Abstract
Morphology and α-amylase production during submerged cultivation were examined in a wild-type strain (A1560) and in strains of Aspergillus oryzae in which chitin synthase B (chsB) and chitin synthesis myosin A (csmA) have been disrupted (ChsB/G and CM101). In a flowthrough cell, the growth of submerged hyphal elements was studied online, making it possible to examine the growth kinetics of the three strains. The average tip extension rates of the CM101 and ChsB/G strains were 25 and 88% lower, respectively, than that of the wild type. The branching intensity in the CM101 strain was 25% lower than that in the wild type, whereas that in the ChsB/G strain was 188% higher. During batch cultivation, inseparable clumps were formed in the wild-type strain, while no or fewer large inseparable clumps existed in the cultivations of the ChsB/G and CM101 strains. The α-amylase productivity was not significantly different in the three strains. A strain in which the transcription of chsB could be controlled by the nitrogen source-regulated promoter niiA (NiiA1) was examined during chemostat cultivation, and it was found that the branching intensity could be regulated by regulating the promoter, signifying an important role for chsB in branching. However, the pattern of branching responded very slowly to the change in transcription, and increased branching did not affect α-amylase productivity. α-Amylase residing in the cell wall was stained by immunofluorescence, and the relationship between tip number and enzyme secretion is discussed.
Filamentous fungi, such as Aspergillus oryzae, are widely used for industrial enzyme production, since they are able to secrete large amounts of proteins (e.g., amylases, proteases, phytases, and lipases) into the medium. Filamentous fungi grow by hyphal extension and branching through processes that are regulated in a way that is still not completely understood. During submerged cultivation, the growing hyphal elements tend to entangle, and this affects the rheology of the cultivation medium and the mixing characteristics in an undesirable fashion. This results in poor mixing and poor mass transfer of the substrate, and it makes the morphologies of filamentous fungi an obvious, yet unanswered, challenge in process optimization. Recently, however, there have been major advances in the tools available for metabolic engineering of aspergilli (reviewed by McIntyre et al. [17]), such as developments in genomics, the increased number of suitable transformation vectors, and powerful image analysis tools that enable rapid quantification of the fungal morphology. This has made metabolic engineering of the morphology for process optimization possible.
Metabolic engineering of morphology requires detailed knowledge of the relationships among productivity, growth, and morphology. The morphologies of filamentous organisms can be classified into macroscopic and microscopic morphologies (21), and techniques for analyzing morphology on both of these levels have advanced rapidly during the last decade (reviewed by Cox et al. [9]). Macroscopic morphology describes the shape, structure, and size of hyphal agglomerates, or pellets. In microscopic morphology, single hyphal elements or hyphal populations are characterized by measuring their dimensions, such as total hyphal length, number of tips, hyphal diameter, number of nuclei per cell (20), and number of membranous organelles per cell (1). Recently, there has also been considerable effort to deduce the kinetics of submerged growth and branching (8, 16, 28, 29), adding important details to the understanding of hyphal growth.
The relationship between the mycelial morphology and the rheological properties of the cultivation medium has been the subject of a number of studies (2, 15, 19, 25). The hyphal length per branch (length of the hyphal growth unit [lHGU]) affects the overall morphology (34); a low lHGU has been associated with the formation of clumps smaller than those formed at a high lHGU (2). Studies of the fungal cell wall suggest that chitin synthesis is an important factor in determining the hyphal morphology. Studies of Aspergillus nidulans have clarified the importance of two chitin synthase gene products in hyphal growth and cell wall formation: ChsB (3, 37) and CsmA (for chitin synthase with a myosin tail) (10, 13, 27). Strains in which chsB has been disrupted are disorganized and hyperbranched, and conidiation efficiency is reduced (3, 14). The csmA disruptants had reduced chitin content (27) and morphological abnormalities in hyphal walls, tips, and septa, and they were sensitive to osmotic stress (27) and chitin-binding dyes (10, 13). Both the myosin and chitin synthase domains of the csmA gene product are needed for formation of normal-shaped hyphae. It has been speculated that the role of the csmA gene product is in septum and conidiophore formation (13). These findings suggest that through control of these two chitin synthases it may be possible to do metabolic engineering of the morphology of Aspergillus in order to optimize the morphology for a fermentation process with a low-viscosity medium and high productivity.
This led us to construct strains of the industrially important fungus A. oryzae containing disrupted chitin synthase genes, chsB and csmA (C. Müller, C. M. Hjort, K. Hansen, and J. Nielsen, in press). Here, we compare the macroscopic and the microscopic morphologies and α-amylase production in a wild-type strain and the chsB and csmA disruption strains during submerged growth in a flowthrough cell, batch cultivation, and chemostat cultivation. In a flowthrough cell (29), the growth of submerged hyphal elements of the three strains was studied online in order to quantify the effect of the disruptions on the tip extension and branching pattern. With a strain in which the transcription of chsB could be controlled, chemostat cultivations were carried out in order to examine whether this regulation could be used to control the morphology during submerged growth. The chemostat cultivation was examined by morphological analysis and Northern blot analysis of the chsB, chsC, and csmA genes. In studying whether the number of tips influenced α-amylase secretion, it was shown by an indirect (two-stage) immunofluorescence method that α-amylase was present in the cell wall of A. oryzae growing submerged.
MATERIALS AND METHODS
Strains.
The A. oryzae strain A1560 (originally named IFO 4177) was donated by Novozymes A/S. The strains with chsB disrupted (ChsB/G) and with csmA disrupted (CM101) were constructed from HowB101, which is a spontaneous pyrG mutant selected from A1560 grown on 5-fluoroorotic acid and uridine. Expression studies of chsB were conducted using the A. oryzae niiA promoter. A niiA-chsB construct was used to transform the ChsB/G strain, forming the NiiA1 strain. For construction of the ChsB/G, CM101, and NiiA1 strains and characterization of the chsB and csmA genes, see Müller et al. (submitted).
Inoculum preparation.
Freeze-dried spores were used to inoculate rice cultures by the method of Carlsen (5). Six to 8 days after inoculation, the rice grains were covered with green or white spores. The spores were harvested by washing the grains with sterile water with 0.1% (wt/wt) Tween and were used as the inoculum for submerged cultivation in a final concentration of 2 × 109 to 6 × 109 spores liter−1.
Biomass determination.
Biomass measurements were made by measuring the grams (dry weight) per kilogram of medium by drying biomass samples filtered on Whatman GF/C filter paper (W&R Balston, Ltd.) for 24 to 48 h at 105°C.
Batch cultivations.
Batch cultivations were carried out in 5-liter in-house bioreactors with a working volume of 4.5 liters. During cultivations, the pH, temperature, agitation, aeration, and off-gas analysis were monitored and controlled. The medium was defined as described previously (6), with an initial glucose (monohydrate) concentration of 25 g liter−1. In all cultivations, the pH was automatically controlled by the addition of either 2 M NaOH or 2 M HCl. The temperature was controlled at 30°C throughout. At inoculation, the aeration rate was 0.2 volume of air culture volume−1 min−1, and the stirrer speed was 100 rpm. These parameters were increased to 1 volume of air culture volume−1 min−1 and 800 to 900 rpm as the fermentations progressed. The pH for inoculation was 3.5, which was increased slowly to 6.0 when the biomass concentration was approximately 1 g (dry weight) kg of medium−1. The pH was increased because α-amylase is stable at pH 6.0.
Chemostat cultivations.
Chemostat cultivations were carried out in Braun M 2-liter glass bioreactors with a working volume of 1.5 liters as described previously (20). The cultivations were carried out with the strains A1560, ChsB/G, and NiiA1 with dilution rates of 0.1 h−1 for the first two strains and with several dilution rates for the NiiA1 strain. During the nitrogen source shift experiment, the feed medium was shifted from medium containing 9.5 mM NaNO3 to medium containing 18.9 mM (NH4)2SO4.
Fluorescence microscopy and image analysis.
A sample taken from submerged cultivation was diluted with 0.1 M phosphate-buffered saline (PBS) buffer (pH 7.4) to an approximate biomass concentration of 0.25 g/kg and mixed in order to disperse the hyphal elements. The diluted sample was then stained with Calcofluor White (0.024 to 0.036 mg ml−1) for 15 min, placed on a microscope slide, covered with a coverslip, and viewed with a fluorescence microscope (Optiphot 2; Nikon) connected to a Quantimet 5501W Image Analyser (Leica) via a Kappa CF 8/1 FMC (Kappa Messtechnik) monochrome video camera. Images were acquired with a 4, 10, or 100× objective on the microscope and with magnifications onscreen of ×220, 550, or 5,500, respectively. With the 4× objective, the relative number of large inseparable clumps in a sample was measured. An inseparable clump was defined as a structure, e.g., a pellet or a clump (with a diameter of more than 40 μm), that could not be separated, that was not dispersed by the dilution, and that had a compactness value (the ratio of the projected area divided by the projected convex area) of more than 0.55. Using the 100× objective, the hyphal diameter, distances from branch point to septa, and the number of branches per apical or subapical compartment were measured. Using the 10× objective, the projected hyphal area and the number of tips per hyphal element with more than four tips were measured. The hyphal growth unit length (lHGU) (4) was then calculated as the projected hyphal area divided by the number of tips counted and divided by the average hyphal diameter in the sample. For all morphological measurements of submerged cultivations, more than 165 hyphae were measured per sample.
Flowthrough cell cultivations.
The cultivation of filamentous fungi in a flowthrough cell (8, 29) made it possible to quantify tip extension rates and branching patterns in extending hyphae of single hyphal elements using image analysis. The flowthrough cell is a small chamber placed under a microscope (Reichert) coupled to an image analysis system (Quantimet 600S; Leica) connected to a charge-coupled device camera (Bischke CCD-5230P). Within the flowthrough cell, fungal spores were attached to the surface with poly-d-lysine, substrate flow was started, and images of the growing hyphal elements were acquired. Images of the developing hyphal elements were acquired every 15 min. All experiments were carried out with a defined medium containing glucose monohydrate (various concentrations), 18.9 mM (NH4)2SO4, 5.6 mM KH2PO4, 4.1 mM MgSO4 · 7 H2O, 17.1 mM NaCl, 6.8 mM CaCl2 · 2 H2O, and 0.5 ml of trace metals liter−1 (5). The design and preparation of the flowthrough cell was described previously (29). The substrate flow rate was adjusted with a peristaltic pump to 52.5 μl min−1, resulting in laminar flow in the cell and a cell residence time of 1.5 min. The medium flowed through a silicone tube equipped with a sterile filter (pore diameter, 0.25 μm) before the flowthrough cell to remove air bubbles. Before each experiment, air was bubbled through the medium in order to ensure oxygen saturation.
α-Amylase activity.
Filtered cultivation liquid was diluted 10 times with 0.1 g of bovine serum albumin liter−1 and 0.1 g of CaCl2 liter−1 and frozen after being sampled. Before analysis, the mixture was thawed and the extracellular α-amylase activity was determined by a modified Boehringer Mannheim method using a Cobas Mira analyzer (Roche Diagnostics) as previously described (7). The unit of α-amylase activity is fungal α-amylase units (FAU) per milliliter (1 FAU is the amount of α-amylase which at 37°C hydrolyzes 5.26 g of starch h−1 [12]). In addition, it has been found that 1 FAU corresponds to approximately 1.63 × 10−4 g of protein (5).
α-Amylase localization. (i) Biomass washing.
Culture liquid (500 μl; approximately 2 g [dry weight] kg−1) was washed with 5 ml of 0.1 M PBS buffer (pH 7.0) in a test tube at 4°C. The solution was shaken and centrifuged for 2 min at 4,400 rpm (Universal 30RF, Hettich-Zentrifugen) at 4°C. The liquid phase was removed, and the biomass was resuspended in 5 ml of 0.1 M PBS buffer and washed again.
(ii) Blocking.
Nonspecific binding was blocked by incubating the biomass in blocking solution (PBS solution with 5% [wt/vol] dehydrated skimmed milk [Difco] and 0.02% [wt/vol] sodium azide) for 2 h in a test tube rotator at 4°C. The blocking solution was removed by centrifugation, and the biomass was resuspended in 5 ml of 0.1 M PBS buffer, shaken, and transferred to a new test tube. The solution was washed two or three times with PBS buffer and centrifuged until no more skimmed milk foam appeared on the liquid surface after it was shaken.
(iii) Primary antibody incubation.
Aliquots of 1 ml were incubated in polyclonal primary antibody solution (10 μl of a 5-mg ml−1 rabbit polyclonal antibody against α-amylase [Novozymes A/S] was added to 5 ml of TBS [0.05 M Tris, 0.15 M NaCl, pH 5.0] solution, giving an antibody concentration of 10 μg ml−1) by adding primary antibody solution to the submerged biomass and incubating it overnight in Eppendorf vials in a test tube rotator at 4°C.
(iv) Washing.
The liquid phase was removed by centrifugation, and the biomass was resuspended in 5 ml of 0.1 M PBS buffer, shaken, transferred to a test tube, and washed five times.
(v) Secondary antibody incubation.
Submerged biomass (100 μl) was transferred to an Eppendorf vial with 30 μl of secondary antibody solution (65 μl of goat anti-rabbit immunoglobulin G FITC-conjugated antibodies [F-0382; Sigma] mixed with 585 μl of 0.1 M PBS buffer, pH 8.6) and 70 μl of PBS buffer. The mixture was incubated in the dark at 4°C for 2 h in the test tube rotator. Afterwards, the solution was centrifuged for 2 min at 4,400 rpm and 4°C, and the biomass was transferred to a new test tube and resuspended in 5 ml of 0.1 M PBS buffer. The solution was centrifuged and washed again.
(vi) Detection.
The washing buffer was taken out, and the biomass fraction was suspended in 5,000 μl of antifade solution (90% [wt/vol] glycerol, 10% 0.1 M PBS buffer, and 0.06 M n-propylgallate [P-3130; Sigma] [1.27 g in 100 ml]). The test tube was shaken and centrifuged for 4 min at 4,400 rpm and 4°C. Samples for obtaining images were taken out with a plastic pipette and transferred to a glass slide. Afterwards, a coverslip was pressed firmly onto the sample and slide, and the edge of the coverslip was sealed with nail polish. The FITC-stained hyphae were viewed in a fluorescent microscope at ×100 magnification using a standard FITC filter block. The intensity of the FITC staining in the hyphae was measured from the tip downwards. Incubation either without primary antibody or with a polyclonal rabbit antibody against Saccharomyces cerevisiae proteinase A (in saline solution with sodium azide [Novozymes A/S]) was used as a control.
Northern blot analysis.
For RNA extraction, biomass was collected from the steady-state culture by filtration (Whatman GF/C filter), washed with cold deionized water, and put in liquid nitrogen within 10 s of being sampled. The frozen cells were kept at −80°C. The mycelium was ground carefully in a mortar with liquid nitrogen and placed in an Eppendorf tube to fill one-third of it. The RNA was then isolated by the TRIzol (Life Technologies) method, and the concentration was determined and adjusted according to the manufacturer's instructions. Electrophoresis and blotting were done according to the method described previously (26), and the RNA was then blotted on Hybond+ nylon membranes (Boehringer Mannheim). The membranes were hybridized to A. oryzae probes as described by Müller et al. (in press) for either chsB, chsC, or csmA and then reprobed with an A. oryzae probe for the triose-phosphate isomerase gene (tpiA) to check that equal amounts of RNA had been loaded in each lane on the gels. The chitin synthase probes (a 469-bp probe for csmA, a 641-bp probe for chsC, and a 629-bp probe for chsB) were chosen from intron-free regions and amplified by PCR, while the ∼800-bp tpi probe was kindly provided by M. Trier, Novozymes A/S. All probes were [α-32P]dCTP labeled using the Readiprime II kit (Amersham), and the probes were purified using QIAquick (Qiagen). The blot was probed at 65°C with [α-32P]dCTP-labeled PCR products. The blots were washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate and then in 0.2× SSC-0.1% sodium dodecyl sulfate before exposure to X-ray film (Biomax MS; Kodak).
RESULTS
Production of α-amylase during batch cultivation.
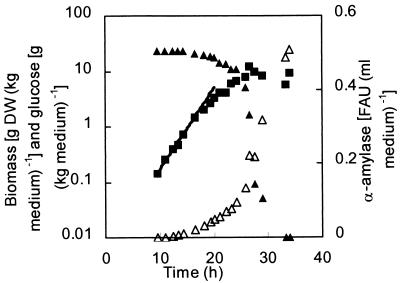
The influence of altering the chitin synthesis on the morphology and α-amylase production of A. oryzae was studied in a total of 15 batch cultivations under identical conditions with the wild-type strain, A1560; the chsB disruption strain, ChsB/G; and the csmA disruption strain, CM101. Glucose, biomass, and α-amylase activity were measured during all batch cultivations, and typical profiles of these variables during cultivation of the wild-type strain are shown in Fig. 1. In the figure, the maximum specific growth rate, μmax, has been determined from 10 to 17 h. After 17 h, the specific growth rate declines progressively, so growth continues with a lower specific growth rate. Extracellular α-amylase activity increased exponentially during the cultivation. For the first exponential phase, the α-amylase yield on biomass was calculated as the specific productivity divided by the maximum specific growth rate. The specific growth rate, the exponential-phase α-amylase yield, and the final yields, calculated as the final α-amylase activity divided by the final biomass concentration, were determined (Table 1). The data show that the differences in specific growth rate and α-amylase yields for the three strains were not statistically significant.
FIG. 1.
Profiles of the biomass (▪), glucose (▴), and α-amylase activity (▵) during a batch cultivation of the wild-type A. oryzae strain, A1560. DW, dry weight.
TABLE 1.
Physiological variables for three A. oryzae strains measured during batch cultivation
| Strain | μmaxa | Exponential-phase yieldb | Final yieldb | No. of batch cultivations |
|---|---|---|---|---|
| A1560 | 0.23 ± 0.03 | 18 ± 4 | 31 ± 6 | 5 |
| ChsB/G | 0.22 ± 0.02 | 14 ± 4 | 38 ± 6 | 7 |
| CM101 | 0.19 ± 0.02 | 16 ± 4 | 40 ± 7 | 3 |
Per hour (± 95% confidence interval).
FAU of α-amylase per gram (dry weight) ± 95% confidence interval. One FAU is the amount of α-amylase which at 37°C hydrolyzes 5.26 g of starch h−1. Dry weight was determined in grams per kilogram of medium by drying biomass samples filtered on Whatman GF/C filter paper for 24 to 48 h at 105°C.
Macroscopic morphology.
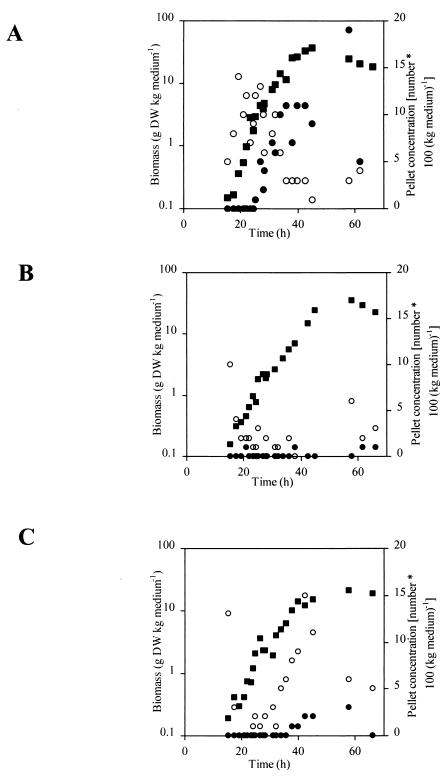
The macroscopic morphology was investigated by examining the relative amounts of large inseparable clumps existing during batch cultivations of the three strains (Fig. 2). Initially, in cultivations containing the A1560 and CM101 strains, there were small inseparable clumps with diameters between 40 and 150 μm. Later, in the wild-type cultivation, these developed into large inseparable clumps with diameters of more than 150 μm. The strain with chsB disrupted (ChsB/G) formed very few large inseparable clumps during branch cultivation and so was essentially a culture of freely dispersed hyphal elements. In comparison, Carlsen (5) modeled pellet formation in A. oryzae A1560 and Wittler et al. (35) examined the dissolved oxygen tension profiles in pellets of Penicillium chrysogenum, and they found the critical pellet diameter (where the oxygen concentration was zero precisely at the center) to be between 130 and 290 μm and between 100 and 400 μm, depending on the pellet structure and operating conditions, respectively. Therefore, it is likely that in our experiments the A1560 strain was oxygen limited in the biomass present in the interior of the large inseparable clumps; this could explain the decrease in specific growth rate observed for A1560 (Fig. 1 and 2). However, possibly due to the small amount of α-amylase produced by A1560 and the derived disrupted strains (1 FAU corresponds to approximately 1.63 × 10−4 g of protein), this did not significantly affect the α-amylase productivity.
FIG. 2.
Formation of inseparable clumps during batch cultivation with A. oryzae. (A) A1560 (wild type); (B) ChsB/G; (C) CM101. ▪, Biomass concentration; ○, inseparable clumps with diameters of <150 μm; •, inseparable clumps with diameters of >150 μm.
Microscopic morphology and branch formation.
The microscopic morphology is the key factor influencing macroscopic morphology (17), and differences among the strains at the microscopic level may explain the differences in the macroscopic morphologies observed here. In order to examine how defects in chitin synthesis affect the microscopic morphology, the degree of branching, the hyphal diameter, and the branching pattern were examined in exponentially growing batch cultivations of the three strains. The degree of branching was measured as the length of the hyphal growth unit, lHGU, and is shown in Table 2. The lHGU value for the A1560 strain (97 ± 7 μm) was consistent with that found previously (105 ± 2 μm [2]) for the same strain, and the lHGU value for the CM101 (csmA disruption) strain was comparable. However, the lHGU for the ChsB/G (chsB disruption) strain was 52% lower than that of the wild type, indicating that the ChsB/G strain was more densely branched. In addition, the hyphal diameter in the ChsB/G strain was significantly higher than in the A1560 strain. The branching pattern was further examined by determining the fractions of the apical and subapical compartments with branches. An eventual effect of the growth rate could be disregarded, since the specific growth rates for the three strains were very similar (Table 1). Table 2 shows that the wild-type (A1560) strain had branches mainly in the apical compartment while the ChsB/G strain had many branches in both the apical (more than one branch per compartment) and subapical compartments. The CM101 (csmA disruption) strain had a lower number of branches in the apical compartment and a higher number of branches in the subapical compartments than the wild-type. In addition, the CM101 cells frequently ballooned subapically, and this resulted in an increased average hyphal diameter compared to the wild type. The CM101 strain was also sensitive to high salt concentrations and the cell wall- and chitin-binding agents Calcofluor White and Congo Red, but wild-type growth could be partly remedied by osmotic stabilizers (data not shown).
TABLE 2.
Microscopic morphology in batch cultivation for three A. oryzae strainsa
| Strain | lHGU (μm tip−1 ± 95% confidence interval) | Hyphal diam (μm ± 95% confidence interval) | Branches/compartment
|
|
|---|---|---|---|---|
| Apical | Subapical | |||
| A1560 | 97 ± 7 | 3.32 ± 0.05 | 0.26 | 0.06 |
| ChsB/G | 51 ± 3 | 3.60 ± 0.09 | 1.40 | 0.79 |
| CM101 | 102 ± 7 | 3.68 ± 0.12 | 0.15 | 0.27 |
Values in batch cultivation were obtained after 23 h of cultivation during exponential growth for A1560, ChsB/G, and CM101. Hyphal diameter was measured in the middle of the second subapical compartment. Branches per compartment is the average number of branches observed in apical compartments (lateral and apical branches excluding the growing tip), and in subapical compartments it is the average number of branches observed in three to seven subapical compartments. More than 165 hyphal elements were measured for each value.
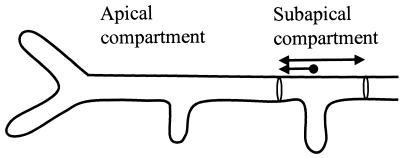
The subapical compartments with branches were further examined in order to investigate whether the formation of a new tip is associated with the locations of the septa and if chitin synthesis has any effect on branch positioning and septal localization. The distance from a branch point to the corresponding compartment end was measured as illustrated in Fig. 3. For all three strains, the subapical compartments had a length of 30 ± 10 μm, and the distance from a subapical branch to the septum closest to the apical compartment was 10 ± 7 μm. This indicates that subapical branches are formed closer to the septum nearest to the apical compartment, and it substantiates the idea that subapical septal placement and subapical branch positioning are processes that are not affected by disruption of chsB or csmA.
FIG. 3.
Diagram of a hypha with two branches in the apical compartment, one lateral and one apical, and one subapical branch. The length of the subapical compartment (double arrow) and position of the branch point in the compartment (arrow) were measured for approximately 100 subapical compartments for each of the three strains.
Growth kinetics of chitin synthase-deficient strains.
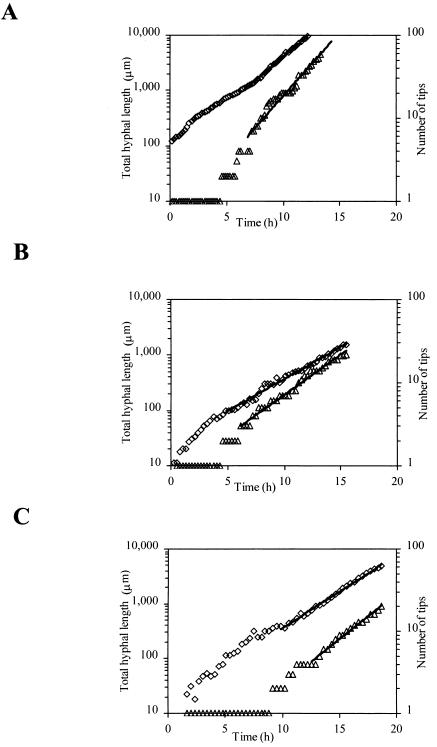
Using a flowthrough cell, the growth kinetics of individual hyphal elements of the strains A1560, ChsB/G, and CM101 were determined. Typical developments of the total hyphal length and the number of tips of the three strains are shown in Fig. 4. From these data, the maximum specific growth rate (determined after 8 to 15 h of growth); the average tip extension rate, qtip (in micrometers per hour); the branching intensity, qbran (in number of tips per hour); and the length of the hyphal growth unit, lHGU (in micrometers per tip) were determined for hyphal elements (for details, see reference 29). The average specific branching rate, kbran, av. (in number of tips per micrometer per hour), for a hyphal element was determined from the equation qbran (lt, z) = kbran, av. (z) · lt, where lt is the total hyphal length (in micrometers) and kbran, av. to a given time could be determined as Δlt divided by [Δt · qbran (lt, z)]. After approximately 10 h of growth, kbran, av. was constant for all strains. Using the data for germination and growth of the primary hyphae (germling), the maximum tip extension rate, ktip, max (in micrometers per hour), was estimated, since it is reasonable to describe growth of the primary hyphae using saturation-type kinetics (8, 29). Table 3 shows a compilation of the average values for a number of measured hyphal elements of the three strains, at two different glucose concentrations in the cases of A1560 and ChsB/G.
FIG. 4.
Development of the total length (⋄) and the number of tips (▵) of single hyphal elements for the three strains growing in defined medium at 1 g of glucose liter−1. The lines represent the fit to exponential growth for the total hyphal length, μl, and the number of tips, μtip, respectively. (A) A1560; μl = 0.39 h−1 and μtip = 0.40 h−1. (B) ChsB/G; μl = 0.24 h−1 and μtip = 0.24 h−1. (C) CM101; μl = 0.30 h−1 and μtip = 0.25 h−1.
TABLE 3.
Flowthrough cell resultsa
| Strain | Glucose (g liter−1) | μmax (h−1) | qtip (μm h−1) | ktip, maxb (μm h−1) | 104 · kbran, av. (no. of tips μm−1 h−1) | lHGU (μm tip−1) | n |
|---|---|---|---|---|---|---|---|
| A1560 | 0.049 | 0.30 ± 0.02 | 69 ± 11 | 164 ± 21 | 14 ± 3 | 233 ± 60 | 5 |
| 4.0 | 0.36 ± 0.07 | 86 ± 12 | 178 ± 15 | 16 ± 3 | 222 ± 36 | 10 | |
| ChsB/G | 0.0025 | 0.18 ± 0.02 | 10 ± 2 | 15 ± 3 | 27 ± 6 | 71 ± 11 | 11 |
| 4.0 | 0.23 ± 0.02 | 10 ± 2 | 21 ± 5 | 46 ± 10 | 52 ± 11 | 15 | |
| CM101 | 4.0 | 0.33 ± 0.04 | 69 ± 12 | 134 ± 26 | 12 ± 2 | 277 ± 52 | 22 |
Average ± standard deviation. n, number of measured hyphal units.
ktip, max was estimated by assuming that the tip extension rate of the primary hyphae can be described using Monod kinetics with respect to the hyphal length.
The specific growth rates found for hyphal growth in the flowthrough cell were higher than those observed in batch cultivation; this could be due to the stress caused by agitation in the batch culture or to fragmentation, as previously noted (29). For the CM101 (csmA disruption) strain, the estimated maximum tip extension rate and the average tip extension rate were reduced by 25 and 20%, respectively, compared to those of the wild-type, indicating that csmA has a role in tip extension. The specific growth rate for ChsB/G was not as high in the flowthrough cell as for the two other strains. The reason for this is unclear. Both the estimated maximum tip extension rate and the average tip extension rate were 88% lower in the ChsB/G strain than in A1560, indicating that this strain has a severe defect in tip extension.
At low glucose concentrations, the branching intensities were lower for the A1560 and the ChsB/G strains, resulting in increased hyphal length per branch. For the A1560 strain, the tip extension rate was decreased at 50 mg liter−1 compared to that at 4.0 g liter−1, but the ChsB/G strain had approximately the same tip extension rate at 2.5 mg of glucose liter−1 and at 4.0 g liter−1. This substantiates the idea that the defect in the chsB disruption strain acts through a limitation in the tip extension rate, not through increased branch formation.
Chitin synthase expression analysis.
Chemostat cultivations were carried out with the NiiA1 strain, which is the ChsB/G strain transformed with an niiA-chsB construct so that the niiA promoter solely controlled the transcription of chsB. This was done in order to examine whether regulation of chsB could be used to control morphology during submerged growth. During chemostat cultivation, the hyphal elements were examined by morphological analysis and Northern blot analysis of three chitin synthase genes, chsB, chsC, and csmA. Specifically, the RNA expression levels of chitin synthases and the microscopic morphology were examined during the shift from a steady state with NO3− as the nitrogen source (induced chsB transcription) to growth on NH4+ (repressed chsB transcription). The expression analysis and the corresponding morphological data are shown in Fig. 5 and Table 4, respectively.
FIG. 5.
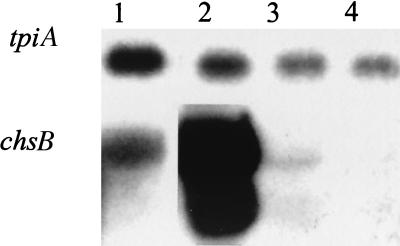
Expression of chsB in A. oryzae strain NiiA1 during chemostat cultivation. The tpiA gene was used as a loading control. Lane 1, control of chsB expression in strain A150 during batch cultivation; lane 2, sample taken at a dilution rate of 0.1 h−1 when the nitrogen source was NO3−; lane 3, sample taken 66.2 h after a shift to NH4+; lane 4, sample taken 406.9 h after a shift to NH4+. Refer to Table 4 for further details.
TABLE 4.
Microscopic morphology during chemostat cultivation of A. oryzae ChsB/G and NiiA1 strainsa
| Parameter | Value for strain:
|
|||||
|---|---|---|---|---|---|---|
| A1560 | ChsB/G | NiiA1 | ||||
| Time after nitrogen source shift (h) | ND | 0b | 66c | 336 | 407d | |
| Time after dilution rate step-up (h) | 0 | 71 | ||||
| Specific growth rate (h−1) | 0.10 | 0.10 | 0.10 | 0.10 | 0.10 | 0.16 |
| Nitrogen source | NH4+ | NH4+ | NO3− | NH4+ | NH4+ | NH4+ |
| lHGU (μm tip−1 ± 95% confidence interval) | 130 ± 14 | 83 ± 6 | 133 ± 9 | 112 ± 9 | 92 ± 7 | 87 ± 6 |
Two control experiments with the wild-type, A1560, and ChsB/G strains show the length of the hyphal growth unit, lHGU, with these strains. For the NiiA1 strain, at time zero the culture was in steady state growing on NO3−. At zero hours, the nitrogen source was shifted to NH4+. At the time points indicated, RNA was extracted, subjected to Northern blotting, and hybridized to probes specific for chsB and the control, tpi.
Northern blot sample 1.
Northern blot sample 2.
Northern blot sample 3.
Lane 2 of Fig. 5 shows that in the presence of NO3−, the niiA promoter is induced, generating considerable amounts of chsB mRNA. There was no difference between wild-type branching patterns or mRNA expression of chitin synthases on NH4+ and on NO3− (data not shown). When growing on NO3−, the NiiA1 strain had an lHGU similar to that of the wild-type strain growing under the same conditions, indicating that chsB expressed with the niiA promoter results in a branching pattern similar to that of the wild-type strain. In the NiiA1 chemostat cultivation, the nitrogen source was then shifted to NH4+, and this made the chsB expression level decrease; however, 66 h after the shift, mRNA of chsB was still present in the cells. This was possibly due to slow washout of NO3− in combination with the presence of multiple copies of the niiA-chsB construct in the genome and only slight repression of the niiA promoter on NH4+. In Fig. 5, there are possibly two bands for chsB in lanes 2 and 3. This may be due to a second, truncated product occurring due to the considerable amounts of chsB mRNA generated. The expression of chsC and csmA was also examined in the experiment, but the level of transcription of these chitin synthase genes did not indicate mutual regulation of chsC, csmA, and chsB. Sixty-six hours after the shift to NH4+, the lHGU had decreased, but even 336 h after the shift to NH4+, the lHGU was still higher than the lHGU for the strain with chsB disrupted (ChsB/G) under similar conditions (Table 4). This indicates that the niiA promoter is not completely silenced during growth on NH4+, as has previously been found for A. nidulans (24).
Location of α-amylase in the cell wall.
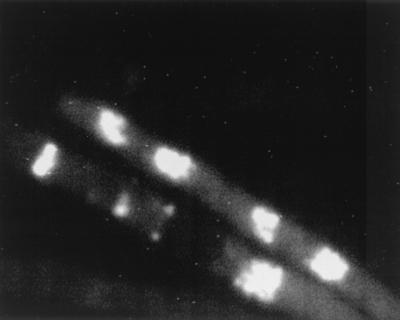
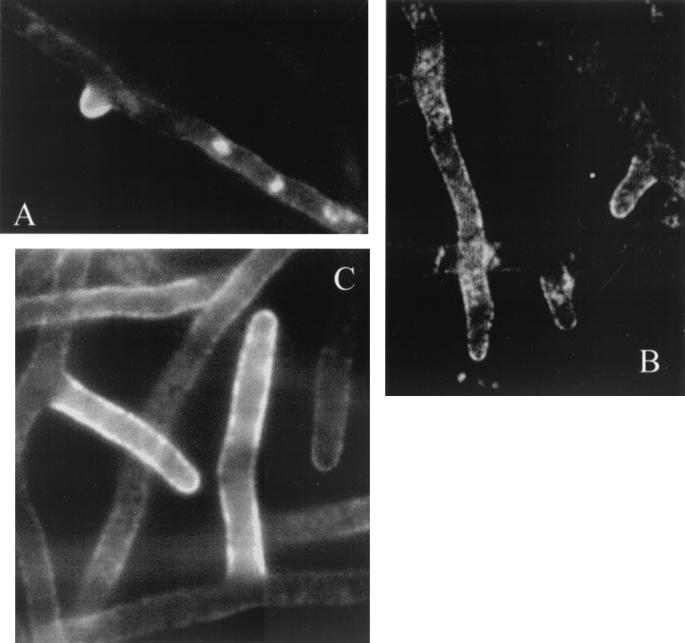
Previously, secretion of enzymes has been demonstrated in hyphal tips on solid medium. Wösten et al. (36) observed primary antibodies against glucoamylase located in agarose surrounding Aspergillus niger hyphal tips, and Spohr et al. (28) observed α-amylase in agarose surrounding growing tips of A. oryzae, supporting the “bulk flow hypothesis” (32). However, during growth on solid medium, secreted proteins will always be located around the hyphae, and therefore these studies are not conclusive. In this study, A. oryzae was grown submerged and α-amylase in the cell wall was detected by an indirect (two-stage) immunofluorescence reaction. The primary antibodies were polyclonal, so specificity was checked in two ways. There was no nonspecific binding of the secondary antibodies to the hyphal cell wall without primary antibodies, and only weak autofluorescence from the interior of the cells could be detected (data not shown). Using polyclonal rabbit antibodies raised against proteinase A from S. cerevisiae (PrA), we examined whether there was nonspecific binding to the fungal structure. Since A. oryzae does not secrete PrA, the areas where binding occurred should be nonspecific sites where nonspecific α-amylase antibodies would also bind. Some nonspecific binding of the antibodies was observed, as shown in Fig. 6. The tiny white dots indicate that the fluorescence has been amplified maximally in the camera (15 s). The fluorescence is mostly autofluorescence, which could also be detected without antibody staining (data not shown). Figure 7 shows the FITC labeling of antibodies directed against α-amylase, suggesting that α-amylase resides in the cell walls of hyphae. Staining was most intense in the parts of the cell wall which had just been synthesized, such as in new tips or in extending hyphae, while older cell walls (more than 100 μm from the tip) were not stained to the same degree.
FIG. 6.
FITC-labeled antibodies against proteinase A in A. oryzae to check specificity of α-amylase antibodies. Note the small dots signifying that the fluorescence has been amplified maximally in the camera by integration for approximately 15 s. The fluorescence is mostly weak intracellular autofluorescence, which can also be detected without FITC. The hyphal diameter is approximately 4 μm.
FIG. 7.
Staining of α-amylase in the cell wall of A. oryzae 1560. (A) Staining in the new tips and in the extending hyphal cell wall is clearly more intense than in the rest of the hyphae. (B) Staining of the cell wall in the new tip is very intense in contrast to the rest of the hyphae. Notice the nonspecific intracellular staining, which is also found when staining with PrA. (C) New tips and extending branch are stained visibly more than older branches. The fluorescence was amplified in the camera by integration for approximately 4 s. The staining intensity is lower when the hypha is not in focus. The hyphal diameter is approximately 4 μm.
DISCUSSION
Involvement of CsmA in branching.
The mechanism of branching in filamentous fungi and the relationship between branching and tip extension, if any, are poorly understood. Our results showed that a disrupted csmA gene leads to an increased number of subapical (but not apical) branches (Table 2). This supports the hypothesis (summarized in reference 18) that subapical branching may be regulated through an interplay between chitinolytic enzymes and chitin synthases. However, it was also found that subapical septal placement and subapical branch positioning are processes that are not affected by disruption of chsB or csmA.
Involvement of ChsB in branching.
It has been suggested that chsB has localized activity at the tips of extending hyphae (3). Our data for tip extension suggest that chsB may represent the main chitin synthase activity involved in apical hyphal extension. In the ChsB/G (chsB disruption) strain, both the estimated maximum tip extension rate and the average tip extension rate were 88% lower than in the A1560 strain (Table 3), indicating a severe defect in tip extension. However, the branching intensity in the ChsB/G strain was 2.9 times higher. As most of these branches were formed in the apical compartment (Table 2), this fits well with the apical-branching hypothesis of Trinci (31), who suggested that apical branching happens because the rate of supply of wall materials exceeds the rate with which the components can be incorporated into the apical-tip wall. The lack of a functional ChsB protein apparently lowers the incorporation rate considerably, mediating formation of an unusual number of apical branches. However, even though there may be an excess of wall materials, the chitin content of the ChsB/G strain is not significantly different from that of the wild-type strain (both had cell walls containing 7% ± 1% [wt/wt] N-acetylglucosamine) (Müller et al., submitted). This signifies that other (not so apically active) chitin synthases incorporate excess chitin monomers. This might also be the reason why the average hyphal diameter is increased in the strain with chsB disrupted (Table 2).
When the hyphae are grown at low glucose concentrations, the tip extension rates and branching frequencies (Table 4) both decrease in the wild-type strain. However, in the strain with chsB disrupted, only the branching frequency decreases, signifying that when chsB is disrupted the direct effect is a limitation on tip extension, with an increased branch formation being a consequence of this (or a secondary effect).
Does protein secretion depend on the number of tips?
For several years it has been debated whether the protein secretion level in fungal cultures depends on the number of tips (2, 17, 23, 33). Conflicting evidence has been presented on the matter, since to date no studies have examined the performance of morphologically genetically engineered strains specifically designed for protein production; only morphological mutants with unknown genotypes had been investigated. When the genotype is unknown, there may also be other, direct or indirect, effects of the mutation on productivity. However, it is clear that protein secretion occurs around the region of the advancing tip in filamentous fungi (11, 36), and this has been further illustrated here (Fig. 7). Our findings support the hypothesis that α-amylase is secreted from the hyphal apex by bulk flow and that some of the α-amylase is retained in the cell wall. In older parts of the cell wall, the α-amylase may have been released into the medium or incorporated into the cell wall by being denatured by, e.g., proteases. Directed genetic changes of, e.g., chsB, which affect the formation of apical branches (Table 2) in a high-yield A. oryzae strain, would be the way to ultimately elucidate whether protein secretion depends on the number of growing tips.
Effect of morphology on submerged cultivations.
The formation of long branched hyphal elements affects the medium rheology because they tend to interact or entwine, forming clumps or pellets (30). In the cases where large clumps are formed, the viscosity of the cultivation liquid is increased, resulting in reduced oxygen mass transfer and an increased fraction of oxygen-limited cells, as well as a decreased productivity and/or production of undesirable metabolites. Although filamentous fungi have been used in the fermentation industry for many years, the high cultivation viscosity is still a major unresolved problem (17, 22), and it makes a filamentous-fungal cultivation more difficult to aerate than single-cell cultivation with the same biomass concentration. Our findings for the ChsB/G (chsB disruption) strain show that it has an altered submerged morphology compared to the wild type; the strain is less prone to form large inseparable clumps (Fig. 2) due to a more branched mycelium. Therefore, strains with chsB disrupted should be easier to aerate and give a lower viscosity, and for these reasons it would be advantageous to use them in industrial fermentations.
The CM101 (csmA disruption) strain, however, had an lHGU similar to that of the wild-type strain (Table 2), but fewer and smaller inseparable clumps were formed with the CM101 strain (Fig. 2). One reason for this may be the disruption, resulting in a weakened cell wall structure, which is likely to cause fragmentation of the elements, especially later in the cultivation. Fragmentation has been observed for this strain (C. Müller, P. Szabo, K. Hansen, and J. Nielsen, unpublished results), and it may have prevented the formation of large pellets. Additionally, it may not only be the number of branches (lHGU) but the positioning of them (apically or subapically) which affects the formation and dimensions of pellets.
If these results prove to be applicable to other fungal strains and processes, manipulation of chitin synthases will represent a simple and inexpensive means to reduce viscosity (clump formation) during industrial filamentous-fungal fermentations.
Acknowledgments
We thank Tanja Andersson for help with the batch cultivations and Torben Christensen for help with the flowthrough cell cultivations.
The work on fungal morphology at the Center for Process Biotechnology is financially supported by Novozymes A/S, Bagsværd, Denmark.
REFERENCES
- 1.Agger, T., A. B. Spohr, M. Carlsen, and J. Nielsen. 1998. Growth and product formation of Aspergillus oryzae during submerged cultivations: verification of a morphologically structured model using fluorescent probes. Biotechnol. Bioeng. 57:321-329. [DOI] [PubMed] [Google Scholar]
- 2.Bocking, S. P., M. G. Wiebe, G. D. Robson, K. Hansen, L. H. Christiansen, and A. P. J. Trinci. 1999. Effect of branch frequency in Aspergillus oryzae on protein secretion and culture viscosity. Biotechnol. Bioeng. 65:638-648. [DOI] [PubMed] [Google Scholar]
- 3.Borgia, P. T., N. Iartchouk, P. J. Riggle, K. R. Winter, Y. Koltin, and C. Bulawa. 1996. The chsB gene from Aspergillus nidulans is necessary for normal hyphal growth and development. Fungal Genet. Biol. 20:193-203. [DOI] [PubMed] [Google Scholar]
- 4.Cadwell, I. Y., and A. P. J. Trinci. 1973. The growth unit of the mould Geotrichum candidum. Arch. Mikrobiol. 88:1-10. [DOI] [PubMed] [Google Scholar]
- 5.Carlsen, M. 1994. α-Amylase production by Aspergillus oryzae. Ph.D. thesis. Technical University of Denmark, Lyngby.
- 6.Carlsen, M., J. Nielsen, and J. Villadsen. 1996. Growth and α-amylase production by Aspergillus oryzae during continuous cultivations. J. Biotechnol. 45:81-93. [Google Scholar]
- 7.Carlsen, M., and J. Nielsen. 2001. Influence of carbon source on α-amylase production by Aspergillus oryzae. Appl. Microbiol. Biotechnol. 57:346-349. [DOI] [PubMed] [Google Scholar]
- 8.Christiansen, T., A. B. Spohr, and J. Nielsen. 1999. On-line study of growth kinetics of single hyphae of Aspergillus oryzae in a flow-through cell. Biotechnol. Bioeng. 63:147-153. [DOI] [PubMed] [Google Scholar]
- 9.Cox, P. W., G. C. Paul, and C. R. Thomas. 1998. Image analysis of the morphology of filamentous microorganisms. Microbiology 144:817-827. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara, M., H. Horiuchi, A. Ohta, and M. Takagi. 1997. A novel fungal gene encoding chitin synthase with a myosin motor-like domain. Biochem. Biophys. Res. Commun. 236:75-78. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, C. L., V. Khalaj, A. F. Ram, D. B. Archer, J. L. Brookman, A. P. J. Trinci, D. J. Jeenes, J. H. Doonan, B. Wells, P. J. Punt, C. A. van den Hondel, and G. D. Robson. 2000. Glucoamylase::green fluorescent protein fusions to monitor protein secretion in Aspergillus niger. Microbiology 146:415-426. [DOI] [PubMed] [Google Scholar]
- 12.Hansen, P. V. 1984. Determination of fungal alpha-amylase by flow injection analysis. Anal. Chim. Acta 158:375-377. [Google Scholar]
- 13.Horiuchi, H., M. Fujiwara, S. Yamashita, A. Ohta, and M. Takagi. 1999. Proliferation of intrahyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 181:3721-3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Horiuchi, H., and M. Takagi. 1999. Chitin synthase genes of Aspergillus species, p. 193-204. In A. A. Brakhage, B. Jahn, and A. Schmidt (ed.), Aspergillus fumigatus. Contributions to microbiology, vol. 2. Karger, Basel, Switzerland. [DOI] [PubMed] [Google Scholar]
- 15.Johansen, C. L., L. Coolen, and J. H. Hunik. 1998. Influence of morphology on product formation in Aspergillus awamori during submerged fermentations. Biotechnol. Prog. 14:233-240. [DOI] [PubMed] [Google Scholar]
- 16.Lejeune, R., J. Nielsen, and G. V. Baron. 1995. Morphology of Trichoderma reesei QM 9414 in submerged cultures. Biotechnol. Bioeng. 47:609-615. [DOI] [PubMed] [Google Scholar]
- 17.McIntyre, M., C. Müller, J. Dynesen, and J. Nielsen. 2001. Metabolic engineering of the morphology of Aspergillus. Adv. Biochem. Eng. Technol. 73:103-128. [DOI] [PubMed] [Google Scholar]
- 18.Merz, R. A., M. Horsch, L. E. Nyhlén, and D. M. Rast. 1999. Biochemistry of chitin synthase. EXS 87:9-37. [DOI] [PubMed] [Google Scholar]
- 19.Metz, B., W. F. Kossen, and J. C. van Suijdam. 1979. The rheology of mould suspensions. Adv. Biochem. Eng. 11:103-156. [Google Scholar]
- 20.Müller, C., A. B. Spohr, and J. Nielsen. 2000. Role of substrate concentration in mitosis and hyphal extension of Aspergillus. Biotechnol. Bioeng. 67:390-397. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen, J. 1992. Modelling of the growth of filamentous fungi. Adv. Biochem. Eng. Biotechnol. 46:187-223. [DOI] [PubMed] [Google Scholar]
- 22.Olsvik, E. S., and B. Kristiansen. 1994. Rheology of filamentous fermentations. Biotechnol. Adv. 12:1-39. [DOI] [PubMed] [Google Scholar]
- 23.Peberdy, J. F. 1994. Protein secretion in filamentous fungi--trying to understand a highly productive black box. Trends Biotechnol. 12:50-57. [DOI] [PubMed] [Google Scholar]
- 24.Punt, P. J., J. Strauss, R. Smit, J. R. Kinghorn, C. A. van den Hondel, and C. Scazzocchio. 1995. The intergenic region between the divergently transcribed niiA and niaD genes of Aspergillus nidulans contains multiple NirA binding sites which act bidirectionally. Mol. Cell. Biol. 15:5688-5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riley, G. L., K. G. Tucker, G. C. Paul, and C. R. Thomas. 2000. Effect of biomass concentration and mycelial morphology on fermentation broth rheology. Biotechnol. Bioeng. 68:160-172. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Specht, C. A., Y. Liu, P. W. Robbins, C. E. Bulawa, N. Iartchouk, K. R. Winter, P. J. Riggle, J. C. Rhodes, C. L. Dodge, D. W. Culp, and P. T. Borgia. 1996. The chsD and chsE genes of Aspergillus nidulans and their role in chitin synthesis. Fungal Genet. Biol. 20:153-167. [DOI] [PubMed] [Google Scholar]
- 28.Spohr, A. B., T. Agger, M. Carlsen, and J. Nielsen. 1998. Quantitative morphology of filamentous microorganisms, p. 373-410. In M. H. F. Wilkinson and F. Schut (ed.), Digital image analysis of microbes: imaging, morphology, fluorometry and motility techniques and applications. John Wiley & Sons, Chichester, United Kingdom.
- 29.Spohr, A. B., C. Dam-Mikkelsen, M. Carlsen, J. Nielsen, and J. Villadsen. 1998. On-line study of fungal morphology during submerged growth in a small flow-through cell. Biotechnol. Bioeng. 58:541-553. [PubMed] [Google Scholar]
- 30.Thomas, C. R., and G. C. Paul. 1996. Applications of image analysis in cell technology. Curr. Opin. Biotechnol. 7:35-45. [DOI] [PubMed] [Google Scholar]
- 31.Trinci, A. P. J. 1979. The duplication cycle and branching in fungi, p. 319-358. In J. H. Burnett and A. P. J. Trinci (ed.), Fungal walls and hyphal growth. Cambridge University Press, London, United Kingdom.
- 32.Wessels, J. G. H. 1990. Cell wall architecture and fungal tip growth, p. 1-29. In I. B. Heath (ed.), Tip growth in plants and fungal cells. Academic Press, San Diego, Calif.
- 33.Wessels, J. G. H. 1993. Wall growth, protein secretion and morphogenesis in fungi. New Phytol. 123:397-413. [DOI] [PubMed] [Google Scholar]
- 34.Wiebe, M. G., and A. P. J. Trinci. 1991. Dilution rate as a determinant of mycelial morphology in continuous culture. Biotechnol. Bioeng. 38:75-81. [DOI] [PubMed] [Google Scholar]
- 35.Wittler, R., H. Baumgartl, D. W. Lübbers, and K. Schürgerl. 1986. Investigations of oxygen transfer into Penicillium chrysogenum pellets by microprobe measurements. Biotechnol. Bioeng. 28:1024-1036. [DOI] [PubMed] [Google Scholar]
- 36.Wösten, H. A., S. M. Moukha, J. H. Sietsma, and J. G. H. Wessels. 1991. Localization of growth and secretion of proteins in Aspergillus niger. J. Gen. Microbiol. 137:2017-2023. [DOI] [PubMed] [Google Scholar]
- 37.Yanai, K., N. Kojima, N. Takaya, H. Horiuchi, A. Ohta, and M. Takagi. 1994. Isolation and characterization of two chitin synthase genes from Aspergillus nidulans. Biosci. Biotechnol. Biochem. 58:1828-1835. [DOI] [PubMed] [Google Scholar]