH (25% [wt/wt]). This was done in two steps and regulated by an adaptive controller. Subsequently, sucrose and yeast extract were added to final concentrations of 83 and 17 mg/liter, respectively. Between days 117 to 124, nutrient concentrations were temporarily increased to 348 mg of sucrose/liter and 69 mg of yeast extract/liter. The neutralized, nutrient-amended wastewater entered the bioreactor at its bottom and left at its top. The volume of the packed bed was 1,000 liters. Pumice granules of 4 to 8 mm (particle size) and 80% porosity (Raab, Luckenau, Germany) were used as carrier material. The flow was bottom to top at 600 (days 0 to 47), 1,200 (days 47 to 128), and 2,000 (days 128 to 240) liters/h. The bioreactor effluent ran top down through an activated carbon filter (particle size, 1 to 3 mm) (Pool Aktivkohle; CarboTech, Essen, Germany) with an 800-liter volume to retain residual mercury. Total mercury concentrations were determined continuously in the wastewater inflow, the bioreactor effluent, and the carbon filter effluent. The bioreactor was protected from adverse wastewater inflow conditions by a bypass that automatically opened at inflow mercury concentrations above 10 mg/liter and a pH value outside the allowed range of 7.0 ± 0.5. A proportion of the factory wastewater entered the pilot plant continuously, and the cleaned or bypassed wastewater reentered the factory wastewater treatment system. Conductivity, redox potential, temperature, and oxygen content were determined by electrode measurement in the bioreactor inflow, and redox potential and oxygen content were also determined in the bioreactor outflow.
Cultivation of inoculum.
The bioreactor was inoculated with seven strains. Four were strains of Pseudomonas putida (Spi3, Spi4, Kon12, Elb2), one was Pseudomonas stutzeri (Ibu8), one was Pseudomonas fulva (Spi11), and one was a Bacillus species (I3F1), which was a contaminant of the Spi11 culture. The inoculum strains were cultivated in seven batch reactors by continuous addition of liquid medium [sucrose, 200 g/liter; yeast extract, 200 g/liter; NaCl, 30 g/liter; Hg(II), 1.0 mg/liter, neutralized to pH 7.0] for 4 days at 35°C. Cultures were stored at 4°C until inoculation of the bioreactor 10 h later.
Inoculation of the bioreactor.
The bioreactor was filled with autoclaved pumice granules, and neutralized wastewater was continuously run through the pilot plant for several days. For inoculation, the wastewater inflow was stopped and the liquid phase was removed by pumping. The seven inoculum culture suspensions (15 liters each) were then introduced into the bioreactor. Neutralized wastewater (750 liters) and 1,875 ml of concentrated medium (sucrose, 167 g/liter; yeast extract, 33 g/liter) were next pumped into the bioreactor until the carrier material was submersed. Inoculum and medium were circulated through the fixed bed for 40 min (300 liters/h). Then, circulation was stopped, and neutralized wastewater inflow started at a flow rate of 500 liters/h and final concentrations of 400 mg of sucrose/liter and 80 mg of yeast extract/liter. The first 1,000 liters of bioreactor effluent were collected in a separate wastewater tank and sterilized before disposal. Subsequent bioreactor effluent was passed through the activated carbon filter. After 1 day, medium addition to the inflowing wastewater was reduced to a final concentration of 83 mg of sucrose/liter and 17 mg of yeast extract/liter.
Mercury assay.
Mercury concentrations were determined continuously by two automated atomic absorption spectrophotometers (Mercury Instruments, Karlsfeld, Germany) by using the cold vapor technique. Sample oxidation was performed by KMnO4 (0.5 g/liter at 90°C), and the subsequent reduction of Hg(II) to Hg(0) was performed by the addition of SnCl2 (20 g/liter). Metallic mercury was volatilized by air (analyzer 1, 80 liters/h; analyzer 2, 20 liters/h ) and determined by automated atomic absorption spectrophotometer spectroscopy at 253.7 nm. Every 60 min, an autozeroing was performed, and every 24 h, an automated calibration check occurred.
Oxygen assay.
Two oxygen sensors (COS 3S; Endress + Hauser, Weil am Rhein, Germany) measured the temperature-corrected oxygen partial pressure in the in- and outflow of the bioreactor, and a transformer (mycom COM151; Endress + Hauser) delivered the values as relative oxygen concentrations.
Data recording and analysis.
Data were continuously recorded by the WIZCON (version 7.5; PC-Soft, Petah Tikrah, Israel) process control software package and stored on disk. Process data were transferred to the data analysis software package ACRON, version 4.52 (DataForum Software, Niesetal, Germany), from which they were exported to Microsoft Excel 97.
Determination of mercury-resistance levels.
A colony of each strain was picked from solid medium (NaCl, 15 g/liter; sucrose, 4 g/liter; and yeast extract, 2 g/liter) and incubated for 1 day at 30°C. The colony was transferred to fresh solid medium containing 1, 5, or 10 mg of Hg(II) (as HgCl2)/liter. Cells were spread by diluting streaks to obtain a cell gradient down to single cells and incubated for 7 days at 30°C.
Identification of strains.
The isolated strains were identified by partial 16S ribosomal DNA sequencing and databank comparison. The isolates Bro12 and Bro30 were identified as Pseudomonas aeruginosa (99.6 and 99.9% identity, respectively), Bro33 was identified as Bacillus sp. (96.7% identity), Bro34 was identified as Sphingomonas sp. (99.7% identity), Bro124 was identified as Aerococcus viridans (98.8% identity), and Bro148 was identified as Microbacterium arborescens (97.8% identity).
Community analysis. (i) Sampling and DNA extraction.
Liquid and solid samples were taken from 4 horizons of the bioreactor (30, 55, 80, and 105 cm above the bottom) and from the effluents of the bioreactor and carbon filter. For isolation of total DNA in the liquid phase, approximately 50 ml was sampled and centrifuged (30 min, 5,000 × g). The pellet was resuspended in 100 μl of Tris-EDTA buffer. For isolation of total DNA in the biofilm, approximately 4 ml of pumice granules was sampled from each bioreactor horizon with a hollow lance. Biofilms were removed from the carrier surface by vigorous mixing in 2 ml of NaCl solution (15 g/liter). The suspension of abraded biofilms and pumice fragments was centrifuged (5 min, 10,000 × g), and the pellet was resuspended in 100 μl of Tris-EDTA buffer. Total cell DNA was extracted by using guanidium thiocyanate as described by Pitcher et al. (21).
(ii) Amplification primers for the 16S-23S ISR.
The primers G1 and L1 were described by Jensen et al. (12). The primer G1 (5′-GAAGTCGTAACAAGG-3′) binds to a highly conserved region of the 16S rRNA gene adjacent to the 16S-23S spacer region. The primer L1 (5′-CAAGGCATCCACCGT-3′) binds to a conserved region of the 23S rRNA gene close to the 16S-23S spacer. The primer R4c (5′-CTCATTTCTGAGCTTTCAGCG-3′) was selected from an alignment of 5 16S-23S interspacer region (ISR) sequences (unpublished data) of P. stutzeri Ibu8, two different P. fulva strains, and two different Pseudomonas fluorescens strains. It binds specifically within the 16S-23S ISR of P. stutzeri Ibu8, 300 nucleotides upstream of the 16S gene.
(iii) PCR conditions.
The amplification assay (20-μl volume) contained 250 μM concentrations of each deoxynucleoside triphosphate (MBI Fermentas, Vilnius, Lithuania), 10% (vol/vol) dimethyl sulfoxide (Fluka, P.A.), 1× PCR buffer (Qiagen, Hilden, Germany), 1 unit of Taq DNA polymerase (Qiagen), 0.5 μM concentrations of the primers G1, L1, and R4c (Gibco BRL, Karlsruhe, Germany), and 0.5 μl of template DNA of various concentrations. To prevent evaporation, the mixture was covered with a drop of Nujol mineral oil (Perkin Elmer, Überlingen, Germany). The PCR was performed in a thermal cycler (TC Varius V; Landgraf, Langenhagen, Germany) with a temperature program consisting of a first DNA denaturation step of 120 s at 94°C followed by 35 cycles of 30 s at 94°C, 60 s at 45°C, and 120 s at 72°C; a final elongation step of 600 s at 72°C; and subsequent cooling at 4°C. The ramping rate between the different temperatures was set to the fastest rate available.
(iv) Electrophoresis.
After the success of DNA amplification had been monitored in an agarose gel, the PCR products were separated in 4 to 20% TBE (Tris base, boric acid, EDTA) acrylamide gradient minigels (Novex, Frankfurt, Germany) as described by Hirsch and Sigmund (10). Three microliters of the PCR products was mixed with 0.5 μl of TBE sample buffer (Novex) and loaded onto the gel. For strain discrimination within the community, a community standard consisting of the PCR products of the prevalent strains (amplified separately and then mixed in equal amounts) was used. The template DNA concentration of each separately amplified strain was 10 μg/ml. The DNA was separated by electrophoresis at 200 V for 100 min in 1× TBE buffer (Novex). The gel was stained for 10 min in ethidium bromide (5 mg/liter) and then destained for 5 min in 1× TBE buffer. Bands were visualized with UV light (254 nm, UV transilluminator 200/2.0; UVP Inc., San Gabriel, Calif.) and digitalized with the Enhanced Analysis system (EASY; Herolab, Wiesloch, Germany).
(v) Analysis of the community fingerprint.
The light intensity (i.e., the number of pixels) of each diagnostic band was calculated with Scion Image (version Beta3b; Scion Corporation, Frederick, Md.). For the determination of the community composition, only one diagnostic band per strain was used. The relative intensities of the diagnostic bands of different gels were normalized to the intensity of the size marker.
Scanning electron microscopy.
Pumice granules containing biofilms from the bioreactor were sampled during operation with a hollow lance, and they were fixed with 2.5% glutaraldehyde in NaCl solution (15 g/liter) for 2 h or several days at 4°C followed by dehydration with an acetone series and critical point drying with liquid CO2 at 41°C and 85 atm. Samples were carbon coated to a 30-nm thickness (sputter coater SCD 040; Balzers Union, Walluf, Germany) and analyzed with a field emission scanning microscope (DSM 982 Gemini; Zeiss, Oberkochen, Germany) at a working distance of 8 mm, an acceleration voltage of 5 kV, and a sampling area of 8 by 8 μm.
RESULTS
Principle of the mercury bioremediation system.
Incoming wastewater at pH 2 to 3 was neutralized to pH 7.0 ± 0.5, supplemented with a small amount of nutrients to provide energy to the bacteria, and then run from bottom to top through the packed-bed bioreactor containing carrier-immobilized biofilms of mercury-reducing bacteria. The effluent from the bioreactor was pumped top down through an activated carbon filter to remove remaining traces of mercury. After adherence of bacteria in the carbon filter, it retained mercury both by physical adsorption and bacterial reduction.
Operation of the mercury bioremediation system and biofilm activity.
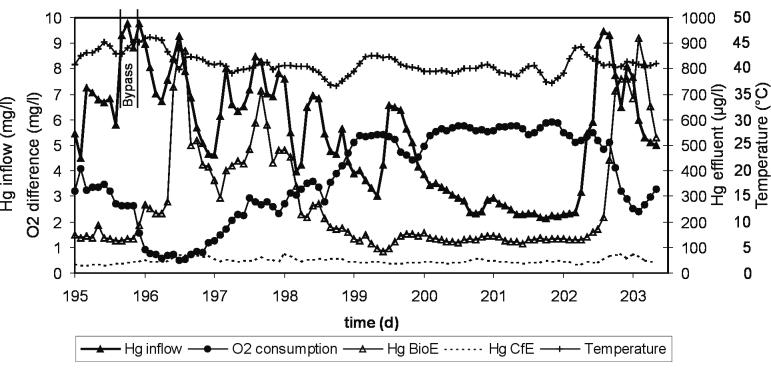
During 240 days of operation, wastewater composition and temperature changed continuously due to chloralkali plant inherent processes. Conductivity varied between 20 and 105 mS/cm, representing chloride concentrations between 8 and 50 mg/liter (31). Mercury concentrations in the wastewater varied between 2 to 10 mg/liter in the bioreactor inflow, with a mean concentration of 4.2 mg/liter. The mean mercury concentration was 415 μg/liter in the bioreactor effluent and 72 μg/liter in the carbon filter effluent, including all periods of suboptimum performance caused by technical problems and temperatures up to 50°C. The inoculum ensured a mercury retention efficiency of 82% right from the beginning, which improved within the first 10 h of operation to 97%. The bioreactor performance did not deteriorate during the tested period of 240 days. Around days 195 to 203, as shown in Fig. 1, it was identical to or even better than the initial performance.
FIG. 1.
Effect of mercury inflow concentration on biofilm activity as determined by the difference between bioreactor inflow and outflow oxygen concentration and mercury retention. The wastewater inflow rate was 2,000 liters/h, and the oxygen inflow concentration was approximately 6 mg/liter. The mercury peak shortly before day 196 was bypassed due to Hg concentrations above 10 mg/liter. Data are 2-hourly mean values calculated from process data recorded every minute. BioE, bioreactor effluent; CfE, carbon filter effluent.
The performance of the bioremediation system and its response to fluctuating wastewater conditions is shown for a representative period of 8 days in Fig. 1. Mercury and, to a much lesser extent, high temperatures affected biofilm activity, which was determined by means of respiration (deduced from oxygen consumption) and mercury reduction (deduced from mercury retention). In the period of days 200 to 202, the wastewater conditions were stable and far from extreme; the biofilms exhibited maximum respiration and consumed all oxygen. The biofilm bacteria efficiently reduced the wastewater mercury, and approximately 97% of 2 to 3 mg/liter were retained in the bioreactor, resulting in a bioreactor effluent concentration of 130 μg/liter. In a second cleaning step, the carbon filter decreased the mercury concentration to 35 μg/liter.
But usually the mercury concentration oscillated daily, as can be seen in the days before and after days 200 to 202, and the bioreactor was periodically subjected to extreme conditions, e.g., more than 9 mg of Hg/liter after day 196. About 2 hours after this mercury peak was detected in the bioreactor inflow, the oxygen consumption decreased. Simultaneously, the mercury concentration of the bioreactor effluent peaked as well. The quickly following mercury peaks of up to 10 mg/liter around day 196 and high temperatures up to 46°C decreased respiration activity by almost 90%. However, the residual bioreactor activity caused 90% retention of the mercury load, and bioreactor effluent concentrations remained below 1,000 μg/liter. The carbon filter retained most of this remaining mercury and showed only a very dampened response to high mercury peaks. After a peak had passed, the biofilm activity recovered spontaneously within a few hours.
Spatially oscillating maximum of mercury reduction.
The spatial distribution of mercury-reducing activity was determined on the basis of mercury concentrations in the bioreactor inflow, the liquid phases of the four bioreactor horizons, the bioreactor effluent, and the carbon filter effluent. The data of four representative days are shown in Table 1. The mercury-reducing activity of the bioreactor horizons depended on both the actual and the preceding mercury inflow concentrations. The time interval from the preceding mercury peak and its height determined the recovery of biofilm activity. The higher the preceding mercury peak, the more time was necessary for complete recovery.
TABLE 1.
Concentration gradients of water-soluble mercury in the bioremediation system and the effect of time lapse since and height of the preceding mercury peak of identical or higher mercury inflow concentrationa
| Day | Preceding Hg peak (μg/liter) | Time lapse (h) | Hg inflow concn (μg/liter) | Concn (μg/liter) of Hg (% Hg retention) in:
|
% of total Hg retention | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Horizon 1 | Horizon 2 | Horizon 3 | Horizon 4 | Bioreactor effluent | Carbon filter effluent | |||||
| 10 | 6,200 | 24 | 3,600 | 1,680 (53) | 320 (38) | 200 (3) | 160 (1) | 160 (0) | 18 (4) | 99.5 |
| 47 | 4,400 | 48 | 4,400 | 1,330 (70) | 520 (18) | 360 (4) | 340 (<1) | 150 (4) | 11 (3) | 99.8 |
| 62 | 8,000 | 10 | 5,800 | 4,930 (15) | 2,600 (40) | 600 (34) | 500 (2) | 270 (4) | 70 (3) | 98.8 |
| 210 | 9,300 | 3 | 4,800 | 4,170 (13) | 3,950 (5) | 3,690 (5) | 3,370 (7) | 2,790 (12) | 34 (57) | 99.3 |
Mercury concentrations were measured in the liquid phase at days 10, 47, 62, and 210.
On days 10 and 47, maximum mercury reduction occurred in the bottom horizon 1 (53 and 70% mercury retention, respectively). The actual mercury inflow concentration was low (3.6 and 4.4 mg/liter, respectively), and the preceding mercury peak had passed at least 24 h earlier. On day 62, the zone of maximum mercury reduction shifted to the middle horizons. Here, the actual mercury concentration was high (5.8 mg/liter), and the time interval from the preceding peak was smaller (10 h). On day 210, directly after a high mercury peak of 9.3 mg/liter had passed, much less mercury was retained in the bottom and middle horizons and maximum mercury reduction occurred in the carbon filter. Thus, the zone of maximum mercury reduction oscillated regularly between the lower and the upper horizons in accordance with the oscillating mercury concentrations in the bioreactor inflow. In all cases, the overall mercury retention rates were about 99%.
Balance of mercury retention.
During 240 days of operation, 7,013,000 liters of chloralkali wastewater were pumped through the bioremediation system. Continuous mercury measurements of bioreactor inflow, bioreactor effluent, and carbon filter effluent showed that a total mercury load of 29.3 kg entered the bioremediation system; 25.8 kg were retained in the bioreactor and another 3.0 kg were retained in the carbon filter. This corresponds to an overall mercury retention of 98.3%. At the end of operation, the bioreactor and carbon filter were dismantled and the mercury content of samples was determined; representative data of the bioreactor horizons are shown in Table 2. The amount of deposited metallic mercury in the bioreactor and carbon filter was calculated on the basis of the samples' mean mercury concentrations. The bioreactor contained at least 8 kg of Hg(0) in the packed bed plus 3 kg in the sludge. The carbon filter contained 9 kg of Hg(0). The metallic mercury deposits exhibited a C-shaped profile with slightly higher concentrations in the bottom and in the top bioreactor horizons. The highest concentration of deposited mercury was found in the sludge of the sedimentation zone near the bioreactor outflow.
TABLE 2.
Amount of mercury deposited in the bioreactor horizons and carbon filter after 240 days of operation
| Location | Amt of Hg (g/liter) |
|---|---|
| Bioreactor horizons | |
| 0 (gravel basic layer) | 8.7 |
| 1 | 8.0 |
| 2 | 6.8 |
| 3 | 5.6 |
| 4 | 8.7 |
| 5 (sludge in sedimentation zone) | 27.5 |
| Carbon filter (avg) | 11.3 |
Spatial variation of mercury-reducing biofilms.
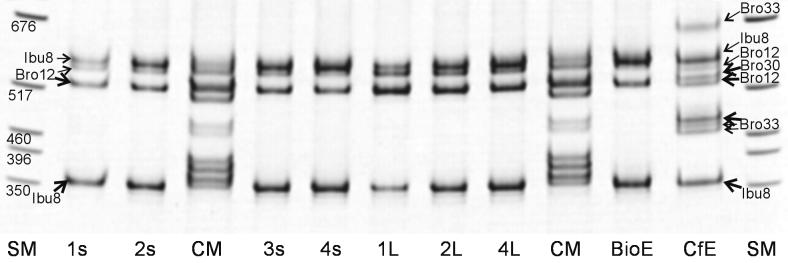
An example of the community PCR RISA fingerprints of the bioremediation system is shown in Fig. 2 for day 127. Compositions of the carrier-bound biofilms (containing planktonic and sessile cells) were identical to their respective liquid-phase communities (containing only planktonic cells). Microbial diversity, i.e., the number of detectable strains, was comparably low in the bioreactor but much higher in the carbon filter community. The spatial bioreactor profiles of the other sampling times (data not shown) were similar: the bioreactor biofilm fingerprints are identical with their respective liquid-phase community fingerprints, and the fingerprints of the four horizons are similar but with an occasionally higher diversity in the upper horizons 3s and 4s.
FIG. 2.
Spatial microbial diversity of the bioremediation system at day 127 as revealed by 16S-23S RISA of the biofilm and liquid-phase communities. The community marker (CM) consists of a mixture of the separately amplified ISR PCR products of the initially prevalent strains (Spi3, Spi4, Elb2, Kon12, Ibu8, I3F1, and Bro12). The present strains exhibited one to four bands, but only one band per strain (indicated with thick arrows) was diagnostically used for community analysis. Nondiagnostic bands are indicated with fine arrows. The numbers at the size marker (SM) bands indicate sizes in base pairs. 1s, bioreactor carrier-bound biofilm 1s (near inflow); 2s, bioreactor carrier-bound biofilm 2s; 3s, bioreactor carrier-bound biofilm 3s; 4s, bioreactor carrier-bound biofilm 4s (near outflow); 1L, bioreactor liquid-phase sample 1L (near inflow); 2L, bioreactor liquid-phase sample 2L; 4L, bioreactor liquid-phase sample 4L (near outflow); BioE, bioreactor effluent; CfE, carbon filter effluent.
Succession of mercury-reducing biofilm communities.
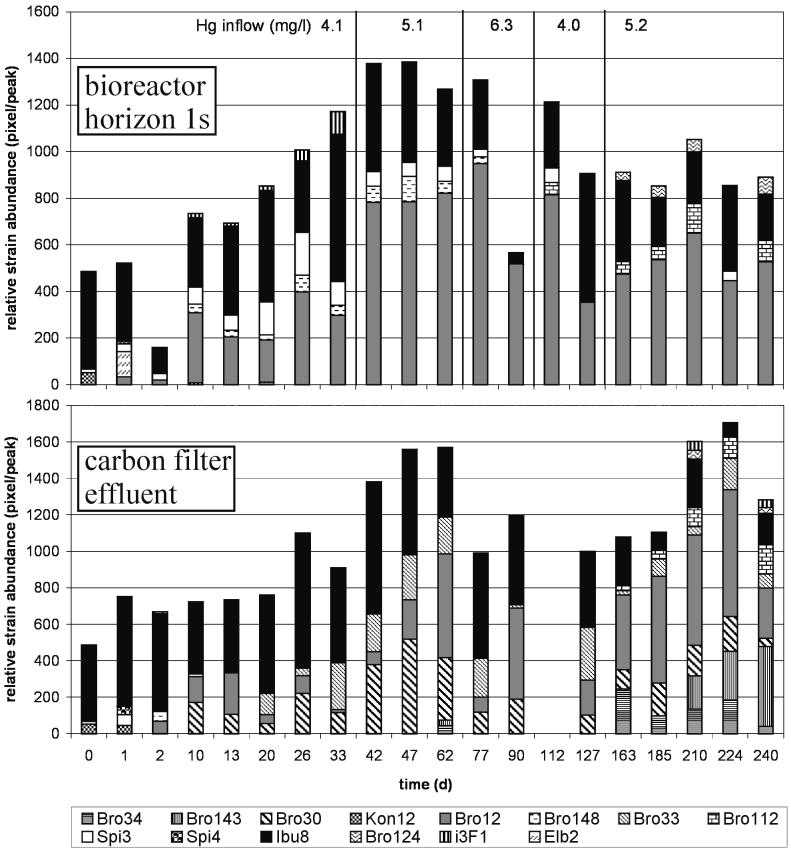
The relative strain abundances in the bottom horizon of the bioreactor and in the carbon filter effluent during 240 days of operation are shown in Fig. 3. Caused by a washout of the planktonic inoculum cells, the number of detectable strains in the bioreactor declined in the first 2 days. In the following days, the numbers of biofilm bacteria increased by 1 to 2 orders of magnitude (data not shown), reflecting biofilm formation. The community composition of the bioreactor biofilms showed a succession of strains but remained stable for the two prevalent strains, Ibu8, an inoculum strain, and the invader Bro12. Most of the other inoculum strains dropped below the detection limit within the first month, but microbial diversity decreased only slightly because the biofilms were subjected to colonization by invading bacteria. A total of four new strains was found in the packed-bed biofilms with various detection periods. Usually the communities consisted of four to five detectable strains. At days 90 and 127, however, the community fingerprints revealed only two strains. At day 90 this was probably caused by continuously high selection pressure by high mercury inflow concentrations with an average of 6.3 mg/liter in the preceding 2 weeks. This was significantly higher than in the time before and after, when mercury concentrations oscillated around mean concentrations of 4 to 5 mg/liter. After day 90, the lower mercury concentration diversity increased spontaneously, revealing that no strain died out but just dropped below the detection limit. The second drastic decrease in diversity at day 127 may have been caused by an enhanced feeding rate and differential growth of the biofilm strains in the preceding week. Culturable cell numbers increased by 2 orders of magnitude between days 112 and 127, and since the detection limit of a strain was about 1% of the community DNA (data not shown), the strains Ibu8 and Bro12 temporarily overgrew the other strains. Biofilm composition and diversity remained stable at increased hydraulic loads after days 47 and 127 as well as at the high temperatures of 45 to 50°C in the last 30 days of operation.
FIG. 3.
Time course of relative strain abundance in the bioreactor biofilm near the inflow and in the carbon filter effluent. The relative abundance of the strains based on the intensity of the diagnostic band of each strain within the community fingerprint in relation to the size marker intensity is shown. Data of the carbon filter community at day 112 are missing due to a failure of electrophoretic separation.
The carbon filter effluent community showed a different dynamic than the bioreactor community. After the initial washout of most of the inoculum strains until day 13, the microbial diversity increased steadily. A total of eight new strains was found in the carbon filter effluent with various detection periods, among which four occurred in the carbon filter but not in the bioreactor. Microbial diversity of the carbon filter effluent was initially lower than in the bioreactor but matched the bioreactor diversity after 1 month and surpassed bioreactor diversity after 2 to 3 months of operation. None of the changes in the community composition of the bioreactor or carbon filter impaired the mercury-reduction efficiency.
Microscopic biofilm structure.
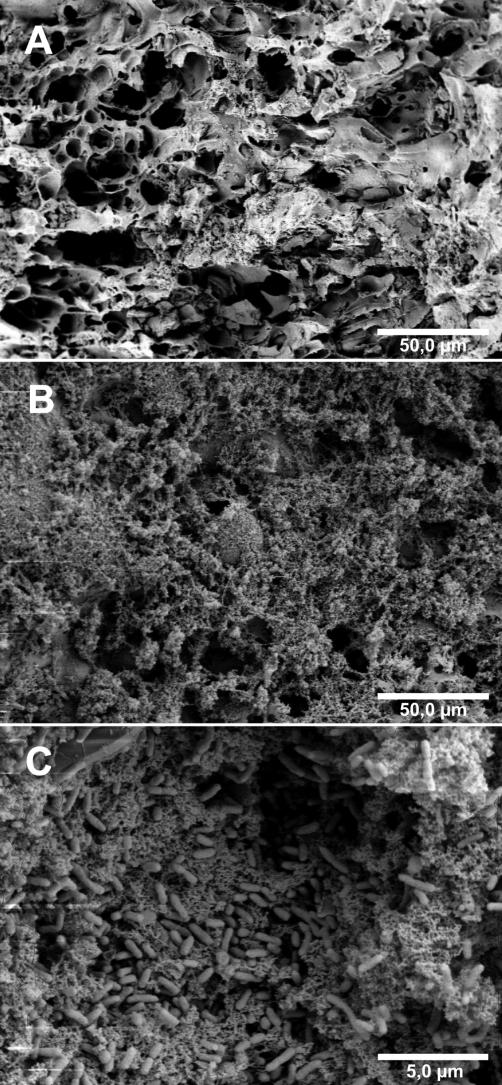
Scanning electron micrographs shown in Fig. 4 illustrate the surface of the packed-bed carriers and the development and microscopic structure of the biofilm. Figure 4A shows the cavernous surface of a carrier granule after 2 days of operation; no biofilm was visible. Figure 4B shows the surface in the same scale but after 224 days of operation. A thick layer of extracellular polysaccharides (EPSs) covered the surface, interspersed with a few single bacteria. In a close-up, shown in Fig. 4C, the structural heterogeneity of the biofilms is revealed. A large population of rod-shaped Pseudomonas cells is stuck in a thick, amorphous EPS matrix. Thus, the biofilm exhibited a variety of ecological niches by mass transport limitations (i.e., transport by diffusion only).
FIG. 4.
Scanning electron micrograph survey of bioreactor pumice granules and biofilm development. (A) After 2 days of operation, the cavernous surface of a pumice granule was almost blank and no biofilms were visible. (B) After 224 days of operation, a dense biofilm covered the surface and filled the caverns. (C) The 224-day-old biofilm consisted of rod-shaped Pseudomonas cells embedded in a thick layer of EPSs.
Levels of mercury resistance.
To test whether the higher diversity of strains in the upper bioreactor horizons and the carbon filter was caused by a lower selection pressure by the lower mercury concentration, their mercury-resistance levels were analyzed. All strains with high resistance levels were detected in both the bioreactor and carbon filter. Strains with a high resistance level, i.e., growth on solid medium containing 5 mg of Hg(II)/liter, were the inoculum strains P. stutzeri Ibu8, P. putida Spi3, P. putida Spi4, P. putida Elb2, P. putida Kon12, and the invader P. aeruginosa Bro12. In addition, weakly resistant strains, i.e., growth at 1 mg/liter but not at 5 mg/liter, such as the inoculum strain Bacillus sp. I3F1 and the invaders Bro112, A. viridans Bro124, and M. arborescens Bro148, occurred in the bioreactor. But they had only low abundances in horizon 1 and higher abundances in horizon 4, indicating that a high resistance level was necessary for a high abundance at high mercury concentrations. In contrast, the weakly resistant strains P. aeruginosa Bro30, Sphingomonas sp. Bro34 and Bro143, and the mercury-sensitive strain Bacillus sp. Bro33 occured at high abundances only in the carbon filter effluent but not in the bioreactor. In the carbon filter community, even strains of low resistance could reach high abundances.
DISCUSSION
Biofilm activity at oscillating mercury concentrations.
Compared to laboratory-scale bioreactors operated under constant conditions, the mercury-reducing microbial biofilms of the technical-scale bioreactor were exposed to oscillating mercury concentrations with peak values of 10 mg/liter and temperatures up to 50°C. In previous laboratory-scale bioreactor experiments, partial breakthrough of mercury at inflow concentrations between 7 and 9 mg/liter was observed (29). Here, we observed that mercury concentrations above 6 mg/liter caused an inhibition of respiration and mercury reduction. If the mercury concentration dropped below this value, the biofilms recovered spontaneously within a few hours up to several days (depending on the extend of biofilm injury). Because high mercury concentrations kill a fraction of a mercury-resistant bacterium population (4), recovery occurred by the growth of surviving cells. However, significantly decreased culturable cell densities (data not shown) were only observed after several days of continuously high mercury concentrations, not after short-time peaks of several hours. Thus, we assume that in the first place, high mercury concentrations inhibited the activity of mercury-resistant cells. Individual cells may have recovered, if the stress lasted only for a few hours. The cells died only if the mercury stress lasted for days.
The spatially oscillating zone of maximum mercury-reducing activity should have resulted in a smooth gradient of metallic mercury deposits, with a maximum at the bioreactor inflow, rather than a strong gradient reported by Brunke et al. (3). However, slightly increased deposits were found not only in the bottom layer of the bioreactor, but also in the top horizons. This may have been effected by the shear force of the flowing water that continuously abraded excess biomass that contained mercury droplets. These mercury-loaded particles were deposited in the top bioreactor section, mostly above the packed bed, due to a reduction in flow velocity and thus the presence of a sedimentation zone. The oscillation of maximum activity through all bioreactor horizons was an advantage for industrial operation because it prevented the bioreactor from being blocked by excess biomass formation and metallic mercury deposits at the bioreactor inflow (a serious problem with small-scale laboratory bioreactors) and thus ensured a prolonged operation time.
Balance of mercury retention.
The difference between yield based on continuous mercury measurements and the actual amount of mercury deposits in the bioreactor and carbon filter was substantial. Twenty-nine kilograms were expected, but only approximately 20 kg were found. This difference may be attributed to the uncertainty about the volume of sludge in the sedimentation zone based on a conservative visual estimate. Since this sludge had an extremely high mercury concentration, small differences in volume would have had a large effect on the total amount of mercury deposits. In addition, the proportion of mercury recovered from the carbon filter was significantly higher than expected. We assume that small biofilm fragments containing metallic mercury, which did not deposit in the sedimentation zone, were continuously washed out of the bioreactor and retained in the narrow matrix of the carbon filter. The mercury monitors analyzed only filtered effluents and thus could not detect the transport of particulate mercury.
The carbon filter retained mercury by three mechanisms. It acted as a physical adsorber of the residual mercury load, it mechanically retained biofilm fragments containing metallic mercury droplets, and it retained ionic mercury by biological reduction. It is not possible to differentiate between the relative impact of each of these mechanisms on the basis of the present data. The maximum physical adsorption capacity of the carbon filter would have been 6.8 kg as estimated by the manufacturer. At the end of operation, 9 kg of mercury was found and no exhaustion of the carbon filter capacity was observed.
RISA.
The patterns produced by RISA are highly reproducible and allow comparison of different populations of microorganisms (6, 8, 10, 22). So far, RISA was used to reveal gross changes in community structures (6, 8, 22). Here, this cultivation-independent technique was used to trace inoculum strains and to detect and trace invaders. The detection limit of RISA for a single strain was approximately 1% of the total community DNA (data not shown), which explains why only 8 invaders were detected in the community fingerprints, although 33 diverse strains were isolated from the effluents. The similarity of biofilm community composition and the respective liquid phases implied that a noninvasive sampling of liquid-phase bioreactor samples alone would have been sufficient to monitor spatial and temporal variations in the microbial community composition.
Osborn et al. demonstrated the effect of decreasing DNA concentrations in a PCR on terminal restriction fragment length polymorphism profiles. Diluting the initial template concentration resulted in a decline in both the number of peaks and the height of each peak present in a profile (19). We have used total DNA of various concentrations. Thus, RISA enabled only a semiquantitative analysis of the relative proportions of prevalent strains. An absolute quantification was also impossible due to the bias inherent in any PCR amplification approach, such as preferential amplification of particular sequences (26) and multicopy target sequences. However, RISA proved to be a powerful tool to trace individual strains in diverse communities for a long period of time.
Selective pressure of mercury mediates microbial diversity.
Mercury-reducing biofilms transform toxic ionic mercury to metallic mercury, which is harmless to bacteria. In the bioreactor, the activity produced a gradient of water-soluble mercury [mainly Hg(II)] with the greatest concentration at the bioreactor inflow and the smallest concentration at the effluent. Along this gradient of decreasing mercury concentrations, microbial diversity increased. Thus, ionic mercury exerted a selective pressure on the microbial communities. High Hg(II) concentrations decreased microbial diversity. The lower selective pressure within the carbon filter permitted the establishment of a variety of weakly resistant strains that occurred only there, although each invader can be assumed to have entered the bioremediation system at the bioreactor inflow and passed through the packed bed. However, some of them were also found in the bioreactor. This can be attributed to the structural heterogeneity of the biofilms (as revealed by scanning electron microscopy), which exhibited microniches and the presence of highly resistant community members; Whiteley et al. reported the survival of toxicant-sensitive bacteria by the protective cell-cell shielding of adjacent cells of a resistant strain (32). This protection by neighbors permitted the establishment of weakly resistant strains in the bioreactor and the establishment of a mercury-sensitive strain in the carbon filter.
Succession of mercury-reducing biofilm communities.
In a previous study of laboratory-scale biofilms, a rather stable community composition was observed (28). Here, only one inoculum strain and one invader were continuously detected. Succession was not only driven by mercury resistance levels, since highly resistant inoculum strains were frequently replaced by less-resistant invaders. We have to assume that the invaders that were able to establish stably in the biofilm community were better adapted by better biofilm formation capability or other traits. Because the mean mercury concentrations were similar (6.2 mg/liter in laboratory-scale bioreactor runs [28] and 4.8 mg/liter in this study), this higher dynamic in the technical scale was probably caused by the different conditions encountered in the up-scaled bioreactor on the site of a factory: a higher flow velocity, repeatedly high mercury peaks, and continuously high temperatures. In both laboratory- and technical-scale bioreactors, no change in community structure altered the mercury-reduction efficiency and no deterioration in activity was observed. A gradual replacement of the inoculum strains was also reported by Stoffels et al. (25) when a fermenter culture adapted to degradation of aromatic compounds was transferred as an inoculum to a trickle-bed bioreactor. In this study, the high mercury-resistance levels of all inoculum strains were no guarantee for their stable establishment in the biofilm, but the main task of the inoculum was to ensure the efficient mercury retention from the beginning and to provide a certain set of heterogeneous strains containing at least one well-adapted strain.
Acknowledgments
We thank K.-H. Ujma, Preussag AG, for establishing cooperation with Preussag AG and for continuous support. We thank R. Böckle and M. Golm, Preussag Wassertechnik, for constructive and creative cooperation during design, construction, and operation of the pilot plant. We are grateful to D. Saβ, ECI, who opened the doors of ECI Elektrochemie Ibbenbüren to this experiment, and to H. Hardemann and K. Bartel of ECI Elektrochemie Ibbenbüren, whose daily support made it possible. H. Lünsdorf is gratefully acknowledged for assistance in scanning electron micrography.
This work was funded by grants LIFE97ENV/D/000463 and BIO4-CT-98-0168 from the European Community.
REFERENCES
- 1.Akagi, H., Y. Fujita, and E. Takabatake. 1977. Methylmercury: photochemical transformation of mercuric sulfide into methylmercury in aqueous solution. Photochem. Photobiol. 26:363-370. [Google Scholar]
- 2.Boudou, A., and F. Ribeyre. 1997. Mercury in the food web: accumulation and transfer mechanisms, p. 289-319. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems: mercury and its effects on environment and biology, vol. 34. Marcel Dekker, Inc., New York, N.Y. [PubMed] [Google Scholar]
- 3.Brunke, M., W. D. Deckwer, A. Frischmuth, J. M. Horn, H. Lunsdorf, M. Rhode, M. Rohricht, K. N. Timmis, and P. Weppen. 1993. Microbial retention of mercury from waste streams in a laboratory column containing merA gene bacteria. FEMS Microbiol. Rev. 11:145-152. [DOI] [PubMed] [Google Scholar]
- 4.Chang, J. S., and J. Hong. 1995. Estimation of kinetics of mercury detoxification from low-inoculum batch cultures of Pseudomonas aeruginosa PU21 (Rip64). J. Biotechnol. 42:85-90. [DOI] [PubMed] [Google Scholar]
- 5.Compeau, G. C., and R. Bartha. 1987. Effect of salinity on mercury-methylating activity of sulfate-reducing bacteria in estuarine sediments. Appl. Environ. Microbiol. 53:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frischmuth, A., P. Weppen, and W.-D. Deckwer. 1993. Microbial transformation of mercury(II). I. Isolation of microbes and characterization of their transformation capabilities. J. Biotechnol. 29:39-55. [Google Scholar]
- 8.Garcia-Martinez, J., S. G. Acinas, A. I. Anton, and F. Rodriguez-Valera. 1999. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J. Microbiol. Methods 36:55-64. [DOI] [PubMed] [Google Scholar]
- 9.Gilmour, C. C., E. A. Henry, and R. Mitchell. 1992. Sulfate stimulation of mercury methylation in freshwater sediments. Environ. Sci. Technol. 26:2281-2287. [Google Scholar]
- 10.Hirsch, C. F., and J. M. Sigmund. 1995. Use of polymerase chain reaction (PCR) fingerprinting to differentiate bacteria for microbial products screening. J. Ind. Microbiol. 15:85-93. [DOI] [PubMed] [Google Scholar]
- 11.Hobman, J. L., and N. L. Brown. 1997. Bacterial mercury-resistance genes, p. 527-568. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems: mercury and its effects on environment and biology, vol. 34. Marcel Dekker, Inc., New York, N.Y. [PubMed] [Google Scholar]
- 12.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langford, N., and R. Ferner. 1999. Toxicity of mercury. J. Hum. Hypertens. 13:651-656. [DOI] [PubMed] [Google Scholar]
- 14.Magos, L. 1997. Physiology and toxicology of mercury, p. 321-370. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems: mercury and its effects on environment and biology, vol. 34. Marcel Dekker, Inc., New York, N.Y. [PubMed] [Google Scholar]
- 15.Mason, R. P., W. F. Fitzgerald, and F. M. Morel. 1994. The biogeochemical cycling of elemental mercury: anthropogenic influences. Geochim. Cosmochim. Acta 58:3191-3198. [Google Scholar]
- 16.Misra, T. K. 1992. Bacterial resistances to inorganic mercury salts and organomercurials. Plasmid 27:4-16. [DOI] [PubMed] [Google Scholar]
- 17.Nriagu, J. O., and J. M. Pacyna. 1988. Quantitative assessment of worldwide contamination of air, water and soils by trace metals. Nature 333:134-139. [DOI] [PubMed] [Google Scholar]
- 18.Osborn, A. M., K. D. Bruce, P. Strike, and D. A. Ritchie. 1997. Distribution, diversity and evolution of the bacterial mercury resistance (mer) operon. FEMS Microbiol. Rev. 19:239-262. [DOI] [PubMed] [Google Scholar]
- 19.Osborn, A. M., E. R. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 20.Pak, K., and R. Bartha. 1998. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl. Environ. Microbiol. 64:1987-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pitcher, D. G., N. A. Saunders, and R. J. Owen.1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 22.Ranjard, L., S. Nazaret, F. Gourbiere, J. Thioulouse, P. Linet, and A. Richaume. 2000. A soil microscale study to reveal the heterogeneity of Hg(II) impact on indigenous bacteria by quantification of adapted phenotypes and analysis of community DNA fingerprints. FEMS Microbiol. Ecol. 31:107-115. [DOI] [PubMed] [Google Scholar]
- 23.Schrope, M. 2001. US to take temperature of mercury threat. Nature 409:124. [DOI] [PubMed] [Google Scholar]
- 24.Silver, S., and L. T. Phung. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 25.Stoffels, M., R. Amann, W. Ludwig, D. Hekmat, and K. H. Schleifer. 1998. Bacterial community dynamics during start-up of a trickle-bed bioreactor degrading aromatic compounds. Appl. Environ. Microbiol. 64:930-939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.von Canstein, H., Y. Li, K. N. Timmis, W.-D. Deckwer, and I. Wagner-Döbler. 1999. Removal of mercury from chloralkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl. Environ. Microbiol. 65:5279-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.von Canstein, H., Y. Li, and I. Wagner-Döbler. 2001. Long-term stability of mercury reducing microbial biofilm communities analyzed by 16S-23S rDNA interspacer region polymorphism. Microb. Ecol. 42:624-634. [DOI] [PubMed] [Google Scholar]
- 29.von Canstein, H., Y. Li, and I. Wagner-Döbler. 2001. Long-term performance of bioreactors cleaning mercury-contaminated wastewater and their response to temperature and mercury stress and mechanical perturbation. Biotechnol. Bioeng. 74:212-219. [DOI] [PubMed] [Google Scholar]
- 30.Wagner-Döbler, I., H. Lünsdorf, T. Lübbehüsen, H. F. von Canstein, and Y. Li. 2000. Structure and species composition of mercury-reducing biofilms. Appl. Environ. Microbiol. 66:4559-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagner-Döbler, I., H. von Canstein, Y. Li, K. N. Timmis, and W.-D. Deckwer. 2000. Removal of mercury from chemical wastewater by microorganisms in technical scale. Environ. Sci. Technol. 34:4628-4634. [Google Scholar]
- 32.Whiteley, M., J. R. Ott, E. A. Weaver, and R. J. McLean. 2001. Effects of community composition and growth rate on aquifer biofilm bacteria and their susceptibility to betadine disinfection. Environ. Microbiol. 3:43-52. [DOI] [PubMed] [Google Scholar]