Abstract
A range of gram-negative bacterial species use N-acyl homoserine lactone (AHL) molecules as quorum-sensing signals to regulate different biological functions, including production of virulence factors. AHL is also known as an autoinducer. An autoinducer inactivation gene, aiiA, coding for an AHL lactonase, was cloned from a bacterial isolate, Bacillus sp. strain 240B1. Here we report identification of more than 20 bacterial isolates capable of enzymatic inactivation of AHLs from different sources. Eight isolates showing strong AHL-inactivating enzyme activity were selected for a preliminary taxonomic analysis. Morphological phenotypes and 16S ribosomal DNA sequence analysis indicated that these isolates probably belong to the species Bacillus thuringiensis. Enzymatic analysis with known Bacillus strains confirmed that all of the strains of B. thuringiensis and the closely related species B. cereus and B. mycoides tested produced AHL-inactivating enzymes but B. fusiformis and B. sphaericus strains did not. Nine genes coding for AHL inactivation were cloned either by functional cloning or by a PCR procedure from selected bacterial isolates and strains. Sequence comparison of the gene products and motif analysis showed that the gene products belong to the same family of AHL lactonases.
N-Acyl-homoserine lactones (AHLs), also known as autoinducers, are widely conserved signal molecules that are present in the quorum-sensing systems of many gram-negative bacteria. The bacteria release, detect, and respond to accumulation of these signal molecules for synchronizing expression of particular sets of genes and for coordinating cellular activities. It has been found that AHLs are involved in regulation of a range of biological functions, including bioluminescence in Vibrio species (4, 13), Ti plasmid conjugal transfer in Agrobacterium tumefaciens (32), induction of virulence genes in Burkholderia cepacia, Erwinia carotovora, Erwinia chrysanthemi, Erwinia stewartii, Pseudomonas aeruginosa, and Xenorhabdus nematophilus (3, 6, 12, 17, 20-23, 25), regulation of antibiotic production in Pseudomonas aureofaciens and E. carotovora (6, 25), swarming motility in Serratia liquifaciens (14), and biofilm formation in Pseudomonas fluorescens and P. aeruginosa (1, 8). More bacterial species are known to produce AHLs, but the relevant biological functions have not been investigated (2, 5, 11).
AHL quorum-sensing signals are a fascinating group of molecular targets for genetic and chemical manipulation. These molecules are highly conserved; they have the same homoserine lactone moiety but differ in the length and structure of the acyl side chain. Although different target genes are regulated by AHLs, the basic mechanisms of AHL biosynthesis and gene regulation seem to be conserved in different bacterial species. The general feature of AHL-mediated gene regulation is cell population-dependent regulation, which is known as quorum sensing. The concentration of an AHL increases along with the growth of bacterial cells. When the AHL concentration reaches a threshold level, it triggers target gene expression (16). The biological functions regulated by AHLs are of considerable scientific and economic importance. New approaches for up or down regulation of bacterial quorum-sensing systems would be of significant interest not only for scientific purposes but also for practical applications.
We recently reported cloning of a novel gene, aiiA240B1 coding for an AHL-inactivating enzyme (AiiA240B1), from the gram-positive bacterium Bacillus sp. strain 240B1 (9). AiiA240B1 inactivates an AHL by hydrolyzing its lactone bond and was designated AHL lactonase (10). Expression of aiiA240B1 in transformed E. carotovora strain SCG1, a pathogen that causes soft rot disease in many plants, significantly reduces release of AHL, decreases extracellular pectrolytic enzyme activities, and attenuates pathogenicity for potato, eggplant, Chinese cabbage, carrot, celery, cauliflower, and tobacco (9). Transgenic plants expressing AHL lactonase exhibit significantly enhanced resistance to E. carotovora infection and delayed development of soft rot symptoms (10). AHL-inactivating mechanisms appear to be widely distributed. An isolate of Variovorax paradoxus has been reported to use AHL molecules as energy and nitrogen sources, indicating that AHL-degrading enzymes are present in this organism (19). In this study, we focused on the biodiversity of AHL-inactivating enzymes and bacterial strains. We identified more than 20 bacterial isolates and strains capable of AHL inactivation obtained from soil and plant samples and from a laboratory bacterial culture collection. Nine genes coding for AHL inactivation (aiiA) were cloned from gram-positive Bacillus species and characterized. Biochemical and molecular analyses showed that the enzymes encoded by these genes belong to the same family of AHL lactonases.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains and plasmids used in this study are described in Table 1. Screening of bacterial isolates capable of inactivating AHL has been described previously (9). All isolates and Bacillus strains were grown at 28°C in Luria-Bertani medium. Escherichia coli strains were grown at 37°C. Ampicillin (100 μg/ml) and tetracycline (15 μg/ml) were added to the medium when they were required. X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Promega) was included in the medium at a concentration of 50 μg/ml for detection of β-galactosidase activity.
TABLE 1.
Bacterial strains and plasmidsa
| Bacterial strain or plasmid | Serotype or related characteristics | Source or reference |
|---|---|---|
| Bacteria | ||
| Bacillus strains | ||
| B1 (B. thuringiensis subsp. thuringiensis) | H1 | BGSC 4A3 |
| B2 (B. thuringiensis subsp. kurstaki) | H3a3b | BGSC 4D1 |
| B12 (B. thuringiensis subsp. aizawai) | H7 | BGSC 4J4 |
| B17 (B. thuringiensis subsp. wuhanensis) | No flagella | Mycogen PSS2A1 |
| B18 (B. thuringiensis) | Lab collection | |
| B20 (B. thuringiensis) | Lab collection | |
| B21 (B. thuringiensis) | Lab collection | |
| B22 (B. thuringiensis subsp. kurstaki) | H3a3b, plasmidless | Lab collection |
| B23 (B. thuringiensis subsp. israelensis) | H14, plasmidless | BGSC 4Q7 |
| B25 (B. cereus) | Lab collection | |
| 14579 (B. cereus) | ATCC 14579 | |
| 6462 (B. mycoides) | ATCC 6462 | |
| 269 (B. fusiformis) | Lab collection | |
| B29 (B. sphaericus) | BGSC 12A4 | |
| Agrobacterium tumefaciens NT1 | traR tra::lacZ749, indicator strain | 24 |
| Escherichia coli DH5α | F−φ80d lacZΔM15 endA1 hsdR17 (rk− mk−) supE44 thi-1 gyrA96 Δ(lacZYA-argF) | 26 |
| Plasmids | ||
| pLARF3 | Tcr | 27 |
| pBluescript SK+ | Apr | Stratagene |
| pGEM-7Zf(+) | Apr | Promega |
| pGEM-T | Apr | Promega |
| pLARF3-aiiACOT1 | Tcr, 24-kb EcoRI fragment from COT1 genomic DNA in pLARF3 | This study |
| pGEM-aiiACOT1 | Apr, 5-kb EcoRI fragment from pLARF3-aiiACOT1 in pGEM-7Zf(+) | This study |
| pBS-aiiACOT1 | Apr, 1.3-kb BamHI fragment from pGEM-aiiACOT1 in pBluescript SK | This study |
BGSC, Bacillus Genetic Stock Centre; ATCC, American Type Culture Collection; Lab collection, laboratory collection strain; Tcr, tetracycline resistance; Apr, ampicillin resistance. Eight new bacterial isolates are described in the text.
AHL bioassay.
To determine AHL-inactivating activity, N-(3-oxohexanoyl)-l-homoserine lactone (OHHL) was added at a final concentration of 20 μM to an overnight bacterial culture which was diluted to an optical density at 600 nm (OD600) of 1.1. The reaction mixture was incubated at 28°C for different times as indicated below. The amount of OHHL remaining in the supernatant or reaction mixture was then determined as previously described (9, 31). A. tumefaciens strain NT1 containing a lacZ fusion in the tra gene of pTiC58 was used as an indicator strain for AHL activity (24). AHL-inactivating activity was expressed as the number of picomoles of OHHL inactivated per hour per unit of OD600 of cell culture.
Cloning of aiiACOT1 gene.
Genomic DNA was purified from bacterial isolate COT1 and digested partially with EcoRI. DNA fragments were ligated to the dephosphorylated EcoRI site of cosmid vector pLAFR3 (27). Ligated DNA was packaged and transfected into E. coli DH5α. Cosmid clones with AHL-inactivating activity were identified by using the bioassay method described above. Subcloning into sequencing vector pGEM-7Zf(+) or pBluescript SK was carried out by routine techniques (26). Sequencing was performed for both strands by using an ABI Prism dRhodamine terminator cycle sequencing Ready Reaction kit (Perkin-Elmer Applied Biosystems).
PCR procedure for cloning the aiiA gene from other Bacillus species.
Genomic DNA samples were used as PCR templates. The primers were designed based on the identical sequences of the 5′ ends of the aiiA240B1 and aiiACOT1 open reading frames (ORFs) and the conserved region 119 nucleotides after the stop codon. The forward primer was 5′-ATG GGA TCC ATG ACA GTA AAG AAG CTT TAT-3′, and the reverse primer was 5′-GTC GAA TTC CTC AAC AAG ATA CTC CTA ATG-3′. Each PCR was performed for 30 cycles consisting of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. PCR products were purified by using a QIAquick PCR purification kit (QIAGEN), and the purified PCR fragments were ligated to the pGEM-T vector (Promega). The clones which conferred AHL-inactivating activity were used for further study. At least two clones of each gene were sequenced to avoid misreading due to PCR error. A database search was performed by using the BLASTA search algorithm. The DNASTAR sequence analysis software package (DNASTAR Inc.) and the GCG sequence analysis software (Genetics Computer Group, Madison, Wis.) were used for sequence analysis at the nucleotide and peptide levels.
Southern blot analysis.
Three bacterial isolates and 15 Bacillus strains, including 12 B. thuringiensis strains, 2 B. cereus strains, and 1 B. sphaericus strain, were used for Southern blot analysis. Genomic DNA (20 μg) that was digested with EcoRI was separated by electrophoresis in a 0.8% agarose gel and transferred onto a Hybond-N+ membrane (Amersham Pharmacia Biotech) according to the manufacturer's instructions. The 1.4-kb BamHI fragment containing the aiiACOT1 coding region was labeled with digoxigenin-11-dUTP and used as a probe for hybridization. After hybridization at 65°C, the membrane was washed twice in 2× SSC-0.1% sodium dodecyl sulfate (SDS) at room temperature for 15 min (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), and this was followed by two washes in 0.1× SSC-0.1% SDS at 65°C for 15 min. A nitroblue tetrazolium-BCIP (5-bromo-4-chloro-3-indolylphosphate) solution was used as the color substrate as recommended by the manufacturer's protocol (Boehringer Mannheim).
Protein extraction and Western blot analysis.
Bacterial cells were grown overnight at 28°C (for Bacillus strains) or 37°C (for E. coli) in Luria-Bertani medium. Cells were harvested by centrifugation at 4°C. Each cell pellet was washed once with phosphate-buffered saline (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4; pH 7.4) and then resuspended in ice-cold phosphate-buffered saline and sonicated on ice. Cell debris was removed by centrifugation. The protein concentration was determined by the Bradford method with reagents obtained from Bio-Rad (Hercules, Calif.). Proteins (25 μg) were separated by SDS-12% polyacrylamide gel electrophoresis, transferred onto a polyvinyidene difluoride membrane (Bio-Rad), and then probed with polyclonal anti-(AHL lactonase) antibodies (10), followed by alkaline phosphatase-conjugated secondary antibodies. Purified AHL lactonase (10 ng) encoded by aiiA240B1 was used as a control in the immunoblot analysis.
Nucleotide sequence accession numbers.
The nucleotide sequences of aiiACOT1, aiiAB1, aiiAB2, aiiAB17, aiiAB18, aiiAB20, aiiAB21, aiiAB22, and aiiAB25 have been deposited in the GenBank database under accession numbers AF350927 to AF350935.
RESULTS
Isolation of bacteria capable of inactivating AHL.
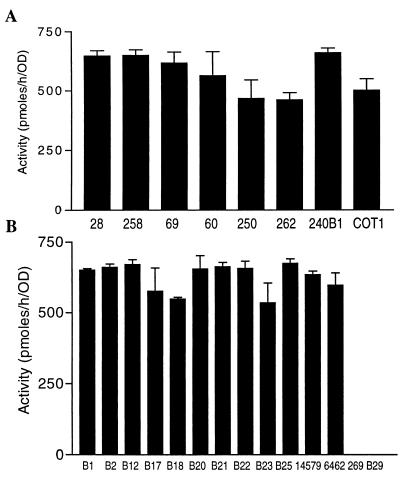
We screened more than 800 bacterial isolates obtained from soil and plants for AHL-inactivating activity. By incubating fresh bacterial cultures with OHHL, we identified more than 20 bacterial isolates that exhibited AHL-inactivating activity during the preliminary screening. Eight of these isolates showing strong activity were selected and characterized at the biochemical and molecular levels. The AHL-inactivating activities were variable, ranging from 480 to 680 pmol/h/unit of OD600 (Fig. 1A). To characterize these isolates, their 16S ribosomal DNA (rDNA) sequences were analyzed by PCR amplification and subsequent sequencing. The results showed that the 16S rDNA sequences of these isolates are highly homologous to the 16S rDNA sequence of B. thuringiensis (data not shown). To confirm the 16S rDNA sequence similarity, we sequenced the 16S rDNA from a known B. thuringiensis strain, B. thuringiensis subsp. thuringiensis BGSC 4A3. A sequence comparison revealed that the levels of identity between the BGSC 4A3 rDNA and the rDNA of these isolates were 97.8 to 98.1%. The other characteristics exhibited by these isolates (gram positive, peritrichous, straight rods, endospore formation) also indicated that they are the members of B. cereus group in the genus Bacillus.
FIG. 1.
AHL inactivation analysis of bacterial isolates and strains. (A) AHL-inactivating activities of newly identified bacterial isolates; (B) AHL-inactivating activities of previously described Bacillus strains (as shown in Table 1). A cell suspension culture (OD600, 1.1) prepared from each bacterial isolate or strain was mixed with an equal volume of 40 μM OHHL and then incubated at 28°C for 30 min. The amount of OHHL remaining in the supernatant was determined as previously described (9, 31). The activities are expressed in picomoles of OHHL inactivated per hour per unit of OD600 of bacterial culture. The values are means ± standard deviations based on four replicates.
All of the B. thuringiensis and closely related strains tested had AHL-inactivating activity.
The B. cereus group includes B. cereus, B. thuringiensis, B. mycoides, and B. anthracis (29). We selected 14 known Bacillus strains, including 9 B. thuringiensis strains, 2 B. cereus strains, 1 B. mycoides strain, 1 B. fusiformis strain, and 1 B. sphaericus strain, to test whether these species have the ability to inactivate AHL. All of the bacterial strains tested except the B. fusiformis and B. sphaericus strains eliminated AHL with levels of enzyme activity (range, 534 to 674 pmol/h/unit of OD600) (Fig. 1B) similar to those of the newly identified AHL-inactivating bacterial isolates (Fig. 1A).
It is known that many B. thuringiensis strains contain plasmids. Our data suggest that AHL-inactivating genes are located in chromosomal DNA and not in plasmids, because B. thuringiensis subsp. kurstaki strain B2 (= BGSC 4D1) and its plasmidless derivative, strain B22, exhibited similar levels of activity. A second plasmidless strain, strain B23 (= BGSC 4Q7) (http://bacillus.biosci.ohio-state.edu/), belonging to B. thuringiensis subsp. israelensis, was also capable of inactivating AHL (Fig. 1B).
Functional cloning of aiiACOT1 gene.
Bacterial cells of strain COT1 eliminated OHHL (20 μM) completely after 2 h of incubation at 28°C, but bacterial cells killed by boiling for 5 min failed to inactivate OHHL (Fig. 2), indicating that there is an enzymatic inactivation mechanism. To identify the gene coding for AHL inactivation in COT1, a cosmid library was constructed and used for functional screening. One clone (pLARF3-aiiACOT1) showing AHL-inactivating activity in the bioassay was identified after we screened about 1,000 cosmid clones. Restriction analysis showed that this clone contained a 24-kb insert. The five fragments generated by complete digestion with EcoRI were subcloned into the pGEM-7 vector. A bioassay with the subclones showed that one subclone, pGEM-aiiACOT1 with an 5-kb insert, conferred AHL-inactivating activity. Further subcloning led to identification of clone pBS-aiiACOT1 containing a 1.3-kb BamHI fragment which codes for AHL inactivation. Complete sequence analysis of clone pBS-aiiACOT1 showed that there was a 750-bp ORF which encoded a 250-amino-acid protein. Expression of this ORF in E. coli confirmed that it encoded a functional AHL-inactivating enzyme, designated AiiACOT1. At the peptide sequence level, AiiACOT1 exhibited 91% identity to AiiA240B1 (9), which has recently been identified as a novel AHL lactonase (10).
FIG. 2.
Enzymatic inactivation of OHHL by suspension culture of COT1. Equal volumes of a cell suspension culture (OD600, 1.1) and 40 μM OHHL were mixed and incubated at 28°C (▴). A boiled culture mixed with 40 μM OHHL was used as a control (▪).
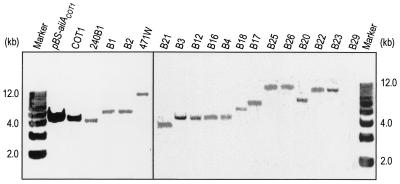
Southern blot detection of aiiACOT1 homologues in Bacillus species.
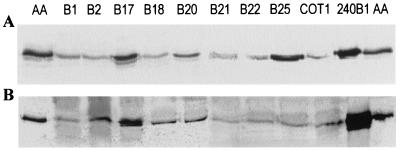
As all of the bacterial isolates and strains capable of inactivating AHL that were tested belong to B. thuringiensis or closely related taxa and the aiiA240B1 and aiiACOT1 genes showed a high level of similarity, it is very likely that the aiiA gene is highly conserved in B. thuringiensis and closely related species. DNA hybridization analysis was performed using the aiiACOT1 gene as a probe. Genomic DNA were isolated from 18 selected bacterial strains and isolates, including 13 B. thuringiensis strains, 1 B. cereus strain, 1 B. sphaericus strain, and 3 isolates. The results (Fig. 3) showed that one hybridizing band was clearly produced by all of the strains tested except B. sphaericus B29, which was unable to inactivate AHL (Fig. 1B). These results indicate that there is an aiiACOT1 homologue in all of the strains of B. thuringiensis and the closely related species B. cereus tested. This is consistent with the bioassay data (Fig. 1). To detect AHL lactonase in these AHL-inactivating bacterial strains, the soluble proteins extracted from the strains were subjected to Western blot analysis using a rabbit anti-AHL lactonase antiserum. As shown in Fig. 4A, immunoblot signals were detected in the strains.
FIG. 3.
DNA hybridization analysis of different bacterial isolates and known Bacillus species. Total DNA was digested with EcoRI, fractionated in a 0.8% agarose gel, blotted, and then probed with a digoxigenin-labeled 1.3-kb fragment containing the aiiACOT1 gene. All of the strains used except B26 (B. cereus) and B29 (B. sphaericus) are B. thuringiensis strains.
FIG. 4.
Western blot analysis. (A) Western blot analysis of the total proteins extracted from bacterial isolates and strains capable of inactivating AHL. Soluble proteins from 10 bacterial isolates and strains and 10 ng of purified AHL lactonase (lanes AA) were separated by SDS-polyacrylamide gel electrophoresis and were visualized by anti-AHL lactonase antibody coupling in a standard alkaline phosphatase immunoassay. (B) Western blot analysis of the total proteins extracted from E. coli strains containing the aiiA gene isolated from different strains.
Cloning of aiiA genes from other selected Bacillus strains.
As the genes for AHL inactivation in Bacillus strains 240B1 and COT1 are highly conserved, a PCR approach was used to clone the AHL lactonase genes from selected B. thuringiensis and B. cereus strains. Genomic DNA isolated from B. thuringiensis strains B1, B2, B17, B18, B20, B21, and B22 and B. cereus strain B25 were used as templates. Purified PCR fragments were cloned into the pGEM-T vector (Promega). The resulting eight clones, which showed AHL-inactivating activity in the bioassay, were used to obtain the sequences of both strands. The sequences of the eight AHL-inactivating genes cloned from strains B1, B2, B17, B18, B20, B21, B22, and B25 all contained a 750-bp ORF which encoded a 250-amino-acid protein. Western blot analysis confirmed that these genes were expressed in E. coli (Fig. 4B).
Motif analysis of AHL-inactivating genes.
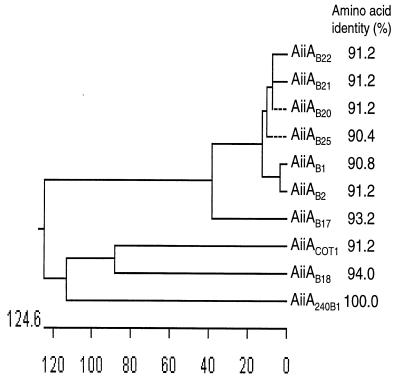
Sequence analysis indicated that 10 aiiA genes cloned from Bacillus species were highly conserved, with amino acid identities ranging from 90.4 to 94.0% compared with the amino acids of AHL lactonase (Fig. 5). As determined by a BLAST CD search of Conserved Domain Databases (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), two conserved regions were found by aligning AHL lactonases with metallo-beta-lactamase (pfam00753, Conserved Domain Databases) and zinc-containing glyoxalase II (Y08357, GenBank) (Fig. 6A). Several histidine and glutamate residues in these two conserved regions are known to be essential for zinc binding and enzyme activity of metallohydrolases (7, 30). Site-directed mutagenesis has shown previously that residues H106, D108, H109, and H169 of AiiA240B1 are necessary for AHL lactonase activity (9). Based on sequence alignment, we performed site-directed mutagenesis with several other conserved amino acid residues, including D191, H235, and D236. We replaced these residues with serine residues. The results showed that the D191S mutation resulted in a complete loss of enzyme activity, whereas the H235S and D236S mutations did not affect the enzyme activity (Fig. 6B). These data established a motif for AHL lactonase, 106HXDH-59 amino acids-H169-21 amino acids-D191, which is essential for enzyme activity.
FIG. 5.
Phylogenetic tree analysis and levels of amino acid identity of AHL lactonases. The phylogenetic tree was produced with the DNASTAR sequence analysis software (DNASTAR Inc.). Distances are shown below the tree. The levels of amino acid identity of AHL lactonases from strains COT1, B1, B2, B17, B18, B20, B21, B22, and B25 to the AHL lactonase from strain 240B1 are shown on the right.
FIG. 6.
Alignment of sequences of the conserved regions and site-directed mutagenesis of AHL lactonase. (A) Alignment of the conserved regions of Bacillus AHL lactonases and portions of two other proteins, the metallo-beta-lactamase (Conserved Domain Databases accession no. pfam00753) amino acid sequence from residue 49 to residue 134 (Lac) and the Arabidopsis glyoxalase II (GenBank accession no. U90928) amino acid sequence from residue 128 to residue 214 (Gly). The regions of the Bacillus AHL lactonases examined included amino acid residues 101 to 195. The amino acids identical to amino acids in AiiA240B are indicated by boldface type. The amino acid residues necessary for AHL lactonase activity are indicated by asterisks. aa, amino acids. (B) Site-directed mutagenesis of AHL lactonase. H, histidine; D, aspartic acid; S, serine. The number in each designation indicates the position of the corresponding amino acid in the AHL lactonase peptide. Histidine and aspartic acid were replaced by serine by site-directed mutagenesis (see reference 9 for an explanation of the method). The values above the bars are the relative activities of the mutants.
DISCUSSION
Nine genes encoding AHL-inactivating enzymes were cloned from gram-positive bacterial isolates and strains. These genes exhibited high levels of homology to aiiA240B1, which encodes an AHL lactonase (9, 10). Similar to the AHL lactonase encoded by aiiA240B1, the putative zinc-binding motif that has catalytic importance is conserved in these newly identified AHL-inactivating enzymes (Fig. 6A). Data suggest that these enzymes are members of the AHL lactonase family. Detailed characterization of the relationship between the structure and activity of these enzymes would be useful for elucidation of the enzymatic mechanism.
Our results showed that the AHL-inactivating bacterial isolates and strains belong to B. thuringiensis, B. cereus, and B. mycoides, whereas two Bacillus species, B. fusiformis and B. sphaericus, did not exhibit detectable AHL-inactivating activity. By searching microbial genome BLAST databases, we found a homologue (gnl/TIGR_1392/banth_2063) in B. anthracis in which 89.1% the nucleotides are identical to those in the aiiACOT1 coding sequence, suggesting that B. anthracis may contain a similar enzyme that inactivates AHLs. It is known that B. anthracis, B. thuringiensis, B. cereus, and B. mycoides are closely related and belong to the B. cereus group in the genus Bacillus (29). The role of AHL lactonase in these Bacillus species is not clear. Unlike most gram-negative bacteria that produce AHL quorum-sensing signals, some gram-positive bacteria produce γ-butyrolactones as quorum-sensing signals, which are structurally quite similar to AHLs (28). It would be interesting to investigate whether AHL lactonases also inactivate the γ-butyrolactones, how widely AHL-inactivating genes are conserved in the microbial community, and the roles of these genes in bacterial interaction and ecology.
B. thuringiensis has been used extensively as a microbial insecticide in last few decades (15, 18). However, most of important insecticidal B. thuringiensis strains have not been exploited for disease control because they usually do not produce antibiotics that are effective against bacteria and fungi. In this study, we found that all of the B. thuringiensis strains tested are capable of inactivating AHL. Because short-side-chain AHLs diffuse easily into bacterial cells, B. thuringiensis could eliminate AHL constantly from its surroundings in order to avoid accumulation of AHLs that activate virulence genes. It is probable that insecticidal B. thuringiensis strains could be used as biocontrol agents against bacterial diseases which are mediated by the AHL quorum-sensing signals.
REFERENCES
- 1.Allison, D., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L., E. P. Greenberg, and A. M. Stevens. 1997. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 179:4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beck von Bodman, S., and S. K. Farrand. 1995. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acyl homoserine lactone autoinducer. J. Bacteriol. 177:5000-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cao, J. G., and E. A. Meighen. 1989. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem. 264:21670-21676. [PubMed] [Google Scholar]
- 5.Cha, C., P. Gao, Y. C. Chen, P. D. Shaw, and S. K. Farrand. 1998. Production of acyl-homoserine lactone quorum-sensing signals by gram-negative plant-associated bacteria. Mol. Plant-Microbe Interact. 11:1119-1129. [DOI] [PubMed] [Google Scholar]
- 6.Costa, J. M., and J. E. Loper. 1997. EcbI and EcbR: homologs of LuxI and LuxR affecting antibiotic and exoenzyme production by Erwinia carotovora subsp. betavasculorum. Can. J. Microbiol. 43:1164-1171. [DOI] [PubMed] [Google Scholar]
- 7.Crowder, M. W., M. K. Maiti, L. Banovic, and C. A. Makaroff. 1997. Glyoxalase II from A. thaliana requires Zn(II) for catalytic activity. FEBS Lett. 418:351-354. [DOI] [PubMed] [Google Scholar]
- 8.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 9.Dong, Y.-H., J.-L. Xu, X.-C. Li, and L.-H. Zhang. 2000. AiiA, a novel enzyme inactivates acyl homoserine-lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dong, Y.-H., L.-H. Wang, J.-L. Xu, H.-B. Zhang, X.-F. Zhang, and L.-H. Zhang. 2001. Quenching quorum sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 11.Dumenyo, C. K. M., A. W. Chun, and A. K. Chatterjee. 1998. Genetic and physiological evidence for the production of N-acyl homoserine lactones by Pseudomonas syringae pv. syringae and other fluorescent plant pathogenic Pseudomonas species. Eur. J. Plant Pathol. 104:569-582. [Google Scholar]
- 12.Dunphy, G., C. Miyamoto, and E. Meighen. 1997. A homoserine lactone autoinducer regulates virulence of an insect-pathogenic bacterium, Xenorhabdus nematophilus (Enterobacteriaceae). J. Bacteriol. 179:5288-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 14.Eberl, L., M. K. Winson, C. Sternberg, G. S. A. B. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 15.Feitelson, J. S., J. Payne, and L. Kim. 1992. Bacillus thuringiensis: insects and beyond. Bio/Technology 10:271-275. [Google Scholar]
- 16.Fuqua, C., and S. C. Winans. 1996. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 178:435-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, S. M., B. Yu, N. J. Bainton, M. Birdsall, B. W. Bycroft, S. R. Chhabra, A. J. R. Cox, P. Golby, P. J. Reeves, S. Stephens, M. K. Winson, G. P. C. Salmond, G. S. A. B. Stewart, and P. Williams. 1993. The Lux autoinducer regulates the production of exoenzyme virulence determination in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 12:2477-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambert, B., and M. Peferoen. 1992. Insecticidal promise of Bacillus thuringiensis. Facts and mysteries about a successful biopesticide. BioScience 42:112-122. [Google Scholar]
- 19.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lewenza, S., B. Conway, E. P. Greenberg, and P. A. Sokol. 1999. Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181:748-756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nasser, W., M. L. Bouillant, G. Salmond, and S. Reverchon. 1998. Characterization of the Erwinia chrysanthemi expl-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol. Microbiol. 29:1391-1405. [DOI] [PubMed] [Google Scholar]
- 22.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 23.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Piper, K. R., S. Beck von Bodman, and S. K. Farrand. 1993. Conjugation factor of Agrobacterium tumefaciens regulates Ti plasmid transfer by autoinduction. Nature 362:448-450. [DOI] [PubMed] [Google Scholar]
- 25.Pirhonen, M., D. Flego, R. Heikinheimo, and E. Palva. 1993. A small diffusible signal molecule is responsible for the global control of virulence and exoenzyme production in the plant pathogen Erwinia carotovora. EMBO J. 12:2467-2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Staskawicz, B. D., N. T. Keen, and C. Napoli. 1987. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol. 169:5789-5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Takano, E., T. Nihira, Y. Hara, J. J. Jones, C. J. Gershater, Y. Yamada, and M. Bibb. 2000. Purification and structural determination of SCB1, a γ-butyrolatone that elicits antibiotic production in Streptomyces coelicolor A3(2). J. Biol. Chem. 275:11010-11016. [DOI] [PubMed] [Google Scholar]
- 29.Turnbull, P. C. B. 1999. Definitive identification of Bacillus anthracis—a review. J. Appl. Microbiol. 87:237-246. [DOI] [PubMed] [Google Scholar]
- 30.Vallee, B. L., and A. Galdes. 1984. The metallobiochemistry of zinc enzymes. Adv. Enzymol. Relat. Areas Mol. Biol. 56:283-430. [DOI] [PubMed] [Google Scholar]
- 31.Zhang, L.-H. 1993. Molecular biology and biochemistry of a novel conjugation factor in Agrobacterium. Ph.D. dissertation. The Adelaide University, Adelaide, Australia.
- 32.Zhang, L.-H., P. J. Murphy, A. Kerr, and M. E. Tate. 1993. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 362:446-447. [DOI] [PubMed] [Google Scholar]