Abstract
Sake, a traditional alcoholic beverage in Japan, is brewed with sake yeasts, which are classified as Saccharomyces cerevisiae. Almost all sake yeasts form a thick foam layer on sake mash during the fermentation process because of their cell surface hydrophobicity, which increases the cells' affinity for bubbles. To reduce the amount of foam, nonfoaming mutants were bred from foaming sake yeasts. Nonfoaming mutants have hydrophilic cell surfaces and no affinity for bubbles. We have cloned a gene from a foam-forming sake yeast that confers foaming ability to a nonfoaming mutant. This gene was named AWA1 and structures of the gene and its product were analyzed. The N- and C-terminal regions of Awa1p have the characteristic sequences of a glycosylphosphatidylinositol anchor protein. The entire protein is rich in serine and threonine residues and has a lot of repetitive sequences. These results suggest that Awa1p is localized in the cell wall. This was confirmed by immunofluorescence microscopy and Western blotting analysis using hemagglutinin-tagged Awa1p. Moreover, an awa1 disruptant of sake yeast was hydrophilic and showed a nonfoaming phenotype in sake mash. We conclude that Awa1p is a cell wall protein and is required for the foam-forming phenotype and the cell surface hydrophobicity of sake yeast.
Sake is a Japanese traditional alcoholic beverage made from steamed rice by parallel fermentation with Aspergillus oryzae, which is a source of saccharification enzymes, and sake yeasts, which produce ethanol from glucose. Sake yeasts are classified as Saccharomyces cerevisiae and are known to produce more than 18% (vol/vol) ethanol in sake mash (16). Almost all sake yeasts form a thick foam layer on sake mash during vigorous fermentation. This foam formation has been used as an indicator of fermentation progress: the foam rises when fermentation becomes strong and it disappears when fermentation becomes weak. However, this characteristic sometimes reduces the efficiency of sake fermentation because a large part of the fermentation tank is occupied by a thick foam layer. Thus, an absence of foam formation during fermentation is a preferable property.
Ouchi and Akiyama (23) developed a method to screen nonfoaming mutants from foaming sake yeast using cell affinity for bubbles, and using this method they isolated a nonfoaming mutant from an industrial sake yeast. This mutant had almost the same characteristics as the parental strain except for its nonfoaming property in fermentation. Thus, this method of screening for nonfoaming mutants was applied to various strains of sake yeasts and the resultant nonfoaming yeasts are now widely used in commercial sake brewing. Comparison of the nonfoaming mutant with its parent revealed that the cell surface of the former is less hydrophobic than that of the latter, suggesting that cell surface hydrophobicity is related to foaming ability (25). However, the detailed molecular mechanism of foaming ability of sake yeast is still unknown. In this study, we have cloned a gene from a foaming yeast that confers the foaming phenotype to its nonfoaming mutant and named it AWA1. Our results indicate that a cell wall protein encoded by AWA1 is responsible for the foam formation ability and cell surface hydrophobicity of sake yeast.
MATERIALS AND METHODS
Strains and culture conditions.
The S. cerevisiae strains used in this study are listed in Table 1. Yeast cells were cultivated in YPAD (1% yeast extract, 2% Bacto Peptone, 0.01% adenine, 2% glucose) medium or YNBD (0.67% yeast nitrogen base, 2% glucose) medium supplemented with the appropriate amount of amino acids and bases depending on the strains. A synthetic pantothenate-deficient medium was purchased from the Brewing Society of Japan. One liter of this medium contains 20 g of glucose, 0.5 g of (NH4)2SO4, 1.5 g of KH2PO4, 0.5 g of MgSO4 · 7H2O, 0.16 g of citric acid, 200 μg of thiamine, 200 μg of riboflavin, 200 μg of nicotinic acid, 1 mg of inositol, 200 μg of p-aminobenzoic acid, 0.2 μg of biotin, 40 μg of β-alanine, and 30 g of agar at pH 5.0. Escherichia coli JM109 was used for DNA manipulation and sequencing. E. coli was cultured in LB (1% tryptone, 0.5% yeast extract, 1% NaCl) medium supplemented with 100 μg of ampicillin per ml.
TABLE 1.
Foam-forming ability of yeast strains used in this studya
| Strain | Genotype | Foam | Source |
|---|---|---|---|
| K7 | Sake yeast MATa/α ecm31/ecm31 | + | ATCC 26422b |
| K701 | Nonfoaming mutant of K7 | − | 23 |
| 7H1 | K7 haploid MATα | + | ATCC 44893 |
| 7H2 | K7 haploid MATα | + | ATCC 44894 |
| 7H3 | K7 haploid MATa | + | ATCC 44882 |
| YPH499 | MATaura3 lys2 ade2 trp1 his3 leu2 | − | 29 |
| YPH500 | MATα ura3 lys2 ade2 trp1 his3 leu2 | − | 29 |
| YHS173 | 7H1 × YPH499 | + | 28 |
| YHS174 | 7H2 × YPH499 | + | 28 |
| YHS175 | 7H3 × YPH500 | + | 28 |
| UT-1 | K701 ura3/ura3 trp1/trp1 | − | 15 |
| YHS180 | UT-1 p4E | + | This study |
| YHS471 | 7H3 awa1::kanr | − | This study |
| YHS233 | UT-1 pRS416-AWA1 | + | This study |
| YHS235 | UT-1 pRS416-3 × HA::AWA1 | + | This study |
Sake was brewed on a small scale using the indicated strain and foam formation was observed.
ATCC, American Type Culture Collection.
Foam formation test by small-scale sake brewing.
Five milliliters of a 48-h culture of yeast cells grown in YPAD medium was inoculated into mash composed of 38 g of dry steamed rice (α-rice), 12 g of dry koji (a culture of A. oryzae on steamed rice), 120 ml of tap water, and 100 μl of 50% (as total acid) lactic acid in a wide-mouth container (8 cm by 12 cm) and incubated at 30°C. Foam formation was observed after 2 days of fermentation.
Gene cloning and DNA manipulation.
Yeast genomic DNA was prepared from K7 as previously described and partially digested with Sau3AI (28). DNA fragments of 5 to 10 kb were collected by 10 to 40% sucrose density gradient centrifugation at 75,000 × g for 24 h. The resultant Sau3AI fragments were ligated to BamHI-digested pPBH1 (28) and used to transform E. coli JM109. The transformants were collected and the plasmids were recovered. This library was used for transformation of UT-1 by the electroporation method (4). The transformed cells were plated on YNBD medium supplemented with tryptophan and were incubated for 4 days at 30°C. The transformants were collected and subjected to a concentration procedure to enrich foam-forming cells. Approximately 108 cells of the transformants were mixed with the same amount of K7 and used for a small-scale sake brewing test. After 2 days of incubation at 30°C, the foam layer was recovered and plated on pantothenate-deficient medium. After incubation at 35°C for 2 days, the colonies that had formed were recovered. After the concentration procedure was repeated three times, the foam-forming activity of individual clones was determined. Plasmid extraction from E. coli cells was performed using a kit (Wizard; Promega). Other DNA manipulations were carried out as described previously (1, 3). DNA sequences were determined with a DNA sequencer (ABI PRISM 310; Perkin-Elmer).
Construction of the plasmid coding for HA-tagged Awa1 protein.
The DNA fragment of p4E containing AWA1 was digested with EcoRV and ligated into the SmaI site of pRS416 (29), resulting in pRS416-AWA1. The sequence for a triple influenza virus hemagglutinin (HA) epitope (5′-TACCCATACGATGTTCCTGACTATGCGGGCTAT CCCTATGACGTCCCGGACTATGCAGGATCCTATCCATATGACGTTCCAGATTACGCT-3′) was amplified from mTn 3XHA::lacZ (5) as a template by PCR with primers 5′-ATACCCATACGATGTTCCTGAC-3′ and 5′-GTAGCGTAATCTGGAACGTCATA-3′. The amplified sequence was cloned into the HpaI site of pRS416-AWA1. The sequence of the inserted DNA was confirmed by DNA sequencing with a primer (5′-TGGGTGTTTACGTCTCCG-3′). The resultant plasmid, pRS416-3XHA::AWA1, was used to transform yeast strains.
Immunological detection of the HA-tagged Awa1 protein.
For immunofluorescence staining of intact yeast cells, yeast cells grown to the exponential phase (optical density at 660 nm [OD660] = 1) were fixed with 3.7% formaldehyde at 30°C for 15 min and washed once with phosphate-buffered saline (PBS; 10 mM Na2HPO4, 2 mM KH2PO4, 0.15 M NaCl, 3 mM KCl, pH 7.4). The washed cells were suspended in PBS containing 1% bovine serum albumin and incubated at 30°C for 30 min, followed by incubation in PBS containing 1 μg of mouse anti-HA monoclonal antibody (Boehringer Mannheim, Mannheim, Germany) per ml at 30°C for 1 h. After three washes with PBS, the cells were incubated with PBS containing fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G antibody (GibcoBRL, Grand Island, N.Y.) at 30°C for 1 h. After three washes with PBS, cells were suspended in PBS and examined under a fluorescence microscope (UFX-II; Nikon).
For Western blotting analysis of HA-tagged Awa1, yeast cell wall proteins were prepared as described previously (27). Briefly, after a 24-h cultivation in YPAD at 30°C with shaking, cells were harvested and disrupted with glass beads. The cell wall fraction was recovered by centrifugation and was treated with 50 mM Tris-HCl buffer (pH 8.0) containing 2% sodium dodecyl sulfate (SDS), 100 mM EDTA, and 40 mM dithiothreitol at 100°C for 10 min to remove noncovalently bound and disulfide bridge-bound proteins. The resultant cell wall fraction was treated with a 5-μg/ml concentration of β-1,6-glucanase purified from westase (Takara), a yeast lytic enzyme prepared from the culture supernatant of Streptomyces rochei (22), in 50 mM sodium acetate buffer (pH 5.0) containing 1 mM phenylmethylsulfonyl fluoride at 30°C for 3 h. The procedures used to purify the enzyme will be published elsewhere. Proteins solubilized from the cell wall were separated by SDS-polyacrylamide gel electrophoresis (2 to 15% gradient) and electrotransferred to a polyvinylidene difluoride membrane (Millipore), followed by immunodetection of the HA tag as described previously (27).
Gene disruption.
Gene disruption of AWA1 was carried out by a PCR-based method (9). A DNA fragment containing the Kanr gene as a selectable marker and 50-bp sequences identical to both flanking regions of the AWA1 open reading frame were amplified with 5′-TTAGTTAGTAAGGCCCACTAAAATGGCGTGATATTTTAAAGGGCCTTTACCAGCTGAAGCTTCGTACGC-3′ and 5′-CTATTCTTACGAGATTTCCCTTGTTCTCGCCGCTCACATCATTGTCGGTAGCATAGGCCACTAGTGGATCTG-3′, in which the underlined sequences were used for amplification of the Kanr gene, as primers and pUG6 as a template. The PCR product was used to transform 7H3, a haploid strain of K7, and a transformant with the disrupted AWA1 gene was screened by PCR. Disruption of the AWA1 gene was confirmed by genomic Southern blot analysis.
CSH assay.
Yeast cell surface hydrophobicity (CSH) was determined by hydrophobic interaction chromatography as described previously (2). Yeast cells grown in YNBD medium for 48 h at 30°C with shaking were harvested, washed twice with 100 mM sodium acetate buffer (pH 4.2), and resuspended in the same buffer to a final concentration of 5% (wet wt/vol). Phenyl Sepharose CL-4B gels (Pharmacia) were packed in a disposable chromatography column (Biospin empty column; Bio-Rad) to a volume of 0.25 ml and equilibrated with 100 mM sodium acetate buffer (pH 4.2) containing 1.0 M NaCl. Yeast cell suspensions (0.1 ml) were then applied to the column and eluted with 3 ml of the buffer containing NaCl. The OD660 of the eluent was measured. CSH was determined by using the following equation: CSH (%) = 100 × (Aapplied − Aeluent)/Aapplied, where Aapplied is the OD660 of 0.1 ml of the cell suspension diluted with 3 ml of elution buffer and Aeluent is the OD660 of the eluent.
Nucleotide sequence accession number.
The nucleotide and deduced amino acid sequences of AWA1 have been submitted to the DDBJ/EMBL/GenBank database under accession number AB071164.
RESULTS
Cloning of a gene complementing the nonfoaming phenotype.
K7 is the most widely used sake yeast in commercial sake brewing and exhibits a foam-forming phenotype. As shown in Table 1, haploid strains (7H1, 7H2, and 7H3) derived from K7 (24) were foam-forming, similar to their parent, while laboratory yeast strains (YPH499 and YPH500) were non-foam-forming. Hence, the K7 haploid strains were mated with the laboratory strains with the opposite mating type. The resultant heterozygous diploids (YHS173, YHS174, and YHS175) were all foam-forming (Table 1), indicating that the foam-forming phenotype of K7 is dominant. Because of poor sporulation efficiency of the heterozygous diploids and poor germination of the formed spores (21), we could not determine whether the foam-forming phenotype of K7 originated from a single nuclear gene. To clone the gene responsible for the foam formation phenotype, we constructed a genomic library from K7 and this library was used to transform a uracil auxotroph (UT-1) of a non-foam-forming mutant (K701) of K7. The plasmid (pPBH1) used for the library construction contained URA3 and ECM31 as selectable markers for transformed cells (28). Twenty thousand transformants were recovered as uracil prototrophs. To enrich the transformants that acquired a foam-forming ability, we made a small-scale mash inoculated with about the same numbers of K7 and transformant cells. After 2 days of culture at 30°C, a thick foam layer developed on the mash. Transformants that became foam-forming should be concentrated in the foam layer, whereas the majority of the transformants should be dispersed in the mash as non-foam-forming yeasts. We recovered yeast cells of the foam layer, spread them on pantothenate-deficient medium, and incubated them at 35°C. K7 and its derivatives are known to be pantothenate auxotrophic at 35°C and this was complemented by ECM31 that was present in pPBH1 (28). Therefore, the K7 cells that were added to the mash could not grow on this medium, whereas the transformants could grow. After repeating this enrichment procedure three times, we identified one foam-forming transformant (Fig. 1). The plasmid was recovered from this transformant and designated p4E. When the plasmid was removed from the transformant by plating on the 5′-fluoroorotic acid-containing medium, yeast cells returned to the non-foam-forming phenotype (data not shown). Moreover, when UT-1 was transformed with p4E, the transformant again showed the foam-forming phenotype (data not shown). These results indicate that the foam-forming phenotype of the transformant is plasmid dependent and that genes in the plasmid are involved in the foam formation of K7.
FIG. 1.
Cloning of the AWA1 gene. K7, UT-1, and YHS180 (UT-1 harboring p4E) were cultured and used for small-scale sake brewing. After 2 days of culture at 30°C, foam formation was observed.
Structural analyses of a gene involved in foam formation and its product.
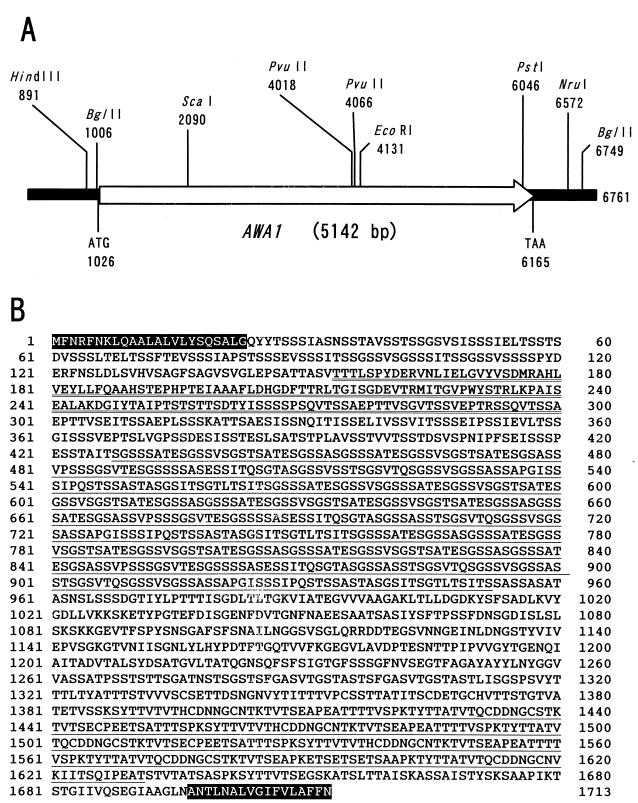
DNA sequence analysis of p4E revealed that this plasmid contained only one open reading frame, of 5,142 bp (Fig. 2A). We named this gene AWA1, after “awa,” the Japanese word for foam. AWA1 encodes a large protein of 1,713 amino acids with a molecular mass of 166,873 Da (Fig. 2B). As described below, Awa1p showed some homologies to other proteins in the S. cerevisiae genome database (http://genome-www.stanford.edu/Saccharomyces/). However, no identical proteins were found in the database, suggesting that Awa1p is specific to sake yeast. Awa1p has two stretches of hydrophobic amino acids, at both the N-terminal and C-terminal regions, as shown in Fig. 3A. These sequences are characteristic of proteins with a glycosylphosphatidylinositol (GPI) anchor, which are cell surface proteins commonly seen in various eukaryotic cells (8). In yeast cells, GPI-anchored proteins are often observed in cell wall proteins (10). Awa1p is very rich in serine (28.9%) and threonine (16.0%), which is also a characteristic of cell wall proteins in yeast cells (12). These results suggest that Awa1p is a cell wall protein localized on the cell surface of K7.
FIG. 2.
Structure of the AWA1 gene. (A) Schematic representation of the AWA1 gene with restriction enzyme sites. (B) Deduced amino acid sequence of Awa1p. The shaded letters indicate putative signal peptide and GPI anchor signal sequences. The underlined letters indicate the SRR and the CTR. The double underlined letters indicate the YJR151c-like sequence.
FIG. 3.
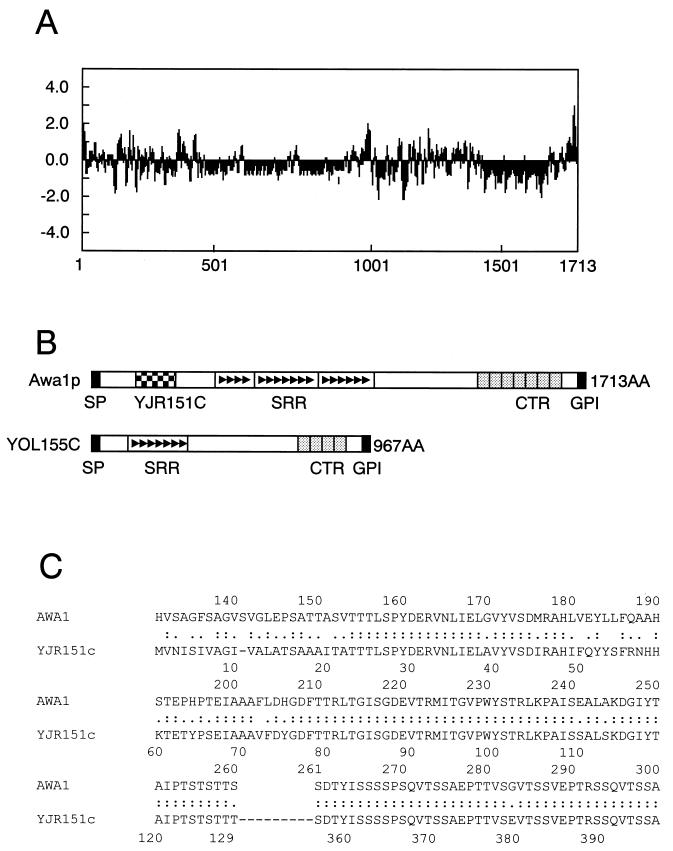
Structural characteristics of Awa1p. (A) Hydropathy plot of Awa1p with a window of 10 residues performed by the method of Kyte and Doolittle (17). (B) Structural comparison of Awa1p and YOL155c. SP, signal peptide (C) Homology between Awa1p and YJR151c. Identical amino acids are denoted by colons, and similar amino acids are denoted by periods.
A search of the database revealed that Awa1p was highly homologous to YOL155c, a putative GPI-anchored protein of S. cerevisiae with unknown function (6). However, a part of the Awa1p sequence (amino acid positions 154 to 300) is not homologous to YOL155c (Fig. 3B). This sequence is homologous to two parts of YJR151c, another putative GPI-anchored protein, as shown in Fig. 3C. This suggests that Awa1p is a chimeric protein derived from different ancestral genes. Following the part of the Awa1p sequence that is homologous to YJR151c, there is a long, complex, serine-rich repeating sequence (SRR; amino acid positions 428 to 952) as shown in Fig. 2B and Fig. 3B. This sequence is comprised of three parts, each of which contains several 13-residue repeats. Although YOL155c has a similar repeating sequence, it has fewer of the smaller repeats than does Awa1p. Before the C-terminal hydrophobic sequence, Awa1p also has a C-terminal repeating sequence (CTR; amino acid positions 1387 to 1631) that includes seven approximately 35-residue repeats as shown in Fig. 2B and Fig. 3B. Although YOL155c also has a similar repeating sequence, the number of repeats is only four.
Localization of Awa1p.
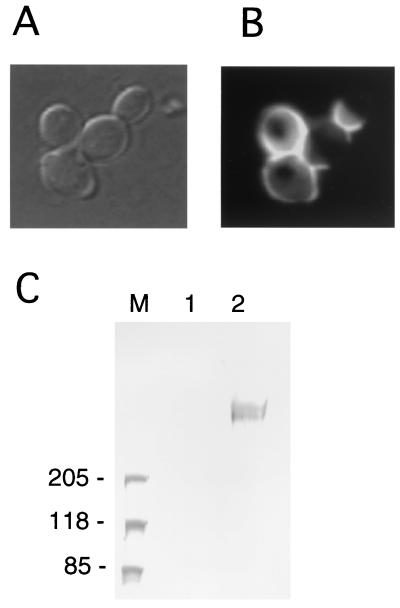
To confirm the localization of Awa1p, we constructed a plasmid encoding an epitope-tagged Awa1p. A DNA fragment corresponding to a triple repeat of the influenza virus HA epitope (3×HA) was inserted into the N-terminal part of the AWA1 gene and the resultant plasmid was used to transform UT-1. The transformant formed foam in a small-scale sake mash, indicating that 3×HA-tagged Awa1p was fully functional (Table 1). After immunofluorescent staining, the transformed cells exhibited intense fluorescence at the peripheral region of the cells (Fig. 4B), indicating that Awa1p is localized on the cell surface.
FIG. 4.
Localization of 3×HA Awa1p. (A) Representative cells of YHS235 (UT-1 harboring pRS416-XHA::AWA1) viewed under a light microscope. (B) The same field viewed with a fluorescence microscope. Fixed yeast cells were probed with the anti-HA antibody and the fluorescein isothiocyanate-labeled anti-mouse immunoglobulin G. (C) Western blotting analysis of the cell wall proteins. The cell wall fraction was prepared from yeast cells and treated with β-1,6-glucanase. Liberated proteins were analyzed by Western blotting on an SDS-2 to 15% polyacrylamide gel electrophoresis gel using the anti-HA antibody. Lane M, molecular size marker; lane 2, β-1,6-glucanase extract of YHS233 (UT-1 harboring pRS416-AWA1); lane 3, the β-1,6-glucanase extract of YHS235 (UT-1 harboring pRS416-XHA::AWA1).
Since Awa1p is a putative GPI-anchored protein, it may be covalently linked to cell wall glucan, although some GPI-anchored proteins are predominantly localized in the plasma membrane (6, 10). Therefore, we analyzed whether Awa1p is covalently bound to the cell wall. The cell wall fraction was prepared from the transformant cells with 3×HA-tagged Awa1p and digested with β-1,6-glucanase, which releases cell wall protein covalently linked to the cell wall glucan (12). A Western blot analysis showed a diffused reactive protein band with mouse anti-HA monoclonal antibody at a high molecular mass (Fig. 4C). This is reasonable, considering that Awa1p has four putative N-glycosylation sites (amino acid positions 34, 1133, 1241, and 1278) and a very high content of serine and threonine, which are putative O-glycosylation sites. These results indicate that Awa1p is a cell wall protein that is covalently bound to the cell wall through β-1,6-glucan.
Gene disruption of the AWA1 gene.
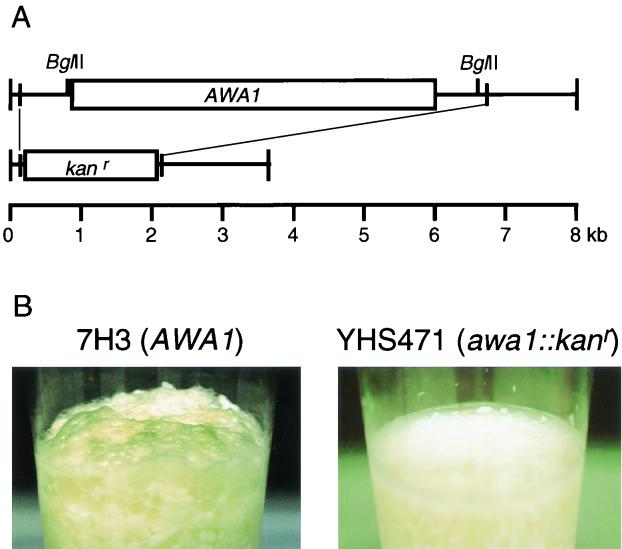
As described above, the haploid cells of K7 were foam-forming. Therefore, we carried out gene disruption of the AWA1 gene of a K7 haploid strain to investigate whether AWA1 is directly involved in foam formation. The AWA1 gene of 7H3 was replaced with a Geneticin (G418) resistance gene by the PCR method (Fig. 5A). The resultant AWA1 disruptant (YHS471) did not form foam in a small-scale sake brewing experiment (Fig. 5B), indicating that the AWA1 gene is necessary for foam formation of sake yeast K7. Growth and fermentation of YHS471 were similar to those of the parent strain (data not shown).
FIG. 5.
Method and effects of disruption of the AWA1 gene. (A) Method of AWA1 gene disruption. The AWA1 gene of 7H3 was replaced by the Kanr gene as a selectable marker by the PCR method. (B) Foam formation test of 7H3 (AWA1) and YHS471 (Δawa1::kanr) using small-scale sake brewing.
AWA1 and CSH.
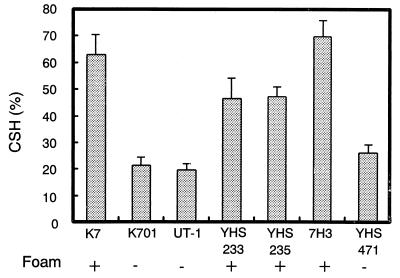
The foam-forming ability of sake yeast was shown to be related to CSH (25). Therefore, we investigated whether the presence of the AWA1 gene affected not only foam-forming ability but also CSH using hydrophobic chromatography. As shown in Fig. 6, there is a clear relationship between CSH and foam-forming ability. Yeast strains with foam-forming ability (K7, YHS233, and 7H3) exhibited higher CSH than strains that do not form foam during fermentation (K701, UT-1, and YHS471). UT-1 transformed with 3×HA-tagged Awa1p (YHS235) also exhibited high CSH, indicating that 3×HA tagging of Awa1p did not affect CSH. These results indicate that Awa1p is involved in both the foam formation and CSH of sake yeast.
FIG. 6.
CSH of sake yeast strains. The CSH value of 48-h yeast culture in synthetic minimal medium was determined. Three independent experiments were performed for each strain of yeast. The values depicted are the means plus standard deviations.
DISCUSSION
Our findings that the AWA1 gene cloned from K7 could confer foaming ability to the nonfoaming mutant of K7 and that an AWA1 deletion mutant of the K7 haploid strain became nonfoaming clearly show that the AWA1 gene is directly involved in the foam formation of sake yeast. Although the laboratory strain does not have the AWA1 gene, it has a closely related homologue, YOL155c. The function of YOL155c is not known since the null mutant does not differ from the wild type in any known phenotypes (18). In addition to having sequences homologous to those of YOL155c, Awa1p contains a region that is similar to two parts of YJR151c, which is not related to YOL155c. Considering that the laboratory strain is not foam-forming, the region that is homologous to the parts of YJR151c might be involved in the foam-forming function of Awa1p.
The primary structure of Awa1p suggested that it is a GPI-anchored protein. Such proteins are widely found in eukaryotes from fungi to mammalian cells. After protein synthesis, these proteins are translocated into the endoplasmic reticulum where N- and C-terminal signal sequences are removed and a GPI anchor is bound to the newly formed C terminus. Then, GPI-anchored proteins are modified by glycosylation through the secretion pathway, after which they are incorporated into the plasma membrane (13). In yeasts, many species of GPI-anchored proteins are transferred from the plasma membrane to the cell wall and are covalently bound to β-1,6-glucan by an unknown mechanism (12). Our results showed that Awa1p is also covalently bound to the cell wall through β-1,6-glucan. The yeast cell wall contains many species of GPI-anchored cell wall proteins and these are divided into two groups by their structures and functions. One is a group of relatively small proteins, such as Cwp1p, Cwp2p, Sed1p, Tip1p, and Tir1p (14, 26, 27, 31), whose functions are not well understood. Some of these proteins are differentially expressed depending on culture conditions and are considered to be structural cell wall proteins that cement other cell wall components together. The other class of proteins includes large cell surface proteins that participate in the interaction between yeast cells and other materials in the environment such as agglutinins (19) and flocculins (32). Awa1p belongs to the second group because its molecular size is larger than the molecular sizes of proteins in the first group and it clearly causes the cells to interact with their environment.
The presence of Awa1p is closely related to CSH, which strongly affects how microorganisms adhere to other materials (7). In the opportunistic pathogenic yeast Candida albicans, CSH affects the virulence of the yeast by controlling adhesion of the organisms to epithelial cells (20). CSH is also important in fermentation technology. CSH of brewer's yeast is one of the major determinants of cell flocculation (30). A high level of CSH in yeast cells may facilitate cell-to-cell contact in the mash solution and lead to a more stable interaction between flocculins and their receptors on the cell surface. Sake yeast cells that form foam are also known to be hydrophobic and this characteristic promotes cell absorption onto carbon dioxide bubbles formed by ethanol fermentation (25). The direct relationship between the presence of AWA1 and hydrophobicity (Fig. 6) suggests that Awa1p has a hydrophobic region that can interact with the bubbles. Considering that Awa1p is anchored to the cell wall through its C terminus and that Awa1p contains a long sequence rich in serine and threonine, Awa1p may form a rod-like structure (11) in which the N-terminal portion of Awa1p extrudes from the cell surface. Similar structures have been reported in Agα1p and Aga1p (19) and in Flo1p (32), which are GPI-anchored proteins and whose N-terminal parts are involved in cell-to-cell interactions. Their lengths are considered to be long enough to interact with cell surface structures of other cells. The tertiary structure of the N-terminal part of Awa1p may have a hydrophobic surface that absorbs to the bubbles.
In conclusion, the AWA1 gene is a unique gene of sake yeast and determines the foam-forming ability of sake yeast by conferring hydrophobicity to the cell surface. We are currently studying the structures of AWA1 genes in various foam-forming and non-foam-forming sake yeasts to investigate the structure and function of Awa1p in detail.
Acknowledgments
We thank H. Wu for analyzing the yeast cell wall proteins and T. Akeno for DNA sequencing. We also thank K. Kitamoto for providing yeast strain UT-1.
REFERENCES
- 1.Adams, A., D. E. Gottschling, C. Kaiser, and T. Stearns. 1997. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 2.Akiyama-Jibiki, M., T. Ishibiki, H. Yamashita, and M. Eto. 1997. A rapid and simple assay to measure flocculation in brewer's yeast. Master Brewers Assoc. Am. Tech. Q. 34:278-281. [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, and D. D. Moore. 1999. Short protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 4.Becker, D. M., and L. Guarente. 1991. High-efficiency transformation of yeast by electroporation. Methods Enzymol. 194:182-187. [DOI] [PubMed] [Google Scholar]
- 5.Burns, N., B. Grimwade, P. B. Ross-Macdonald, E. Y. Choi, K. Finberg, G. S. Roeder, and M. Snyder. 1994. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 8:1087-1105. [DOI] [PubMed] [Google Scholar]
- 6.Caro, L. H., H. Tettelin, J. H. Vossen, A. F. Ram, H. Van Den Ende, and F. M. Klis. 1997. In silicio identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477-1489. [DOI] [PubMed] [Google Scholar]
- 7.Doyle, R. J., and M. Rosenberg. 1990. Microbial cell surface hydrophobicity. American Society for Microbiology, Washington, D.C.
- 8.Ferguson, M. A. 1999. The structure, biosynthesis and functions of glycosylphosphatidylinositol anchors, and the contributions of trypanosome research. J. Cell Sci. 112:2799-2809. [DOI] [PubMed] [Google Scholar]
- 9.Guldener, U., S. Heck, T. Fielder, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamada, K., H. Terashima, M. Arisawa, and K. Kitada. 1998. Amino acid sequence requirement for efficient incorporation of glycosylphosphatidylinositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 273:26946-26953. [DOI] [PubMed] [Google Scholar]
- 11.Jentoft, N. 1990. Why are proteins O-glycosylated? Trends Biochem. Sci. 15:291-294. [DOI] [PubMed] [Google Scholar]
- 12.Kapteyn, J. C., H. Van Den Ende, and F. M. Klis. 1999. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426:373-383. [DOI] [PubMed] [Google Scholar]
- 13.Kinoshita, T., and N. Inoue. 2000. Dissecting and manipulating the pathway for glycosylphosphatidylinositol-anchor biosynthesis. Curr. Opin. Chem. Biol. 4:632-638. [DOI] [PubMed] [Google Scholar]
- 14.Kitagaki, H., H. Shimoi, and K. Itoh. 1997. Identification and analysis of a static culture-specific cell wall protein, Tir1p/Srp1p in Saccharomyces cerevisiae. Eur. J. Biochem. 249:343-349. [DOI] [PubMed] [Google Scholar]
- 15.Kitamoto, K., K. Oda, K. Gomi, and K. Takahashi. 1990. Construction of uracil and tryptophan auxotrophic mutants from sake yeasts by disruption of URA3 and TRP1 genes. Agric. Biol. Chem. 54:2979-2987. [Google Scholar]
- 16.Kodama, K. 1993. Sake-brewing yeast, p. 129-168. In A. H. Rose and J. S. Harrison (ed.), The yeasts, vol. 3. Academic Press, London, United Kingdom. [Google Scholar]
- 17.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 18.Lafuente, M. J., and C. Gancedo. 1999. Disruption and basic functional analysis of six novel ORFs of chromosome XV from Saccharomyces cerevisiae. Yeast 15:935-943. [DOI] [PubMed] [Google Scholar]
- 19.Lipke, P. N., and J. Kurjan. 1992. Sexual agglutination in budding yeasts: structure, function, and regulation of adhesion glycoproteins. Microbiol. Rev. 56:180-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Masuoka, J., and K. C. Hazen. 1997. Cell wall protein mannosylation determines Candida albicans cell surface hydrophobicity. Microbiology 143:3015-3021. [DOI] [PubMed] [Google Scholar]
- 21.Nakazawa, N., T. Ashikari, N. Goto, T. Amachi, R. Nakajima, S. Harashima, and Y. Oshima. 1992. Partial restoration of sporulation defect in sake yeast, Kyokai no. 7 and Kyokai no. 9, by increased dosage of the IME1 gene. J. Ferment. Bioeng. 73:265-270. [Google Scholar]
- 22.Nishi, A., K. Ohbuchi, M. Hamachi, and C. Kumagai. 1999. Breeding of a constitutive-producing strain of Streptomyces rochei PHA-34 that produces the lytic enzyme for Phaffia rhodozyma and extraction of astaxanthin using the enzyme. Seibutsu-kogaku Kaishi 77:60-65. [Google Scholar]
- 23.Ouchi, K., and H. Akiyama. 1971. Non-foaming mutant of sake yeast: selection by cell agglutination method and by froth floatation method. Agric. Biol. Chem. 35:1024-1032. [Google Scholar]
- 24.Ouchi, K., and H. Akiyama. 1976. Breeding of useful killer sake yeasts by repeated back-crossing. J. Ferment. Technol. 54:615-623. [Google Scholar]
- 25.Ouchi, K., and Y. Nunokawa. 1973. Non-foaming mutants of sake yeasts: their physico-chemical characteristics. J. Ferment. Technol. 51:85-95. [Google Scholar]
- 26.Shimoi, H., Y. Iimura, and T. Obata. 1995. Molecular cloning of CWP1: a gene encoding a Saccharomyces cerevisiae cell wall protein solubilized with Rarobacter faecitabidus protease I. J. Biochem. 118:302-311. [DOI] [PubMed] [Google Scholar]
- 27.Shimoi, H., H. Kitagaki, H. Ohmori, Y. Iimura, and K. Ito. 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 180:3381-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimoi, H., M. Okuda, and K. Ito. 2000. Molecular cloning and application of a gene complementing pantothenic acid auxotrophy of sake yeast Kyokai no. 7. J. Biosci. Bioeng. 90:643-647. [DOI] [PubMed] [Google Scholar]
- 29.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smit, G., M. H. Straver, B. J. Lugtenberg, and J. W. Kijne. 1992. Flocculence of Saccharomyces cerevisiae cells is induced by nutrient limitation, with cell surface hydrophobicity as a major determinant. Appl. Environ. Microbiol. 58:3709-3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Van der Vaart, J. M., L. H. Caro, J. W. Chapman, F. M. Klis, and C. T. Verrips. 1995. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 177:3104-3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Watari, J., Y. Takata, M. Ogawa, H. Sahara, S. Koshino, M. L. Onnela, U. Airaksinen, R. Jaatinen, M. Penttila, and S. Keranen. 1994. Molecular cloning and analysis of the yeast flocculation gene FLO1. Yeast 10:211-225. [DOI] [PubMed] [Google Scholar]