Abstract
We investigated the application of an oligonucleotide microarray to (i) specifically detect Cryptosporidium spp., (ii) differentiate between closely related C. parvum isolates and Cryptosporidium species, and (iii) differentiate between principle genotypes known to infect humans. A microarray of 68 capture probes targeting seven single-nucleotide polymorphisms (SNPs) within a 190-bp region of the hsp70 gene of Cryptosporidium parvum was constructed. Labeled hsp70 targets were generated by PCR with biotin- or Cy3-labeled primers. Hybridization conditions were optimized for hybridization time, temperature, and salt concentration. Two genotype I C. parvum isolates (TU502 and UG502), two C. parvum genotype II isolates (Iowa and GCH1), and DNAs from 22 non-Cryptosporidium sp. organisms were used to test method specificity. Only DNAs from C. parvum isolates produced labeled amplicons that could be hybridized to and detected on the array. Hybridization patterns between genotypes were visually distinct, but identification of SNPs required statistical analysis of the signal intensity data. The results indicated that correct mismatch discrimination could be achieved for all seven SNPs for the UG502 isolate, five of seven SNPs for the TU502 isolate, and six of seven SNPs for both the Iowa and GCH1 isolates. Even without perfect mismatch discrimination, the microarray method unambiguously distinguished between genotype I and genotype II isolates and demonstrated the potential to differentiate between other isolates and species on a single microarray. This method may provide a powerful new tool for water utilities and public health officials for assessing point and nonpoint source contamination of water supplies.
Cryptosporidium parvum parasites can cause acute, self-limiting diarrhea that lasts 7 to 10 days in healthy individuals but can be fatal for AIDS and cancer chemotherapy patients (3). There are two well-documented genotypes of C. parvum known to infect humans, although other genotypes have been proposed (12, 14, 15). Although natural infections in rhesus monkeys and experimental infection in gnotobiotic pigs have been observed (21), genotype I isolates appear to be transmitted primarily between humans. Outbreaks attributed to this genotype tend to predominate in urban areas. Genotype II isolates, however, can also be transmitted from animals to people. Outbreaks attributed to this genotype appear to predominate in rural areas. In recent years, documented waterborne disease outbreaks caused by this apicomplexan parasite have generated significant interest in developing rapid and specific methods for the detection and characterization of C. parvum isolates in the environment (4, 6, 11, 12, 14, 15, 16, 17, 19, 20, 21, 22, 23).
Because each user's needs for C. parvum detection are different, few attempts have been made to bridge the gap between detection for regulatory needs (20) and genotype differentiation for risk characterization of watersheds and outbreak investigations (12, 19, 21, 22, 23). Nearly all investigators who use molecular methods employ PCR amplification as the first step for identification and differentiation. PCR is generally required for C. parvum detection because the number of target organisms in a sample can be quite low (e.g., one oocyst per 10 or more liters of water), and no direct nucleic acid detection method has been developed to detect extremely low copy numbers of these target organisms in a sample. However, many investigators confirm PCR products of specific genes by using hybridization (6, 16) or other molecular methods (4, 6, 11, 15, 19, 22, 23).
DNA sequencing is clearly the highest-resolution technique for differentiating between strains and isolates of C. parvum (19, 22, 23), but it is still not a rapid analysis method that can be used for routine detection and identification by public utilities because the isolate may first require enzymatic amplification, cloning, and characterization prior to sequencing. DNA microarrays, however, represent an elegant “sequencing by hybridization” method (7, 10, 13, 24) that may be able to simultaneously satisfy the rapid detection requirement for regulatory compliance, epidemiology, public health, and risk assessment investigations. Therefore, the purpose of this study was to design a C. parvum oligonucleotide microarray that could specifically detect Cryptosporidium spp., differentiate between the principle genotypes known to infect humans, and demonstrate the potential to differentiate between other closely related isolates and species through single-nucleotide mismatch discrimination.
MATERIALS AND METHODS
Cryptosporidium isolates and nontarget DNA.
Purified genomic DNA preparations for genotype I isolates TU502 and UG502 and genotype II isolate GCH1 were a generous gift from Saul Tzipori at Tufts University School of Veterinary Medicine. Genomic DNA from the Tufts isolates was prepared as described by Feng et al. (6).
Purified (CsCl2) Iowa isolate oocysts (genotype II) were obtained from Marilyn M. Marshall at the University of Arizona School of Veterinary Science. Genomic DNA from the Iowa isolate was prepared by freezing the oocysts in liquid nitrogen for 5 min, followed by boiling for 5 min at 95°C in a heat block. This process was repeated two times, and the DNA was frozen at −20°C until use.
Nontarget DNAs from 22 organisms were obtained from the Metropolitan Water District of Southern California. DNA was extracted from these organisms as previously described (16). The test organisms were Aeromonas hydrophila (ATCC 7965), Citrobacter freundii, Clostridium perfringens, Comamonas testosteroni, Enterobacter cloacae, Escherichia coli (ATCC 2592), E. coli O157:H7, Klebsiella pneumoniae (ATCC 13883), Nitrosomonas europaea, Pseudomonas fluorescens, Salmonella enterica serovar Typhimurium, Serratia marcescens, Shigella sonnei, Yersinia enterocolitica, Giardia lamblia, Candida albicans, Saccharomyces cerevisiae, Ourococcus sp., Chlorella sp., Synechococcus sp. (ATCC 27145), Pseudoanabaena sp., and Anabaena sp. All of these test organisms were negative for both PCR amplification and hybridization to the array (not shown).
PCR primers and microarray capture probes.
Several candidate genes were reviewed for capture oligonucleotide selection, including the 18S rRNA, hsp70, COWP, TRAPC1, TRAPC2, β-tubulin, and acetyl coenzyme A genes. At the time this array was constructed, fairly complete sequence data were available only for the 18S rRNA gene and the hsp70 gene. Based on the published literature (19, 22) and sequence availability in a public database (GenBank), the array was targeted at seven variable positions within a 190-bp fragment of the hsp70 gene between base positions 1360 and 1550 (sequence numbering based on a C. parvum human isolate sequence; GenBank accession number AF221535 [19]). All available Cryptosporidium hsp70 sequences were aligned with the ClustalW program (Baylor College of Medicine, Houston, Tex.), and variable bases were identified by visual inspection. The single-nucleotide-mismatch probes deduced in this manner encompassed variable nucleotides at base positions 1368 and 1371 (14 probes), 1404 (9 probes), 1419 and 1422 (13 probes), 1464 (7 probes), 1479 (9 probes), 1533 (8 probes), and 1542 (8 probes) (Table 1). Oligonucleotides (15-mers) were designed to hybridize with the DNA coding strand or mRNA, with the diagnostic base (single-nucleotide polymorphism [SNP]) as close to the center of the oligonucleotide as possible (7). From Table 1, the m1 probe (e.g., 1368 m1 or 1404 m1, etc.) for each diagnostic position is the perfect match for the published sequence of C. parvum human isolate AF 221535 (19). Depending on the proximity of the nucleotide mismatches along the 190-bp fragment, the remaining probes were all possible single-nucleotide variants of the m1 probe, many of which were perfect matches for the hsp70 sequence found in other C. parvum isolates or Cryptosporidium species.
TABLE 1.
Sequences of the microarray capture probes
| Probe name | Probe sequencea | Tm (°C)b | Sequence host specificity |
|---|---|---|---|
| 1368 m1 | 5′-TCA-AAA-GTA-ACT-TCA-3′ | 38 | C. parvum genotype I (AF 221535 sequence) |
| 1368 m2 | 5′-TCA-AAT-GTA-ACT-TCA-3′ | 38 | SNP variant at position 1371 |
| 1368 m3 | 5′-TCA-AAC-GTA-ACT-TCA-3′ | 40 | SNP variant at position 1371 |
| 1368 m4 | 5′-TCA-AAG-GTA-ACT-TCA-3′ | 40 | SNP variant at position 1371 |
| 1368 m5 | 5′-TCA-AAA-GTT-ACT-TCA-3′ | 38 | C. muris (AF 221542 sequence) |
| 1368 m6 | 5′-TCA-AAA-GTG-ACT-TCA-3′ | 40 | C. parvum pig, monkey, and ferret isolates; C. wrairi, C. meleagridis |
| 1368 m7 | 5′-TCA-AAA-GTC-ACT-TCA-3′ | 40 | C. muris (AF 221543 sequence) |
| 1368 m8 | 5′-TCA-AAT-GTT-ACC-TCA-3′ | 40 | C. baileyi |
| 1368 m9 | 5′-TCA-AAA-GTT-ACC-TCA-3′ | 40 | C. serpentis |
| 1368 m10 | 5′-TCA-AAG-GTG-ACC-TCA-3′ | 44 | C. parvum dog isolate |
| 1368 m11 | 5′-TCA-AAA-GTA-ACC-TCA-3′ | 40 | Cryptosporidium sp. desert monitor isolate |
| 1368 m12 | 5′-TCG-AAG-GTT-ACC-TCG-3′ | 46 | C. felis |
| 1368 m13 | 5′-TCA-AAG-GTG-ACT-TCA-3′ | 42 | C. parvum genotype II isolates |
| 1368 m14 | 5′-TCA-AAA-GTC-ACT-TCA-3′ | 40 | C. muris (AF 221543 sequence) |
| 1404 m1 | 5′-CAG-CAG-ATA-CAT-TCA-3′ | 42 | C. parvum genotype I, pig, mouse, and ferret isolates; C. wrairi, C. meleagridis |
| 1404 m2 | 5′-CAG-CAG-AAA-CAT-TCA-3′ | 42 | SNP variant at position 1404 |
| 1404 m3 | 5′-CAG-CAG-AGA-CAT-TCA-3′ | 44 | SNP variant at position 1404 |
| 1404 m4 | 5′-CAG-CAG-ACA-CAT-TCA-3′ | 44 | C. parvum genotype II isolates |
| 1404 m5 | 5′-CTG-CTG-ATA-CAT-TTA-3′ | 40 | C. muris (AF 221542 sequence) |
| 1404 m6 | 5′-CAG-CAG-ACA-CAT-TTA-3′ | 42 | C. felis |
| 1404 m7 | 5′-CTG-CCG-AAA-CAT-TTA-3′ | 42 | C. serpentis |
| 1404 m8 | 5′-CAG-CAG-AAA-CAT-TTA-3′ | 40 | C. baileyi |
| 1404 m9 | 5′-CGG-CGG-AAA-CAT-TCA-3′ | 46 | C. parvum dog isolate |
| 1419 m1 | 5′-CCA-GTG-CTT-TTA-TCA-3′ | 42 | C. parvum genotype I isolates |
| 1419 m2 | 5′-CCA-GTA-CTT-TTA-TCA-3′ | 40 | SNP variant at position 1422 |
| 1419 m3 | 5′-CCA-GTC-CTT-TTA-TCA-3′ | 42 | SNP variant at position 1422 |
| 1419 m4 | 5′-CCA-GTT-CTT-TTA-TCA-3′ | 40 | SNP variant at position 1422 |
| 1419 m5 | 5′-CCA-GTG-CTA-TTA-TCA-3′ | 42 | SNP variant at position 1419 |
| 1419 m6 | 5′-CCA-GTG-CTC-TTA-TCA-3′ | 44 | C. meleagridis |
| 1419 m7 | 5′-CCA-GTG-CTG-TTA-TCA-3′ | 44 | SNP variant at position 1419 |
| 1419 m8 | 5′-CCA-GTA-CTT-TTA-TCT-3′ | 42 | C. parvum pig isolate |
| 1419 m9 | 5′-CCG-GTG-CTG-TTG-TCA-3′ | 48 | C. parvum dog isolate |
| 1419 m10 | 5′-CCA-GTG-CTC-TTG-TCA-3′ | 46 | C. felis |
| 1419 m11 | 5′-CCG-GTA-CTC-TTG-TCA-3′ | 46 | C. muris (AF 221542 and AF 221543 sequences) |
| 1419 m12 | 5′-CCG-GTA-CTC-TTA-TCA-3′ | 44 | C. serpentis |
| 1419 m13 | 5′-CCA-GTA-CTC-TTA-TCA-3′ | 42 | C. parvum genotype II and ferret isolates; C. baileyi, C. wrairi, desert monitor isolate |
| 1464 m1 | 5′-ATA-ATC-TAC-CCT-TAT-3′ | 38 | C. parvum genotype I and II isolates, C. parvum pig and ferret isolates, C. meleagridis, desert monitor isolate |
| 1464 m2 | 5′-ATA-ATC-TTC-CCT-TAT-3′ | 38 | SNP variant at position 1464 |
| 1464 m3 | 5′-ATA-ATC-TGC-CCT-TAT-3′ | 40 | C. parvum mouse isolate |
| 1464 m4 | 5′-ATA-ATC-TCC-CCT-TAT-3′ | 40 | SNP variant at position 1464 |
| 1464 m5 | 5′-AGA-GCC-TTC-CCT-TGA-3′ | 46 | Does not match any Cryptosporidium sequences based on this alignment |
| 1464 m6 | 5′-ATA-AAC-GAC-CTT-TAT-3′ | 38 | C. muris (AF 221542 and AF 221543 sequences), C. serpentis |
| 1464 m7 | 5′-ATA-AAC-GAC-CCT-TAT-3′ | 40 | C. baileyi |
| 1479 m1 | 5′-CAA-TAT-CGT-CCT-TTG-3′ | 42 | C. parvum genotype I, II, and ferret isolates; C. wrairi, C. meleagridis |
| 1479 m2 | 5′-CAA-TAT-CAT-CCT-TTG-3′ | 40 | SNP variant at position 1479 |
| 1479 m3 | 5′-CAA-TAT-CTT-CCT-TTG-3′ | 40 | C. muris (AF 221542 and AF 221543 sequences), C. serpentis |
| 1479 m4 | 5′-CAA-TAT-CCT-CCT-TTG-3′ | 42 | SNP variant at position 1479 |
| 1479 m5 | 5′-CGA-TAT-CAT-CCT-TTG-3′ | 42 | C. parvum pig isolate |
| 1479 m6 | 5′-CGA-TAT-CGT-CCT-TGG-3′ | 46 | C. parvum dog isolate |
| 1479 m7 | 5′-CAA-TAT-CTT-CTT-TAG-3′ | 38 | C. baileyi |
| 1479 m8 | 5′-CGA-TAT-CTT-CCT-TAG-3′ | 42 | C. felis |
| 1479 m9 | 5′-CAA-TAT-CAT-CCT-TAC-3′ | 40 | Does not match any Cryptosporidium sequences based on this alignment |
| 1533 m1 | 5′-GTC-TGT-TTT-GCT-CAT-3′ | 42 | C. parvum human (AF 221535 sequence), mouse, and ferret isolates; C. wrairi |
| 1533 m2 | 5′-GTC-TGT-TAT-GCT-CAT-3′ | 42 | SNP variant at position 1533 |
| 1533 m3 | 5′-GTC-TGT-TGT-GCT-CAT-3′ | 44 | C. parvum monkey isolate (AF 221534) |
| 1533 m4 | 5′-GTC-TGT-TCT-GCT-CAT-3′ | 44 | C. parvum genotype II isolates |
| 1533 m5 | 5′-GTT-TAT-TTT-GTT-CAT-3′ | 36 | Cryptosporidium sp. (desert monitor isolate) |
| 1533 m6 | 5′-GTC-TAT-TTT-GTT-CAT-3′ | 38 | C. parvum pig isolate |
| 1533 m7 | 5′-TAC-GAT-TTT-GTT-CAT-3′ | 36 | C. baileyi |
| 1533 m8 | 5′-CAC-GAT-TCT-GTT-CAT-3′ | 42 | C. muris (AF 221542 and AF 221543), C. serpentis |
| 1542 m1 | 5′-CAA-TCT-TGA-GTC-TGT-3′ | 42 | C. parvum genotype I, mouse, and ferret isolates; C. wrairi, C. meleagridis |
| 1542 m2 | 5′-CAA-TCT-TAA-GTC-TGT-3′ | 40 | C. parvum genotype II isolates |
| 1542 m3 | 5′-CAA-TCT-TTA-GTC-TGT-3′ | 40 | SNP variant at position 1542 |
| 1542 m4 | 5′-CAA-TCT-TCA-GTC-TGT-3′ | 42 | SNP variant at position 1542 |
| 1542 m5 | 5′-CAA-TCT-TGA-GTT-TAT-3′ | 38 | Cryptosporidium sp. (desert monitor isolate) |
| 1542 m6 | 5′-CAA-TCT-TGA-TAC-GAT-3′ | 40 | C. baileyi |
| 1542 m7 | 5′-CAA-TCT-TTT-CAC-GAT-3′ | 40 | C. muris (AF 221542 sequence), C. serpentis |
| 1542 m8 | 5′-CAA-TCT-TCT-CAC-GAT-3′ | 42 | C. muris (AF 221543 sequence) |
Boldface indicates sequence polymorphisms between isolates and species.
Calculated by the 2(A+T) + 4(G+C) method.
PCR primers were designed to flank the 190-bp variable region of the human sequence isolate (positions 1360 to 1550) but were also predicted to amplify hsp70 sequences from C. parvum isolates. The forward primer was biotin- or Cy3 labeled so that detectable hybridization would be expected to occur between the forward DNA sequence of the amplicon and the capture probe(s) on the array. The sequences of the PCR primers were as follows: forward primer, 5′-biotin or Cy3-ACCAAGAGGTGTACCACAAA-3′; reverse primer, 5′-CTCCAAAGAGTTCTTAGCCT-3′. Both the capture oligonucleotides and PCR primers were obtained from BioSource International. All primers and probes were synthesized at the 0.2-μmol scale and purified by using Biosource's Econopure method. PCR primers were purified by high-pressure liquid chromatography.
Microarray fabrication.
Unmodified oligonucleotides were printed on 12-well epoxy silane-derivatized (3-glycidoxypropyltrimethoxysilane; Aldrich, Milwaukee, Wis.) (2%, vol/vol in methanol) Teflon-masked slides (Erie Scientific, Portsmouth, N.H.) as previously described (2, 18). Oligonucleotide capture probes were resuspended in reagent-grade water, and the concentration of each was measured, in triplicate, using spectrophotometry (Smartspec 3000; Bio-Rad, Hercules, Calif.). Subsequently, capture probes were reconstituted at 80 to 100 μM in 0.01% sodium dodecyl sulfate-50 mM NaOH print buffer. Probes were printed with a 417 arrayer (Affymetrix, Santa Clara, Calif.), with a complete hsp70 array contained within each well of the Teflon-masked slide. In addition to the capture probes, each array contained four spots of a biotinylated quality control oligonucleotide (5′-biotin-TTGTGGTGGTGGTGTGGTGGTGGGGTTGGG TGGTGG-3′), which served as a positional reference and a positive control for enzymatic signal generation. After printing, the slides were baked for 30 min at 130°C and stored at room temperature.
Generation of labeled targets by PCR.
Labeled hsp70 targets were generated by using PCR. Reagents were from the HotStar Taq kit (Qiagen, Valencia, Calif.), except for the deoxynucleoside triphosphates (Amersham Pharmacia Biotechnology, Piscataway, N.J.). PCR amplification was carried out in a total volume of 100 μl, utilizing a Tetrad thermal cycler and 96-well plates (MJ Research, Watertown, Mass.). Final reaction conditions were as follows: a minimum of 104 oocyst equivalents of genomic DNA (in 10 μl), 1× PCR buffer, 1× Qiagen Q solution, 2.5 mM Mg2+, 200 μM deoxynucleoside triphosphates, 2.5 U of Taq polymerase, and a 0.2 μM concentration of each primer. Reagent-grade water was used as a negative control. Thermal cycling conditions were as follows: 95°C for 15 min, followed by 40 cycles of denaturation for 1 min at 95°C, primer annealing for 2 min at 46.7°C, and extension for 3 min at 72°C. PCR was terminated by a final extension period of 5 min at 72°C and then quenching of the reaction by cooling to 4°C. PCR amplification was confirmed by running 20-μl aliquots of the amplification reaction products on a 2% Tris-acetate-EDTA agarose gel. Biotinylated PCR products were hybridized directly to the array without further manipulation, but Cy3-labeled PCR products were further purified with an AutoSeq G-50 spin column per the instructions of the manufacturer (Amersham Pharmacia Biotechnology) prior to microarray hybridization.
Optimization of array hybridization conditions.
The Iowa isolate (a genotype II strain) was used for the array optimization studies because of its availability at the time this project began. Biotinylated amplicons served as the labeled DNA for the initial microarray optimization studies but were subsequently confirmed by using Cy3-labeled amplicons. A range of salt concentrations (1× to 4× SSC [1× SSC is 0.15 M NaCl plus 0.015 M trisodium citrate, pH 7.0]), temperatures (4 and 22°C) and hybridization times (2 h, 4 h, and overnight) were investigated to optimize hybridization of the labeled amplicons to the array. For all tests, 20 μl of the PCR mixture was diluted to 70 μl in concentrated hybridization buffer, such that the final hybridization reaction contained 5× Denhardt's solution (1 g of Ficoll 400 per liter, 1 g of polyvinylpyrolidone per liter, and 1 g of ultrapure bovine serum albumin per liter) and the desired SSC concentration (1× to 4×). Amplicons were heat denatured for 10 min at 95°C and then snap cooled on ice for 5 min. Thirty-five-microliter aliquots of the target solution were applied to replicate arrays and incubated under the desired time and temperature regimen within a humidified reaction chamber.
Signal development and detection of hybridization reactions.
Microarrays that were hybridized with biotin-labeled amplicons were developed according to the procedures described by Small et al. (18). Cy3-labeled PCR amplicons did not require extensive posthybridization processing. After hybridization, the slides were instead thoroughly rinsed with ice-cold 1× SSC and then dried immediately with compressed N2. Both the biotin and Cy3 microarrays were imaged at 10.1-μm resolution for 0.01 and 0.3 s per panel, respectively, using an ArrayWoRx microarray scanner (Applied Precision, Issaquah, Wash.) (excitation and emission wavelengths of 365 and 535 nm for biotin and 548 and 595 nm for Cy3). Images for publication were captured as jpeg files, and spot intensity data were exported as tab-delimited files for statistical analysis with Excel 2000 and MatLab release 12.1.
Discriminating between SNPs.
Optimized hybridization conditions (deduced as described above) were used as the initial protocol for SNP discrimination. Because PCR amplification of genotype I isolates with biotinylated primers was not successful and because the formal statistical analysis (described below) indicated that the biotin labeling strategy was ultimately suboptimal for SNP analysis, the biotin labeling strategy was tested only on genotype II isolates. However, all four C. parvum isolates were evaluated with Cy3-labeled PCR amplicons. Twenty microliters of PCR products was resuspended in a total volume of 70 μl in optimized hybridization buffer (3× SSC and 5× Denhardt's solution [final concentrations]). The amplicons were heat denatured as described above and hybridized to the array overnight (ca. 17 h) at 4 and 22°C. Slides were imaged and data files were exported to Excel and MatLab as described above.
Statistics and data analysis.
For each isolate and hsp70 PCR product, the perfectly matched probe for each of the seven variable positions was expected to have significantly greater signal intensity than the mismatched probes. Each probe set (for a diagnostic position, e.g., 1368 probes, 1404 probes, etc.) is represented once in each array, and at least two replicate arrays were examined for each isolate-probe set combination. Although similar hybridization profiles within a probe set were observed for each of the hsp70 targets analyzed during this study, the signal intensity between replicate hybridizations could vary considerably from array to array.
The following analysis of variance and error effects model, which allows testing the significance of hybridization signal intensity differences between probes for a given diagnostic position (e.g., differences in signal intensities for probes within the 1368 probe set, the 1404 probe set, etc.), was developed and adapted from that of Kerr et al. (9) for this study to account for the observed variability between arrays: ln(Yij) = μ + si + wj + ɛij, where Yij is the ratio of the spot intensity to the local background fluorescence [e.g., ln(Cy3 spot intensity) − ln(Cy3 background), defined by the ArrayWoRx imager software], μ represents the overall mean fluorescence for all probe spots over all replicate microarrays (n = 2 to 4), si is the effect of the probe (over all arrays) on the overall mean fluorescence, wj is the effect of array j on the overall mean fluorescence, and ɛij is the corresponding random error in the fluorescence measurements. A natural log transformation was applied to all of the factor effects, considered to be multiplicative, to be rewritten as the additive model shown above (9).
The model assumes that the array effect is uniform across all probe spots. The background fluorescence measurements have been used to correct for local background effects. While other approaches to correct for local background were investigated, this correction, combined with natural log transformation of the data, allowed the best approximation to a normal distribution of the data (i.e., linearity of normal probability plots and random scatter of residual plots).
This model was then fit for each probe set (e.g., 1368 probes, 1404 probes, etc.), and Tukey's multiple-comparison procedure (experiment-wide α = 0.05) was then used to assess the significance of observed mean differences between the perfectly matched and mismatched probes for a given probe suite.
RESULTS
Specificity and array optimization studies.
Only DNAs from the four isolates of C. parvum that we tested could be PCR amplified and subsequently detected on the array. In addition, PCR products from no-template controls failed to hybridize to any of the capture probes on the array, indicating that primer dimers or other labeled amplification artifacts (if present) did not contribute to the hybridization signal on the array. Thus, the ability of the microarray method (PCR plus array hybridization) to differentiate between Cryptosporidium spp. and other, nontarget genera was demonstrated.
Optimization of conditions for hybridization to the array initially indicated that the best combination of time, temperature, buffer, and probe labeling strategy was overnight hybridization (ca. 17 h) at 4 or 22°C, 3× SSC-5× Denhardt's buffer, and the use of either biotin- or Cy3-labeled amplicons as probes (data not shown). However, formal analysis of the SNP data by using the statistical models developed for this study (discussed below) indicated that the Cy3 reporter system was ultimately more robust than the biotin reporter system. (Robust in this context means the ability to reproducibly generate labeled targets and, more importantly, to correctly identify SNPs.) Consequently, Cy3-labeled amplicons were used to assess the specificity of the microarray method for C. parvum detection and SNP genotyping.
Differentiation of C. parvum genotype I isolates from genotype II isolates.
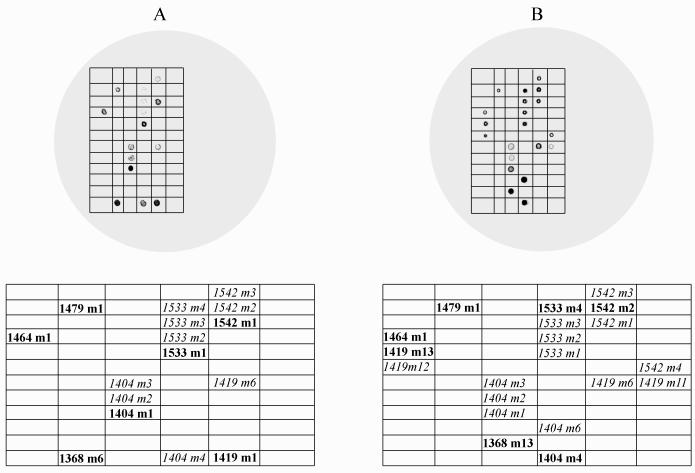
Figure 1 illustrates the hybridization patterns for Cy3-labeled hsp70 amplicons generated from genotype I and genotype II isolates. Figure 1A shows the typical hybridization pattern for genotype I isolates (results for the UG502 isolate are shown) and Fig. 1B shows the hybridization patterns observed for genotype II isolates (results for the Iowa isolate are shown). The table below each image is mapped to the grid which overlays the image and shows the locations of the perfectly matched probes for each genotype and hybridized mismatched probes.
FIG. 1.
Typical microarray hybridization patterns for C. parvum hsp70 amplicons. (A) Genotype I isolate UG502; (B) genotype II isolate Iowa. The grids below the patterns indicate the hybridized probes and their relative positions on the array. Boldface type indicates the perfectly matched probes for each isolate, and italic type indicates the mismatched probes that also resulted in a positive signal. Images were enhanced for publication by using Adobe Photoshop 6.0, but the underlying data for statistical analysis remained unchanged.
The patterns of probe hybridization within a genotype were similar even when the signal intensity varied between arrays. For example, in Fig. 1A the 1404 m1 probe (the perfectly matched probe for genotype I isolates) appears to have the strongest hybridization signal of all four probes within the 1404 probe suite. In a replicate hybridization, the signal intensities of all of the 1404 probes, as well as the signal intensities of all other probes on the array, might decrease. However, this visual difference in signal intensity was always proportional, such that the 1404 m1 probe reproducibly generated the strongest signal intensity relative to the three other mismatched probes. The underlying premise of the statistical model was to quantitatively determine if the perfectly matched probes for a genotype had the strongest signal intensity relative to the mismatched probes, given the variability of the signal intensity between arrays. Therefore, statistical analysis of the image intensity data was required to determine if SNPs had been achieved for the perfectly matched probes for each isolate.
Although the array was designed based on the published sequence for a human isolate (AF 221535 [19]), certain probes were determined to be specific for genotype II isolates to the exclusion of genotype I isolates, other closely related C. parvum isolates, or different Cryptosporidium species. Thus, an SNP code for genotype I versus genotype II could be defined, where the genotype II SNP code is composed of capture probes 1368 m13, 1404 m4, 1533 m4, and 1542 m2 (Table 1). For single-nucleotide mismatch discrimination (SNP analysis) and unambiguous identification of genotype II isolates, we would expect each of the genotype II SNP code probes to have a statistically greater hybridization signal than all other mismatched probes at a diagnostic position. In contrast, we would expect genotype I isolates to generate significantly greater hybridization with the m1 probes within the genotype II SNP code. Given the predicted differential hybridization result for the genotype II SNP code, the simple 68-probe microarray is therefore (in principle) able to distinguish genotype I from genotype II isolates. We explicitly tested this hypothesis with four genotype I and genotype II isolates.
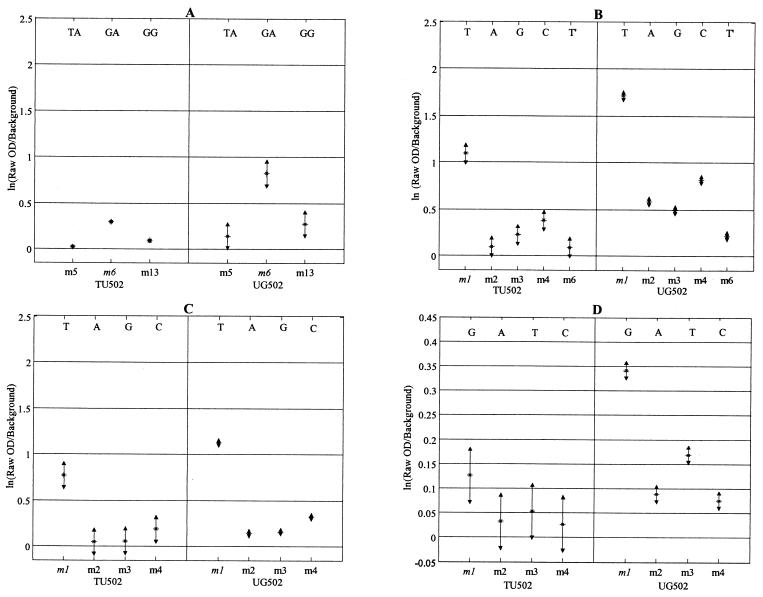
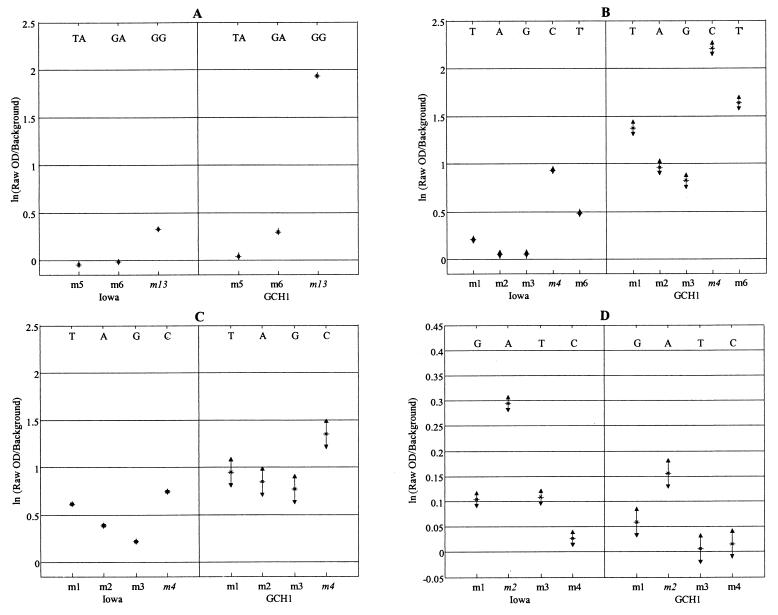
Figures 2 and 3 illustrate the statistical analysis of the single-nucleotide mismatch studies for base positions 1368, 1404, 1533, and 1542 for genotypes I and II, respectively, using the Cy3 labeling strategy. For base position 1368 probes, there are actually two polymorphisms within the capture probes: one at base position 1368 and the other at 1371 (AF 221535 numbering [19]). Thus, genotype I isolates may hybridize strongly to two possible probes: 1368 m1, with the published sequence for the human isolate (AF 221535 [19]), and 1368 m6, with the published sequence for the monkey isolate (AF 221534 [19]). Figure 2A and statistical analysis of all probes for position 1368 showed that when both isolates TU502 and UG502 (genotype I) were hybridized to the array, the 1368 m6 probe had the greatest mean hybridization signal of all position 1368 probes (Tukey's multiple comparison of means, α = 0.05). As predicted and illustrated in Fig. 3A, the 1368 m13 probe was diagnostic for genotype II isolates Iowa and GCH1. The 1368 m13 probe was quite robust for the genotype II isolates, in that the same hybridization result and statistical conclusion were achieved regardless of hybridization temperature, time, salt concentration, and labeling strategy (Table 2).
FIG. 2.
Detection of SNPs at selected diagnostic positions for genotype I isolates TU502 and UG502. The letters above the probe positions indicate the polymorphic nucleotides for each probe combination. Probes in italics are the perfectly matched probes for genotype I isolates (see Table 1 for the complete probe sequences). Results were based on a minimum of two replicates for each isolate. Error bars represent ±2 standard errors of the mean. All hybridizations were conducted by using the Cy3 labeling strategy and overnight hybridization at 4°C. (A) Positions 1368 and 1371. For clarity, only the probes with the three greatest values are shown. (B) Position 1404. T′ is a single-base-pair mismatch between the genotype II probe, m4, and the probe for C. felis. This polymorphism occurs at position 1398 (numbering based on GenBank accession number AF221535). (C) Position 1533. (D) Position 1542. OD, optical density.
FIG. 3.
Detection of SNPs at selected diagnostic positions for genotype II isolates Iowa and GCH1. The letters above the probe positions indicate the polymorphic nucleotides for each probe combination. Probes in italics are the perfectly matched probes for genotype II isolates (see Table 1 for the complete probe sequences). Results were based on a minimum of two replicates for each isolate. Error bars represent ±2 standard errors of the mean. All hybridizations were conducted by using the Cy3 labeling strategy and overnight hybridization at 4°C, except for panel D, where the hybridization temperature was room temperature (ca. 22°C). (A) Positions 1368 and 1371. For clarity, only the probes with the three greatest values are shown. (B) Position 1404. T′ is a single-base-pair mismatch between the perfectly matched probe, m4, and the probe for C. felis. This polymorphism occurs at position 1398 (numbering based on GenBank accession number AF221535). (C) Position 1533. (D) Position 1542. OD, optical density.
TABLE 2.
Results of Tukey's multiple-comparison test of the ability of the microarray to achieve single-nucleotide mismatch discrimination for all labeling and hybridization conditions evaluateda
| Isolate | Treatment
|
Result for diagnostic positionb:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Signaling strategy | Hybridization temp (°C) | 1368c | 1404 | 1419d | 1464 | 1479 | 1533 | 1542 | |
| Iowa | Biotin | 4 | + | − | − | − | − | − | − |
| RTe | + | − | − | − | − | − | − | ||
| Cy3 | 4 | + | + | − | + | + | + | − | |
| RT | + | + | − | + | + | + | + | ||
| GCH1 | Biotin | RT | + | − | − | + | + | − | − |
| Cy3 | 4 | + | + | + | + | + | + | − | |
| RT | + | − | − | − | − | + | + | ||
| TU502 | Cy3 | 4 | + | + | + | + | − | + | − |
| RT | − | + | + | − | − | − | − | ||
| UG502 | Cy3 | 4 | + | + | + | + | + | + | + |
| RT | − | + | + | − | − | − | − | ||
Tukey's multiple-comparison test was performed after the analysis-of-variance model determined that significant differences existed within a suite of probes for a given diagnostic position. The experiment-wide error rate (probability of observing an experiment with one or more pairwise comparisons falsely declared significant) was controlled at α = 0.05.
If the perfectly matched probe for the isolate tested had a significantly greater hybridization signal than all other probes within the suite, as determined by Tukey's multiple-comparison test, the SNP was achieved at the diagnostic position (+). If the mean hybridization signal for the perfectly matched probe was not significantly different than that for one or more mismatched probes within a suite for a diagnostic position, the SNP was not achieved (−).
For this analysis, the diagnostic positions 1368 and 1371 were grouped together.
For this analysis, the diagnostic positions 1419 and 1422 were grouped together.
RT, room temperature.
Likewise, statistical analysis of the microarray data demonstrated clear SNP discrimination between genotype I and genotype II isolates at diagnostic position 1404, where 1404 m1 is the predicted sequence for genotype I and 1404 m4 is specific for genotype II (Fig. 2B and 3B for genotypes I and II, respectively). The 1404 probe suite was interesting in that the m6 probe (Cryptosporidium felis) was identical to the m4 probe (diagnostic for genotype II), except for an SNP at the 3′ end (base position 1398, AF 221535 numbering [19]). Despite the occurrence of an SNP at the 3′ end of the capture probe, a situation that is undesirable for SNP analysis (7), the array reproducibly and statistically demonstrated that type II isolates preferentially hybridized to the 1404 m4 probe versus the 1404 m6 probe under our optimized hybridization protocol (Cy3 labeling strategy and overnight hybridization at 4°C) (Table 2). Unambiguous results were also obtained for genotype I isolates at the 1404 position.
For labeled PCRs hybridized to the 1533 probes (Fig. 2C and 3C for genotype I and genotype II isolates, respectively), single-nucleotide mismatch discrimination between genotype I and genotype II isolates was also possible. In terms of differences between mean signal intensities displayed in Fig. 2C and 3C, better mismatch discrimination was achieved for type I isolates than for type II isolates, in that the genotype II amplicons displayed higher levels of cross-reactivity with mismatched probes than did the genotype I amplicons.
Probe 1542 m1 has the predicted sequence for genotype I isolates. Mismatch discrimination for isolate TU502 was not achieved at the 1542 diagnostic position, because the labeled amplicons hybridized poorly to any of the 1542 probes (Fig. 2D). The failure of the method was due, in part, to poor hsp70 product yield during the PCR amplification. However, mismatch discrimination could be achieved for isolate UG502 with overnight hybridization at 4°C. As shown in Table 2 and Fig. 3D, mismatch discrimination for genotype II isolates for base position 1542 was dependent upon both the hybridization temperature and the labeling strategy. Figure 3D shows the results for the Iowa and GCH1 isolates after overnight hybridization at room temperature using Cy3-labeled hsp70 amplicons. At 4°C, the greatest mean hybridization signal was generated at the m2 probe, as predicted, but Tukey's multiple-comparison test of the analysis-of-variance model failed to show a significant difference between the mean hybridization signals of probes 1542 m1 to 1542 m4 (α = 0.05). When the analysis was repeated with hybridization at room temperature, however, not only did the correct (m2) probes show the greatest mean signal intensity, but Tukey's multiple-comparison test did show a significant difference.
Unlike genotype II isolates, where four specific probes comprise a unique SNP code, only one probe is absolutely unique for genotype I isolates (Table 3) (1419 m1, specific for both human and monkey sequence isolates AF 221535 and AF 221534 [19]). The capture probes for base position 1419, like those for position 1368, have two polymorphisms: one at position 1419 and one at position 1422. Probe 1419 m1 is the perfect match for the published sequences from human and monkey isolates, and probe 1419 m13 is the perfect match for genotype II isolates. As shown in Fig. 4, statistically significant mismatch discrimination was achieved for both the TU502 and UG502 genotype I isolates. Mismatch discrimination could not be reliably achieved for type II isolates, however (Table 2). The perfectly matched probe 1419 m13 and the 1419 m6 probe had statistically equivalent mean hybridization signals for most conditions tested. These two probes differ by a single base at position 1422 (Table 1); potential explanations for this result are described in detail below.
TABLE 3.
SNP signatures for C. parvum isolatesa
| Accession no. | Isolate | Nucleotide at sequence position (AF221535 numbering)b:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1368 | 1371 | 1404 | 1419 | 1422 | 1464 | 1479 | 1533 | 1542 | ||
| AF221535 | Genotype I human isolate | A | A | T | T | G | A | G | T | G |
| AF221534 | Genotype I monkey isolate | G | A | T | T | G | A | G | G | G |
| U71181 | Genotype II bovine isolate | G | G | C | C | A | A | G | C | A |
| AF221532 | C. parvum ferret isolate | G | A | T | C | A | A | G | T | G |
| AF221536 | C. wrairi | G | A | T | C | A | A | G | T | G |
| AF221537 | C. meleagridis | G | A | T | C | G | A | G | C | G |
| AF221538 | C. felis | T | G | C | C | G | T | T | C | G |
| AF221539 | C. baileyi | T | T | A | C | A | A | T | T | G |
FIG. 4.
Comparison of hybridization results for genotype I isolates TU502 and UG502 at position 1419. No hybridization signal was detected for 1419 m12 and m13, which are omitted from this graph for clarity. Probe 1419 m1 is the perfectly matched probe for genotype I isolates and the only position of relevance for the genotype I SNP code. Letters above the probes indicate the polymorphic nucleotides at positions 1419 and 1422, respectively. Probes m8 to m13 are outlier probes containing multiple polymorphisms at positions other than and in addition to 1419 and 1422. Results were based on a minimum of two replicates for each isolate. Error bars represent ±2 standard errors of the mean. Hybridizations were conducted by using the Cy3 labeling strategy and overnight hybridization at 4°C. Single-nucleotide mismatch could not be reliably achieved for genotype II isolates when the m13 probe was the perfectly matched probe for these isolates. OD, optical density.
DISCUSSION
Defining method specificity.
In the microbiological view of Cryptosporidium detection, method specificity is usually defined as the ability to distinguish between Cryptosporidium sp. DNA and non-Cryptosporidium sp. DNA. For SNP analyses on the microarray, however, specificity is defined as achieving correct single-nucleotide mismatch discrimination within a probe suite. Method specificity, in this sense, therefore applies to multiple levels of biological inquiry encompassing the analysis of Cryptosporidium species and isolates. We used a standard, stepwise approach to optimize hybridization conditions for SNP discrimination and utilized the concept of specific SNP codes to address the multiple levels of biological analyses, including identifying C. parvum versus non-C. parvum species and discriminating between C. parvum genotypes (e.g., genotype I versus genotype II).
Optimized microarray method.
While differences in hybridization patterns between genotypes could be visually ascertained, the statistical model was required to quantify and accurately identify SNPs. Even though Cy3 amplicons resulted in a lower overall fluorescent signal than the biotinylated amplicons, statistical analysis showed that SNPs could be more easily and definitively identified with a Cy3-labeled amplicon than with a biotinylated amplicon. There are several possible explanations for this observation. First, the signal development processes for the two labeling methods are different. The Cy3 microarray is imaged directly after hybridization, and each labeled PCR molecule has (in theory) one fluorescent molecule hybridized to one capture probe. In contrast, the signal development process for the biotin-labeled target involves signal amplification. In theory, the amount of chemiluminescent precipitate that is deposited on the hybridized probe is also directly proportional to the level of probe hybridization. However, signal amplification created excessive background precipitate in the microarray image that complicated spot intensity measurements, such that the signal intensity for the perfectly matched probes was often statistically the same as that for the mismatched probes.
Second, the imaging software and user interface may result in two fundamentally different images and data sets depending upon the reporter system, which may lead to false-negative (or false-positive) SNP identifications. After scanning the slide, the user creates a reference file that tells the image analysis software where the array probes are located. The user defines a circular spot diameter and square region of interest outside the defined spot. The image analysis software then analyzes the spot intensity within the defined circle and uses the region of interest to define the local background. Based on the spot intensity values and background values, the image analysis software makes a determination of whether a probe is hybridized or not. Thus, from the image analysis perspective, it was better to have the weaker signal intensities, “doughnut” images, and virtually minimal background observed with the Cy3 system than the strong spot intensities and high local background observed with the biotin labeling strategy. We have since discovered that Cy3 signal intensities can be increased by increasing the exposure time from 0.3 s/panel to 4 s/panel, which increases both the overall signal intensity of the hybridized probes and the local background but does not change the signal-to-noise ratio.
Third, SNP identification depends explicitly upon the microarray format and the statistical model itself. The model developed and utilized for this study implicitly assumes that the local background for each spot within a probe suite, within an array, and between arrays is fairly uniform. For arrays hybridized to Cy3 amplicons, this assumption was true. For arrays hybridized to biotinylated amplicons, this assumption was violated because the local background was highly variable between probe spots within a probe suite, within an array, and between arrays. We therefore developed additional statistical models in an attempt to achieve SNP discrimination with the biotin labeling and reporter strategy. These models included (i) using only the signal intensities of the hybridized probe spots, with and without log transformation, and (ii) imaging slides by using two fluorescent channels (Cy3 and Cy5 channels), where the hybridized probe spots should not be visible in the second channel. Neither of these models was sufficient to identify SNPs based on the biotinylated targets. Thus, resolving SNPs with a biotinylated target will require extensive optimization of the chemiluminescent reporter system and the development of new statistical models.
Statistically, the most consistent mismatch discrimination for all four isolates was achieved when the hsp70 amplicon was labeled with Cy3 and hybridized overnight at 4°C (Table 2). Using these hybridization conditions and labeling strategy, perfect mismatch discrimination could be achieved for all seven diagnostic SNPs for UG502, five of seven SNPs for TU502, six of seven SNPs for GCH1, and five of seven SNPs for the Iowa isolate. Successful hybridization and single-nucleotide mismatch discrimination at 4°C are counterintuitive conditions based on a traditional understanding of solution- or solid-phase hybridization kinetics (1). However, Drmanac et al. (5) provide an excellent theoretical discussion and practical demonstration of cold hybridizations and sequence-specific discrimination (as defined above) using hexa- or octamer probes. In particular, their theoretical analysis showed that the discriminatory ability of short-oligonucleotide hybridization either is temperature independent or decreases with increasing temperature (5). In practice, reproducible hybridization and detection validated the theoretical predictions. We believe that the oligonucleotide microarray is fundamentally similar to the short-oligonucleotide hybridization experiments described by Drmanac et al. (5) rather than to traditional blot hybridization techniques and that cold hybridization temperatures are the preferred baseline condition for optimizing short-oligonucleotide microarrays. Indeed, Guschin et al. (8) and Small et al. (18) also required cold hybridization conditions for the analysis of 16S rRNA sequences on gel element and planar oligonucleotide arrays, respectively.
While the microarray results with the Iowa isolate showed that overnight hybridization at room temperature slightly improved our ability to statistically identify SNPs (the SNP at position 1542 was achieved), this result could not be reproducibly achieved for all other isolates tested. Therefore, we instituted a standard method consisting of Cy3-labeled targets hybridized overnight at 4°C in 3× SSC-5× Denhardt's solution followed by 4°C rinses in 1× SSC. The statistical model and Tukey's multiple-comparison test were applied to all subsequent analyses, regardless of the overall signal intensity observed on replicate arrays.
Differentiating between principle genotypes, isolates, and species.
Because the hsp70 gene is highly conserved among all Cryptosporidium spp., an SNP can be used to differentiate between genotype I, genotype II, other genotypes, and other species in the genus. In this study, we designed and tested an oligonucleotide microarray that examined seven SNPs within the hsp70 gene. The seven SNPs result in a diagnostic signature for each species, as illustrated in Table 3. However, single-nucleotide mismatch discrimination at all seven diagnostic positions is not required in order to address important regulatory or epidemiological questions. For example, the unambiguous genotype II SNP code is based on only four diagnostic positions (1368 m13, 1404 m4, 1533 m4, and 1542 m2), whereas the unambiguous genotype I SNP code is a single SNP at position 1419. Therefore, the 68-probe microarray encompasses enough polymorphic positions within the hsp70 gene sequence that SNP codes can be defined and used to identify Cryptosporidium spp., specific C. parvum isolates, or specific genotypes, all with the same microarray. The ability of the microarray method to differentiate between the principle genotypes known to be pathogenic to humans (genotypes I and II), other C. parvum isolates, and other Cryptosporidium spp. is implied from probe sequence data from Table 1 and the predicted SNP signatures shown in Table 3 and is demonstrated by the SNP code results for the diagnostic base positions and four isolates discussed above.
Since a microarray is only as good as the initial sequences from which probes are designed, failure to achieve perfect mismatch discrimination at all loci in all cases may be partly due to erroneous sequence information in public databases. For example, the GenBank sequence for genotype I human isolates predicts that the strongest hybridization signal for the 1368 probe suite is m1 (19). However, probe 1368 m6, with the published sequence for the monkey isolate (AF 221534 [19]), consistently yielded the (significantly) highest signal for the two genotype I isolates tested in this study. Our results suggest that either (i) the GenBank entry contains an error at position 1368, (ii) the isolates we tested are polymorphic at position 1368 (A and G), or (iii) the SNP was not achieved. We believe that our data are best explained by a GenBank error for several reasons. First, we have tested a third genotype I isolate (UHPS) and found that the 1368 m6 probe is the most strongly hybridizing probe for all probes within the 1368 probe suite. Second, for genotype II isolates, only the 1368 m13 perfectly matched probe yields a detectable hybridization signal with the genotype II isolates tested in this study. Thus, the hybridization conditions employed in this study are sufficient to achieve single-nucleotide mismatch discrimination for this probe suite.
Ambiguous hybridization results were also observed for genotype II isolates and the 1419 probe suite. The perfectly matched probe in this case is 1419 m13. Tukey's multiple-comparison analysis showed that the SNP could not be consistently differentiated between 1419 m13 and 1419 m6; in fact, these two probes were consistently a statistical “tie.” Failure to achieve single-base mismatch discrimination at this position may therefore be due to a degeneracy in the genotype II hsp70 target at base position 1419.
Summary and conclusions.
Investigators who use PCR for rapid Cryptosporidium detection usually perform a hybridization experiment for verification and quality control (16). Not only do microarrays provide this type of verification of amplification products, but the multiplexing capabilities of a microarray provide confirmation at multiple diagnostic sites within the amplicon sequence. The hsp70 array used in this study was designed primarily to detect SNPs of C. parvum from human genotype I isolates, but the microarray method developed in this study was able to address several questions related to rapid detection. First, the method (PCR plus microarray) was shown to be specific to Cryptosporidium spp., a result that is of direct relevance for regulatory compliance and rapid detection. Second, the reproducibility of the optimized microarray method and results from four isolates indicate that the method can distinguish between the principle genotypes, other isolates, and other members of this genus, a result with implications for both regulatory compliance and epidemiology. Finally, the microarray unequivocally differentiated between the two principle genotypes of C. parvum known to infect humans, a result that is of direct relevance to epidemiologists. In practice, it becomes very important to recognize the subtle distinction between the expected microarray signature arising from all interrogated diagnostic positions (Table 3) and the specific SNP code that applies to the different levels of biological inquiry (e.g., genotype II SNP code). In this study, for example, we did not achieve perfect mismatch discrimination for all isolates at all diagnostic loci (as described above), but we did achieve perfect mismatch discrimination for the SNP codes corresponding to discrimination between genotype I and genotype II. We can continue to improve the detection and genotyping ability of the microarray by interrogating a longer region of the hsp70 gene or analyzing other conserved genes, resulting in even more robust SNP codes. Nevertheless, this work demonstrates important first steps in bridging the gap between rapid detection of C. parvum for regulatory purposes and strain typing for epidemiological investigations.
Acknowledgments
This research was supported by the U.S. Environmental Protection Agency's Science to Achieve Results (STAR) program. Pacific Northwest National Laboratory is operated for the U.S. Department of Energy by Battelle Memorial Institute under contract DE-AC06-76RLO 1830.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Call, D. R., D. P. Chandler, and F. Brockman. 2001. Fabrication of DNA microarrays using unmodified oligonucleotide probes. BioTechniques 30:368-379. [DOI] [PubMed] [Google Scholar]
- 3.Clark, D. P. 1999. New insights into human cryptosporidiosis. Clin. Microbiol. Rev. 12:554-563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng, M. Q., and D. O. Cliver. 1998. Differentiation of Cryptosporidium parvum isolates by a simplified randomly amplified polymorphic DNA technique. Appl. Environ. Microbiol. 64:1954-1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drmanac, R., Z. Strezoska, I. Labat, S. Drmanac, and R. Crkvenjakov. 1990. Reliable hybridization of oligonucleotides as short as six nucleotides. DNA Cell Biol. 7:527-534. [DOI] [PubMed] [Google Scholar]
- 6.Feng, X., S. M. Rich, D. Akiyoshi, J. K. Tumwine, A. Kekitiinwa, N. Nabukeera, S. Tzipori, and G. Widmer. 2000. Extensive polymorphism in Cryptosporidium parvum identified by multilocus microsatellite analysis. Appl. Environ. Microbiol. 66:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo, Z. G., A. Guilfoyle, A. J. Thiel, R. Wang, and L. M. Smith. 1994. Direct fluorescence analysis of genetic polymorphisms by hybridization with oligonucleotide arrays on glass supports. Nucleic Acids Res. 22:5456-5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guschin, D. Y., B. K. Mobarry, D. Proudnikov, D. A. Stahl, B. E. Rittmann, and A. D. Mirzabekov. 1997. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 63:2397-2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comp. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 10.Lockhart, D. J., H. Dong, M. C. Byrne, M. T. Follettie, M. V. Gallo, M. S. Chee, M. Mittmann, C. Wang, M. Kobayashi, H. Horton, and E. L. Brown. 1996. Expression monitoring by hybridization to high density oligonucleotide arrays. Nat. Biotechnol. 14:1675-1680. [DOI] [PubMed] [Google Scholar]
- 11.Morgan, U. M., C. C. Constantine, P. O'Donoghue, B. P. Meloni, P. A O'Brien, and R. C. Thompson. 1995. Molecular characterization of Cryptosporidium isolates from humans and other animals using random amplified polymorphic DNA analysis. Am. J. Trop. Med. Hyg. 52:559-564. [DOI] [PubMed] [Google Scholar]
- 12.Morgan, U. M., L. Xiao, R. Fayer, A. A. Lal, and R. C. A. Thompson. 1999. Variation within Cryptosporidium: towards a taxonomic revision of the genus. Int. J. Parasitol. 29:1733-1751. [DOI] [PubMed] [Google Scholar]
- 13.Pease, A. C., D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes, and S. P. A. Fodor. 1994. Light-generated oligonucleotide arrays for DNA sequence analysis. Proc. Natl. Acad. Sci. USA 91:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pedraza-Diaz, S., C. Amar, G. L. Nichols, and J. McLauchlin. 2001. Nested polymerase chain reaction for amplification of the Cryptosporidium oocyst wall protein gene. Emerg. Infect. Dis. 7:49-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peng, M. M., L. Xiao, A. R. Freeman, M. J. Arrowood, A. A. Escalante, and A. C. Weltman. 1997. Genetic polymorphisms among Cryptosporidium parvum isolates: evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 3:567-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rochelle, P. A., R. DeLeon, M. H. Stewart, and R. L. Wolfe. 1997. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl. Environ. Microbiol. 63:106-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shianna, K. V., R. Rytter, and J. G. Spanier. 1998. Randomly amplified polymorphic DNA PCR analysis of bovine Cryptosporidium parvum strains isolated from the watershed of the Red River of the North. Appl. Environ. Microbiol. 64:2262-2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Small, J., D. R. Call, F. J. Brockman, T. M. Straub, and D. P. Chandler. 2001. Direct detection of 16S rRNA in soil extracts by using oligonucleotide microarrays. Appl. Environ. Microbiol. 67:4708-4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sulaiman, I. M., U. M. Morgan, R. C. Thompson, A. A. Lal, and L. Xiao. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70-kilodalton heat shock protein (hsp70) gene. Appl. Environ. Microbiol. 66:2385-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.U.S. Environmental Protection Agency. 2001. Method 1623: Cryptosporidium and Giardia in water by filtration/IMS/FA. EPA-821-R-01-025. U.S. Environmental Protection Agency, Washington, D.C.
- 21.Widmer, G., D. Akiyoshi, M. A. Buckholt, X. Feng, S. M. Rich, K. M. Deary, C. A. Bowman, P. Xu, Y. Wang, X. Wang, G. A. Buck, and S. Tzipori. 2000. Animal propagation and genomic survey of a genotype I isolate of Cryptosporidium parvum. Mol. Biochem. Parasitol. 108:187-197. [DOI] [PubMed] [Google Scholar]
- 22.Xiao, L., L. Escalante, C. Yang, I. Sulaiman, A. A. Escalante, R. J. Montali, R. Fayer, and A. A. Lal. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 65:1578-1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiao, L., J. Limor, U. M. Morgan, I. M. Sulaiman, R. C. A. Thompson, and A. A. Lal. 2000. Sequence differences in the diagnostic target region of the oocyst wall protein gene of Cryptosporidium parasites. Appl. Environ. Microbiol. 66:5499-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yershov, G., V. Barsky, A. Belgovskiy, E. Kirillov, E. Kreindlin, I. Ivanov, S. Parinov, D. Guschin, A. Drobishev, S. Dubiley, and A. Mirzabekov. 1996. DNA analysis and diagnostics on oligonucleotide microchips. Proc. Natl. Acad. Sci. USA 93:4913-4918. [DOI] [PMC free article] [PubMed] [Google Scholar]