Abstract
To develop a better understanding of respiration by sulfate-reducing bacteria, we examined transcriptional control of respiratory genes during growth with lactate or hydrogen as an electron donor. RNA extracts of Desulfovibrio desulfuricans subsp. aestuarii were analyzed by using random arbitrarily primed PCR. RNA was reverse transcribed under low-stringency conditions with a set of random primers, and candidate cDNAs were cloned, sequenced, and characterized by BLAST analysis. Putative differentially expressed transcripts were confirmed by Northern blot analysis. Interestingly, dissimilatory bisulfite reductase was upregulated in the presence of hydrogen. To link these transcriptional changes to the physiology of sulfate-reducing bacteria, sulfide was measured during growth of several strains of Desulfovibrio on hydrogen or lactate, and this revealed that hydrogen-grown cells produced more sulfide per unit of cell mass than lactate-grown cells. Transcription of other redox proteins was characterized by Northern blotting to determine whether or not they were also transcribed to higher levels in hydrogen-grown cells. Growth on lactate produced greater transcription of [NiFe] hydrogenase. H2-grown cells transcribed the adenylylsulfate reductase b subunit and HmcA to higher levels. The results we describe here provide new insight into the continuing debate over how Desulfovibrio species utilize redox components to generate membrane potential and to channel electrons to sulfate, the final electron acceptor.
Sulfate-reducing bacteria (SRB) are a diverse group of microorganisms found in a variety of anaerobic environments, and all members possess the ability to use sulfate as a terminal electron acceptor. SRB can use a variety of organic electron donors, including hydrogen, lactate, formate, malate, fumarate, pyruvate, alcohols, and even environmental contaminants (13). While progress has been made in understanding the biochemistry of proteins involved in respiratory processes, to date little is known about how such proteins are utilized by the SRB to gain energy for cell growth.
For years, a debate over the mechanism by which lactate is oxidized in SRB has existed. In 1981, Odom and Peck proposed the hydrogen-cycling model for growth on lactate (12). In this model, electrons from lactate are used by a cytoplasmic hydrogenase to generate hydrogen that can diffuse out across the cell membrane to be utilized by periplasmic dehydrogenases. Membrane potential is generated as protons remain in the periplasm while electrons are transferred across the cell membrane to reduce sulfate. When only hydrogen is utilized as an electron donor, it is likely oxidized in the periplasm by hydrogenases, but it may use different electron carriers in the reduction of sulfate than electrons generated during lactate oxidation.
We know that lactate dehydrogenase, pyruvate-ferredoxin oxidoreductase, phosphotransacetylase, and acetate kinase convert lactate to acetate and that ATP sulfurylase, pyrophosphatase, adenosine-5′-phosphosulfate (APS) reductase, and bisulfite reductase are responsible for linking electrons produced with sulfate reduction (13). Unfortunately, the identity of electron carriers involved in lactate and hydrogen metabolism remains elusive and has prompted a few biochemical and molecular studies in recent years in hopes of better defining this process. Voordouw et al. (18) examined the distribution of cytoplasmic and periplasmic hydrogenases for 22 Desulfovibrio species and determined that only the genes for periplasmic [NiFe] hydrogenase were present in all species surveyed. This finding challenged the hydrogen-cycling model, which requires SRB to possess a cytoplasmic hydrogenase (possibly an [NiFeSe] hydrogenase) in addition to a periplasmic hydrogenase. Discovery of the hmc operon in Desulfovibrio vulgaris Hildenborough offered a solution as to how electrons in the periplasm could reach the cytoplasmic sulfate reduction enzymes (14, 15). By using antibodies to HmcA and HmcF, expression of the hmc operon in D. vulgaris was found to be highest during growth on hydrogen (8). Deletion of the hmc operon in D. vulgaris (Hildenborough) impaired growth on hydrogen but not that on lactate or pyruvate, confirming the importance of the Hmc complex in electron transport from hydrogen in the periplasm to sulfate in the cytoplasm (4).
This study was originally intended to evaluate the applicability of random arbitrarily primed PCR (RAP-PCR) to environmental bacteria that reduce sulfate, but initial findings led us to probe further into differential expression of SRB redox proteins. We used a combination of RAP-PCR and Northern blotting to identify genes that were differentially transcribed under conditions of growth with either hydrogen or lactate as an electron donor. After the observation that bisulfite reductase was transcribed to higher levels in hydrogen-grown cells than in lactate-grown cells, differential transcription of other known redox proteins, including [NiFe] hydrogenase and HmcA, was characterized by Northern blotting.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Desulfovibrio desulfuricans Essex 6 (freshwater strain) and D. desulfuricans subsp. aestuarii (marine strain) were from the culture collection of Michael McInerney. Desulfovibrio sp. strain ASR was obtained from Bradley Tebo at Scripps Institute. Cells were grown in mineral medium containing 10 mM Na2SO4, 0.2% yeast extract, 0.0001% resazurin as a redox indicator, and vitamins, minerals, and metals solutions prepared as described by Krumholz et al. (9). D. desulfuricans subsp. aestuarii was used in RAP-PCR experiments, while Essex 6 and ASR were used in a sulfide production assay. For the marine strains, 342 mM NaCl and 14.76 mM MgCl2 were added to the mineral medium. Media were prepared using the techniques of Hungate (6) as modified by Balch et al. (1). The headspace was N2-CO2 (4:1). After boiling and cooling, 42 mM sodium bicarbonate and 1 mM sodium sulfide were added from sterile stock solutions. For comparison of hydrogen versus lactate as an electron donor, 25 mM lactate was added to the medium before autoclaving or 10 ml of H2 per 10 ml of medium was added to the headspace after autoclaving. Growth curves were determined at 30 and 37°C, with measurements of optical density at 600 nm taken periodically with a spectrophotometer. Sulfide was quantitated with the dimethyl-phenylene diamine assay (3).
RNA extraction.
Total RNA was isolated as described by Shepard and Gilmore (17), with minor modifications to the procedure. Mid-log-phase cells for D. desulfuricans strain Essex 6 and late log cells for D. desulfuricans subsp. aestuarii were harvested by centrifugation for 5 min at 7,000 × g. The pellet was resuspended in 1 ml of Tri Reagent (Sigma) and transferred to sterile tubes containing 0.5 ml of 100-μm-diameter zirconia-silicon beads. Cells were broken in a Mini-Beadbeater (Biospec Products) for 1 min. The supernatant was extracted with 300 μl of chloroform and placed on ice for 15 min. After centrifugation for 10 min (12,000 × g), nucleic acids in the aqueous phase were precipitated with 750 μl of isopropyl alcohol and placed on ice for 10 min. Following centrifugation and a 75% ethanol wash, pellets were resuspended in 200 μl of diethyl pyrocarbonate (DEPC)-treated water. Contaminating genomic DNA was removed by addition of 22 μl of Multi Core Restriction Enzyme Buffer (Promega) and 5 U of RNase-free DNase (Stratagene) with incubation at 37°C for 15 min. Following this treatment, 500 μl of phenol-chloroform-isoamyl alcohol was added and centrifuged (12,000 × g, 10 min). The aqueous phase was removed, and the phenolic phase was extracted a second time with 250 μl of DEPC-treated water. After centrifugation, this second aqueous phase was mixed with the first, and RNA was precipitated with 1 ml of 100% ethanol for at least 30 min at −80°C. RNA was pelleted by centrifugation (12,000 × g, 30 min), washed with 75% ethanol, resuspended in 0.1 mM EDTA, and stored at −80°C. Integrity of RNA was determined by 0.8% agarose electrophoresis in Tris-borate-EDTA buffer, and the concentration was determined by measuring the A260/A280 ratio spectrophotometrically (16).
RAP-PCR.
RAP-PCR was performed as described by Shepard and Gilmore (17). For each reaction, 14.5 μl containing 1 μg of total RNA, diluted in DEPC-treated water when necessary, was heated to 70°C for 10 min and placed on ice. After a 1-min incubation on ice, 2 μl of reverse transcription buffer (Fisher), 40 U of RNase Block RNase inhibitor (Stratagene), a 1.25 mM concentration of each deoxynucleoside triphosphate, and a 1.25 μM concentration of arbitrary primer (Stratagene) were added. After the contents were mixed, the reaction mixture was held at 37°C for 5 min. After equilibration, 25 U of Moloney murine leukemia virus reverse transcriptase (RT) (Stratagene) was added. First-strand cDNA synthesis occurred at 37°C for 1 h, and then the reaction mixture was heated to 90°C for 5 min to inactivate the RT and placed on ice for 10 min. For second-strand synthesis, 10 μl of a 1:10 dilution of first-strand cDNA product was mixed with 39.8 μl of standard PCR mix containing Taq buffer without MgCl2 (Sigma), 3 mM MgCl2, a 50 μM concentration of each deoxynucleoside triphosphate, 10 μCi of [α-33P]dCTP, and a 1 μM concentration of the same arbitrary primer used in first-strand synthesis for a final reaction volume of 50 μl. The reaction mixture was heated to 96°C for 10 min following overlay with 50 μl of light mineral oil. After incubation at 36°C for 15 min, 1 U of Taq polymerase (Sigma) was added to each reaction mixture. The reaction continued to equilibrate for another 15 min at 36°C, followed by incubation at 72°C for 5 min. For the remaining 39 cycles of PCR, the following parameters were used: 94°C (1 min), 50°C (1 min), 72°C (2 min), and a final extension at 72°C for 10 min. The reaction mixture was stored at 4°C. Products were resolved on a 6% polyacrylamide gel (Life Technologies) prepared in Tris-borate-EDTA buffer and run at 1,500 V until the xylene cyanol dye migrated to the bottom of the gel. The gel was transferred to 3MW paper (Midwest Scientific, Valley Park, Mo.) and dried under vacuum at 70°C for 45 min. The gel was exposed to Kodak BioMax MR film for 12 h at room temperature. Putative differentially transcribed bands were excised from the gel, eluted from the filter paper with elution buffer (0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, 0.1% sodium dodecyl sulfate), precipitated with 100% ethanol, and resuspended in 10 μl of sterile water. The cDNA was then reamplified with the PCR parameters used in second-strand synthesis and resolved on a 6% polyacrylamide gel with the original RAP-PCR product for comparison to ensure that the correct band was isolated. The candidate PCR product was ligated into pCR4-TOPO vector and transformed into chemically competent One Shot TOP10 Escherichia coli (Invitrogen). Plasmid was isolated from 2 ml of liquid cultures of candidate clones grown in Luria broth plus ampicillin (50 μg/ml) by using a miniprep plasmid isolation kit (Qiagen) according to the manufacturer's directions. DNA sequencing was carried out at the Oklahoma Medical Research Foundation Core Sequencing Facility. Typically three clones were sequenced per transformation. Candidate inserts ranged in size from 328 to 1,026 bases. GenBank sequence comparison was performed by both nucleotide and protein BLAST searches (19).
Confirmation of differential gene transcription.
RNA probes were prepared from plasmids isolated from RAP-PCR clones. Plasmids were first digested with NotI or PstI to linearize DNA prior to transcription. Linearized DNA (0.2 μg) in RNA polymerase buffer (Ambion); 4 mM (each) ATP, CTP, and GTP; and [α-32P]UTP was incubated with either T3 DNA-dependent RNA polymerase or T7 DNA-dependent RNA polymerase (depending on orientation) at 37°C for 60 min. RNase-free DNase (1 U) was added and left for an additional 30 min to remove plasmid and template DNAs. The mixture was diluted 20-fold, and unincorporated radiolabeled nucleotide was separated from the probe with a Sephadex G-50 column. At least 106 cpm of labeled RNA probe was added to each hybridization mixture. For the RNA blots, 20 μg of total RNA from both growth conditions was loaded onto a 1.5% agarose gel containing 15% formaldehyde and 1× MOPS [3-(N-morpholino)propanesulfonic acid]. RNA was electrophoresed for 2 h at 150 V with 0.5× MOPS as the running buffer. The gel was transferred overnight with a Turboblotter to Nytran SuPerCharge nylon membranes according to the directions of the manufacturer (Schleicher and Schuell). The membrane was cross-linked to RNA with UV at 125 mJ/cm2 for 1 min. The membrane was then exposed to a standard prehybridization buffer for 6 h at 65°C and then to hybridization buffer and probe for 12 h at 65°C. Washings were performed with 1× SSPE (0.18 M NaCl, 10 mM NaH2PO4, and 1 mM EDTA [pH 7.7]) with 0.1% sodium dodecyl sulfate at room temperature (three times for 5 min each) and at 65°C (three times for 30 min each). Blots were imaged with a Molecular Dynamics Storm PhosphorImager and a Packard Instant Imager.
RESULTS
RNA extraction and RAP-PCR.
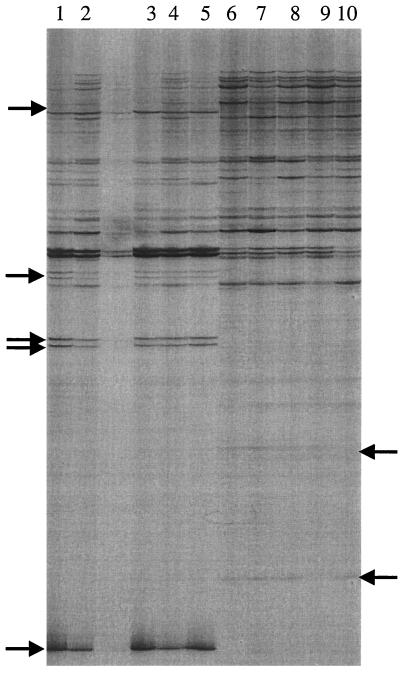
The RNA extraction yielded intact RNA which appeared as three bands representing 23S, 16S, and 5S rRNAs following electrophoresis (data not shown). Random primers A3 and A1 (Stratagene) created several RAP-PCR products that were unique to conditions of growth with either hydrogen or lactate (Fig. 1). As expected, a majority of bands were shared by both growth conditions. Products which were unique to either condition occurred reproducibly from at least five independent cultures grown with hydrogen and five independent cultures grown with lactate. Products also did not match bands derived from contaminating genomic DNA in the no-RT control reaction (data not shown). When products were first isolated, reamplified, and run on a gel again to verify that the correct band had been removed, several bands of various sizes appeared in each lane, suggesting that nearby contaminating bands had been excised in addition to the band of interest. The correct band, determined by comparison to the original RAP-PCR product mix run in an adjacent lane, was once again isolated from this second gel and used in transformation of competent E. coli cells.
FIG. 1.
DNA products derived from RAP-PCR of total RNA from five independent cultures of D. desulfuricans subsp. aestuarii grown with either hydrogen (lanes 1 to 5) or lactate (lanes 6 to 10) as the electron donor. Arrows indicate cDNA products of differentially transcribed mRNA. A paired control reaction without RT in the first-strand synthesis reaction indicated that no differentially transcribed products were derived from contaminating genomic DNA.
Sequence identification and confirmation of RAP-PCR bands.
Table 1 lists results where cDNA showed sequence similarity in GenBank by nucleotide and protein BLAST searches. Cloned insert sizes ranged from 328 to 1,026 bp. Putative differentially transcribed genes had significant similarity to genes for dissimilatory bisulfite reductase, F1F0 ATP synthase, and ATP sulfurylase from D. vulgaris in hydrogen-grown cells and to genes for Bacillus subtilis folate biosynthesis protein, Clostridium perfringens molybdopterin biosynthesis protein, and Campylobacter jejuni malate dehydrogenase in lactate-grown cells.
TABLE 1.
DNA sequence homology of differentially transcribed bands obtained by the RAP-PCR procedure
| Electron donor | Band size (bp) | Similar gene sequence (accession no.) | Lowest-sum probability score | Type of BLAST search | Confirmed by Northern blotting | % Increase in transcriptiona |
|---|---|---|---|---|---|---|
| Hydrogen | 1,026 | D. vulgaris F1F0 ATPase subunit b (AB022018) | 8 × 10−24 | BLASTX | No | |
| Lactate | 606 | B. subtilis folate biosynthesis protein (F37854) | 1 × 10−23 | BLASTX | Yes | 104 |
| Lactate | 986 | C. jejuni malate dehydrogenase (H81336) | 2 × 10−57 | BLASTX | Yes | 634 |
| Hydrogen | 328 | D. desulfuricans subsp. aestuarii dissimilatory bisulfite reductase (AJ289157) | 1 × 10−168 | BLASTN | Yes | 203 |
| Hydrogen | 507 | Entamoeba histolytica ATP sulfurylase (AB013399) | 1 × 10−30 | BLASTX | No | |
| Lactate | 507 | C. perfringens molybdopterin biosynthesis protein (BAA76927) | 8 × 10−24 | BLASTX | Yes | 451 |
The observed differences between growth conditions with lactate and hydrogen were found to be statistically significant at the 5% level using Student's t test.
Growth experiments.
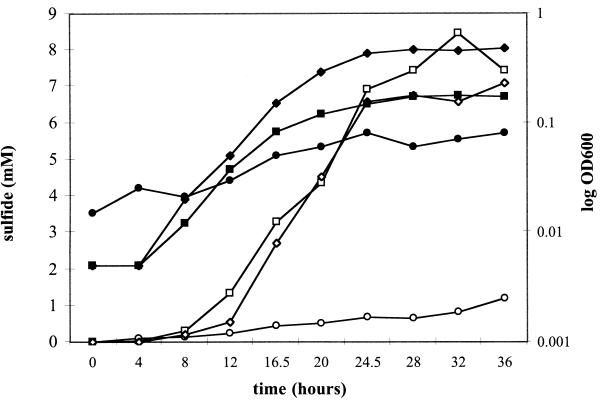
To provide further support that the bisulfite reductase gene and perhaps other respiratory protein genes were more highly transcribed during growth on hydrogen than on lactate, sulfide assays were performed over the growth curves of several D. desulfuricans strains. Figure 2 shows that hydrogen-grown D. desulfuricans subsp. aestuarii produced more sulfide relative to cell concentration (determined by optical density measurements) than lactate-grown cells, suggesting that transcription of the bisulfite reductase gene could have been increased, in turn increasing expression of the bisulfite reductase protein. Similar results were observed when sulfide production was monitored over a growth curve for D. desulfuricans strain Essex 6 and Desulfovibrio sp. strain ASR (data not shown). Through these growth curves, yeast extract (0.2%) included in the medium to support growth appeared to serve as an electron donor to a small extent, resulting in the production of some sulfide in the absence of either H2 or lactate (Fig. 2).
FIG. 2.
Sulfide production in D. desulfuricans subsp. aestuarii throughout the growth curve in mineral medium with 10 mM sulfate added. Growth was monitored spectrophotometrically (optical density at 600 nm [OD600]) with H2 (▪), lactate (⧫), or neither (•) as the electron donorl. Sulfide production was monitored by dimethyl-phenylene diamine assay throughout growth on hydrogen (□), lactate (◊), or neither electron donor (○). Yeast extract (0.2%) was included in all media.
Northern blotting.
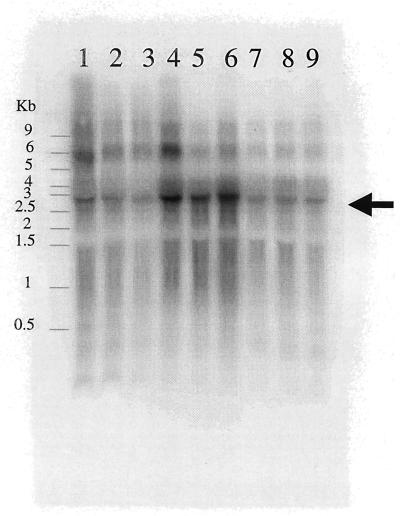
In order to confirm differential transcription of genes identified during RAP-PCR, Northern blotting was performed. Total RNA extracts from D. desulfuricans subsp. aestuarii were incubated with a 32P-labeled RNA probe constructed from the cDNA clone resulting from RAP-PCR. Figure 3 shows the Northern blot for the RAP-PCR band with sequence similarity to dissimilatory bisulfite reductase gene. The results demonstrate that cells grown with hydrogen as the electron donor (lanes 4 to 6) produced higher levels of mRNA for this gene than cells grown without hydrogen. Northern blots also confirmed that malate dehydrogenase, molybdopterin biosynthesis protein, and folate biosynthesis protein (2-amino-4-hydroxy-6-hydroxymethyldihydropteridine pyrophosphokinase) were more highly transcribed under growth conditions with lactate (data not shown). We were not able to confirm differential transcription of F1F0 ATPase and ATP sulfurylase by Northern blotting.
FIG. 3.
Northern blot confirmation of differential transcription of dissimilatory bisulfite reductase mRNA. Each lane represents 20 μg of total RNA extracted from three independent cultures of D. desulfuricans subsp. aestuarii grown with neither electron donor (lanes 1 to 3), with hydrogen (lanes 4 to 6), or with lactate (lanes 7 to 10). The blot was incubated with a 32P-labeled RNA probe constructed from a cDNA clone resulting from RAP-PCR and corresponding to the gene sequence for dissimilatory bisulfite reductase.
Differential expression of D. desulfuricans strain Essex 6 redox proteins.
Differential transcription of bisulfite reductase and growth results showing increased sulfate reduction activity during growth on H2 led us to examine whether or not mRNAs encoding other proteins involved in redox reactions were also differentially transcribed. Northern blotting was carried out with total RNA isolated from cells grown with either hydrogen or lactate. Probes were synthesized from known gene sequences to strain Essex 6 redox proteins shown in Table 2. Adenylylsulfate reductase b subunit and HmcA mRNAs were more highly transcribed in hydrogen-grown cells, while mRNA for the small subunit of [NiFe] hydrogenase was more highly transcribed in lactate-grown cells. Flavodoxin was not differentially transcribed under the conditions tested.
TABLE 2.
Differential transcription of D. desulfuricans strain Essex 6 redox proteins with either hydrogen or lactate serving as an electron donor
| Redox protein (accession no.) | Probe size (bp) | Growth condition of higher transcription | % Increasea |
|---|---|---|---|
| Flavodoxin (X59438) | 430 | None | |
| HmcA (AF127653) | 820 | Hydrogen | 84 |
| Ni-Fe hydrogenase small subunit (AF216303) | 680 | Lactate | 132 |
| Adenylylsulfate reductase b subunit (AF226708) | 480 | Hydrogen | 55 |
The observed differences between growth conditions with lactate and hydrogen were found to be statistically significant at the 5% level using Student's t test.
DISCUSSION
This study was originally intended to evaluate the applicability of RAP-PCR to environmental bacteria that reduce sulfate. The conditions tested were chosen because we were interested in examining mRNA involved in growth on different electron donors. In the past, biochemical and genetic studies with mutants have provided some insight into how Desulfovibrio species use these as electron donors. Our study shows that several mRNA species involved in either redox reactions or carbon metabolism are differentially transcribed under these conditions. Sequences for malate dehydrogenase have strong similarity to those for lactate dehydrogenase (2, 5, 10). Since the sequence for the SRB lactate dehydrogenase is not available in GenBank, the RAP-PCR band that was larger in lactate-grown cells could be from mRNA encoding lactate dehydrogenase, rather than a malate dehydrogenase. The other proteins more highly transcribed during lactate metabolism may come as a result of using lactate as a carbon source as opposed to the CO2 or yeast extract components utilized by hydrogen-grown cells. Molybdopterin is a component of molybdenum cofactors that participate in redox reactions (11), and its increased transcription in lactate-grown cells may come as a result of increased expression of the redox protein for which it serves as a cofactor. Known molybdopterin-containing enzymes in SRB include aldehyde oxidoreductases, formate dehydrogenase, and nitrate reductase (11). Less-well-characterized molybdenum proteins have been isolated but not classified (11). The genes for these proteins may or may not represent the gene for the redox protein more highly transcribed in lactate-grown D. desulfuricans subsp. aestuarii. In the yeast Pichia canadensis, molybdopterin biosynthesis was found to branch from the folic acid biosynthetic pathway at dihydrohydroxymethylpterin (7). As molybdopterin biosynthesis likely occurs in a similar manner in D. desulfuricans subsp. aestuarii, increased transcription of genes for folic acid and molybdopterin biosynthesis proteins during growth on lactate may be linked.
Increased transcription of the F1F0 ATP synthase and ATP sulfurylase during growth with H2 could not be confirmed by Northern blot analysis. However, we did confirm that dissimilatory bisulfite reductase and adenylylsulfate reductase are transcribed to higher levels in hydrogen-grown cells. It is likely that ATP sulfurylase, one of the four cytoplasmic enzymes involved in sulfate reduction by an eight-electron transfer, and F1F0 ATPase were differentially transcribed but that the Northern blotting was not effective either because of mRNA instability or because of lack of sensitivity. Transcripts with slight levels of differential transcription that RAP-PCR can detect but Northern blots cannot may require more sensitive techniques, such as real-time PCR, for confirmation.
As a follow-up to our findings with dissimilatory bisulfite reductase, we tested whether other redox proteins would be differentially transcribed under conditions of growth with hydrogen. Sequence information for genes encoding several redox proteins was available for D. desulfuricans strain Essex 6. Specific primer pairs were designed for flavodoxin, HmcA, [NiFe] hydrogenase small subunit, and adenylylsulfate reductase b subunit. We were unable to amplify these sequences from D. desulfuricans subsp. aestuarii genomic DNA but were able to amplify and construct probes from Essex 6 DNA. Because the same phenomenon of increased sulfide production during growth on H2 versus lactate occurred in Essex 6 as in the marine strain, other redox proteins can be expected to behave in the same manner in D. desulfuricans Essex 6 as in D. desulfuricans subsp. aestuarii. Northern blots with D. desulfuricans Essex 6 RNA demonstrated that not all putative electron transport-related proteins were transcribed to higher levels when hydrogen was used as an electron donor (Table 2). These results may indicate that certain redox proteins are not involved in hydrogen- or lactate-driven electron transport or that these proteins may not be carrying out rate-limiting reactions. Higher transcription of periplasmic [NiFe] hydrogenase in lactate-grown cells could suggest that this protein plays an important role in metabolism of H2 or in the production of H2 by D. desulfuricans Essex 6 from lactate, lending support to the hydrogen-cycling model. HmcA expression to higher levels during growth on H2 was also demonstrated by Keon et al. (8) through Western blotting.
This study helps in revealing the extent to which different redox proteins play a role in either hydrogen or lactate metabolism. Coupled with previous genetic and biochemical studies, these results may help us come to understand the different pathways employed in the metabolism of lactate or hydrogen in these organisms. From the success of this study, we also feel that RAP-PCR can be applied to other questions of environmental significance, such as the identification of genes that are upregulated in the presence of environmental contaminants.
Acknowledgments
This research was funded by the Natural and Accelerated Bioremediation Research (NABIR) program, Biological and Environmental Research (BER), U.S. Department of Energy (US-DOE DE-FG03-99ER62866); the Department of Defense and the Office of Naval Research through the DOD Epscor program; and the Oklahoma Regents for Higher Education.
We thank Chris Bausch and Darren Smalley for providing help with Northern blotting. We also thank Brett Shepard and Michael Gilmore for help with RAP-PCR technique.
REFERENCES
- 1.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charnock, C. 1997. Structural studies of malate dehydrogenases (MDHs): MDHs in Brevundimonas species are the first reported MDHs in Proteobacteria which resemble lactate dehydrogenases in primary structure. J. Bacteriol. 179:4066-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cline, J. D. 1969. Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol. Oceanogr. 14:454-458. [Google Scholar]
- 4.Dolla, A., B. K. J. Pohorelic, J. K. Voordouw, and G. Voordouw. 2000. Deletion of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough hampers hydrogen metabolism and low-redox-potential niche establishment. Arch. Microbiol 174:143-151. [DOI] [PubMed] [Google Scholar]
- 5.Goward, C. R., and D. J. Nicholls. 1994. Malate dehydrogenase: a model for structure, evolution, and catalysis. Protein Sci. 3:1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hungate, R. E. 1969. A roll tube method for the cultivation of strict anaerobes. Methods Microbiol. 3B:117-132. [Google Scholar]
- 7.Irby, R. B., and W. L. Adair, Jr. 1994. Intermediates in the folic acid biosynthetic pathway are incorporated into molybdopterin in the yeast Pichia canadensis. J. Biol. Chem. 269:23981-23987. [PubMed] [Google Scholar]
- 8.Keon, R. G., F. Rongdian, and G. Voordouw. 1997. Deletion of two downstream genes alters expression of the hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough. Arch. Microbiol. 167:376-383. [DOI] [PubMed] [Google Scholar]
- 9.Krumholz, L. R., and M. P. Bryant. 1986. Eubacterium oxidoreducens sp. nov. requiring H2 or formate to degrade gallate, pyrogallol, phloroglucinol and quercetin. Arch. Microbiol. 144:8-14. [Google Scholar]
- 10.Madern, D. 2000. The putative l-lactate dehydrogenase from Methanococcus jannaschii is an NADPH-dependent l-malate dehydrogenase. Mol. Microbiol. 37:1515-1520. [DOI] [PubMed] [Google Scholar]
- 11.Moura, I., A. S. Pereira, P. Tavares, and J. J. G. Moura. 1999. Simple and complex iron-sulphur proteins in sulfate-reducing bacteria. Adv. Inorg. Chem. 47:361-419. [Google Scholar]
- 12.Odom, J. M., and H. D. Peck, Jr. 1981. Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria Desulfovibrio sp. FEMS Microbiol. Lett. 12:47-50. [Google Scholar]
- 13.Odom, J. M., and R. Singelton, Jr. 1993. The sulfate-reducing bacteria: contemporary perspectives. Springer-Verlag, New York, N.Y.
- 14.Pollock, W. B. R., M. Loutfi, M. Bruschi, B. J. Rapp-Giles, J. D. Wall, and G. Voordouw. 1991. Cloning, sequencing, and expression of the gene encoding the high-molecular-weight cytochrome c from Desulfovibrio vulgaris Hildenborough. J. Bacteriol. 173:220-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossi, M., W. B. R. Pollock, M. W. Reij, R. G. Keon, R. Fu, and G. Voordouw. 1993. The hmc operon of Desulfovibrio vulgaris subsp. vulgaris Hildenborough encodes a potential transmembrane redox protein complex. J. Bacteriol. 175:4699-4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Shepard, B., and M. Gilmore. 1999. Identification of aerobically and anaerobically induced genes in Enterococcus faecalis by random arbitrarily primed PCR. Appl. Environ. Microbiol. 65:1470-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voordouw, G., V. Niviere, F. G. Ferris, P. M. Fedorak, and D. W. S. Westlake. 1990. Distribution of hydrogenase genes in Desulfovibrio spp. and their use in identification of species from the oil field environment. Appl. Environ, Microbiol. 56:3748-3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren, G., and D. J. States. 1993. Identification of protein coding regions by database similarity search. Nat. Genet. 3:266-272. [DOI] [PubMed] [Google Scholar]