Abstract
The specificity of Bacillus stearothermophilus TRS40 neopullulanase toward amylose and amylopectin was analyzed. Although this neopullulanase completely hydrolyzed amylose to produce maltose as the main product, it scarcely hydrolyzed amylopectin. The molecular mass of amylopectin was decreased by only one order of magnitude, from approximately 108 to 107 Da. Furthermore, this neopullulanase selectively hydrolyzed amylose when starch was used as a substrate. This phenomenon, efficient hydrolysis of amylose but not amylopectin, was also observed with cyclomaltodextrinase from alkaliphilic Bacillus sp. strain A2-5a and maltogenic amylase from Bacillus licheniformis ATCC 27811. These three enzymes hydrolyzed cyclomaltodextrins and amylose much faster than pullulan. Other amylolytic enzymes, such as bacterial saccharifying α-amylase, bacterial liquefying α-amylase, β-amylase, and neopullulanase from Bacillus megaterium, did not exhibit this distinct substrate specificity at all, i.e., the preference of amylose to amylopectin.
We previously found a new type of pullulan-hydrolyzing enzyme, neopullulanase (EC 3.2.1.135) from Bacillus stearothermophilus TRS40 (15), and showed that it catalyzes the hydrolysis of α-1,4- and α-1,6-glucosidic linkages (6), as well as transglycosylation to form α-1,4- and α-1,6-glucosidic linkages (27). The replacement of several amino acid residues that constitute the active center of the neopullulanase showed that one active center of the enzyme participated in all four of the reactions described above (17). Based on this series of experimental results using the neopullulanase and the structural similarities of the enzymes that catalyze these four reactions, we proposed and defined a general idea for one enzyme family, an α-amylase family (27). Based on the concept of the α-amylase family (13), we controlled the substrate preference and transglycosylation activity of the neopullulanase (14), and Preiss and his coworkers (16) analyzed the regions that determined the specificity of maize branching enzyme (EC 2.4.1.18) isoforms.
The neopullulanase hydrolyzes pullulan to produce panose (Glc pα1-6Glc pα1-4Glc) as the main product. Maltose and glucose are also produced as the final products. The final molar ratio of panose, maltose, and glucose is about 3:1:1 (6). Neopullulanases have been reported for other bacteria, such as Bacteroides thetaiotaomicron 95-1 (25), alkalophilic Bacillus (5), and Bacillus polymyxa CECT155 (33). Most of the α-amylases that have been investigated so far could not hydrolyze pullulan. However, neopullulanase-type α-amylase has also been reported for Thermoactinomyces vulgaris (31). Maltogenic amylase (EC 3.2.1.133) from Bacillus licheniformis ATCC 27811 also hydrolyzed pullulan to produce panose (9).
Starch constitutes most of the dry matter in certain crops, and it therefore is not only the primary source of calories in the human diet but can also be regarded as a renewable resource that can be used in many industrial applications. It is a mixture of two macromolecules: amylose, which is essentially composed only of α-1,4-linked glucose polymers, and amylopectin, which is composed of α-1,4-linked glucose polymers branched by α-1,6 linkages. The possibility that amylolytic enzymes may have different specificities towered amylose and amylopectin has been discussed previously (8, 15).
We describe here a unique macromolecule recognition by B. stearothermophilus neopullulanase in comparison with other amylolytic enzymes. The similar substrate preferences of a cyclomaltodextrinase (EC 3.2.1.54) and a maltogenic amylase are also reported.
MATERIALS AND METHODS
Chemicals.
Synthetic amyloses with average molecular masses of 70, 110, 320, and 1,000 kDa (amyloses AS-70, -110, -320, and -1000, respectively) were purchased from Nakano Vinegar Co., Ltd. (Aichi, Japan). Amylose EX-I (average degree of polymerization, 17), amylose EX-III (average degree of polymerization, 117), and pullulan (average molecular mass, 98 kDa) were purchased from Hayashibara Biochemical Laboratories Inc. (Okayama, Japan). Highly branched cyclic dextrin was prepared in our laboratory (28). Amylose and amylopectin from potato starch were purchased from Sigma Chemical Co. (St. Louis, Mo.). Potato starch and waxy maize starch were obtained from Nihon Shokuhin Kako Co. Ltd. (Tokyo, Japan). Cyclomaltodextrins, soluble starch, and other chemical reagents were purchased from Wako Pure Chemical Industries Ltd. (Osaka, Japan).
Enzymes.
The neopullulanase was purified from the culture broth of Bacillus subtilis ANA-1 (21) (arg-15 hsdR hsdM ΔaprA3 amyE npr) carrying pPP10 (Tcr nplT+ [structural gene of the neopullulanase from B. stearothermophilus TRS40]) as described previously (15). Bacterial liquefying α-amylase from Bacillus liqueniformis was purchased from Sigma. Bacterial saccharifying α-amylase from B. subtilis was from Nagase Chemtex (Osaka, Japan). Maltogenic α-amylase from B. stearothermophilus (Novamyl) (1) was obtained from Novozymes (Tokyo, Japan). β-Amylase from soybean was purchased from Nagase Chemtex. Isoamylase from Pseudomonas amyloderamosa was purchased from Hayashibara Biochemical Laboratories. Cyclomaltodextrinase, B. licheniformis maltogenic amylase, and Bacillus megaterium neopullulanase were prepared according to the procedures described below.
Enzyme assays.
The activities of the amylolytic enzymes were assayed using potato amylose as a substrate. The reaction mixture (200 μl) containing 0.5% potato amylose in buffer (described below) and the enzyme was incubated at 40°C (cyclomaltodextrinase, B. licheniformis maltogenic amylase, and B. megaterium neopullulanase) or 50°C (B. stearothermophilus neopullulanase, bacterial saccharifying α-amylase, bacterial liquefying α-amylase, Novamyl, and β-amylase) for 10 min. The buffer was 100 mM sodium acetate buffer (pH 5.5) for bacterial liquefying α-amylase, bacterial saccharifying α-amylase, and β-amylase; 100 mM sodium acetate buffer (pH 5.0) for Novamyl; 100 mM sodium 2-(N-morpholino)ethanesulfonic acid (MES-Na) buffer (pH 6.0) for B. stearothermophilus neopullulanase and B. megaterium neopullulanase; and 100 mM sodium phosphate buffer (pH 6.5) for cyclomaltodextrinase and B. licheniformis maltogenic amylase. The reducing sugar after the enzyme reaction was assayed based on the 3,5-dinitrosalicylic acid method as described previously (22). One unit of enzyme activity was defined as the amount of enzyme needed to produce 1 μmol of reducing sugar as glucose per min from potato amylose under the assay conditions described above.
Expression of the cyclomaltodextrinase gene in Escherichia coli.
The cyclomaltodextrinase gene was amplified by PCR from chromosomal DNA of alkalophilic Bacillus sp. strain A2-5a (23). The forward primer 5′-TGGCCATGGTAAAAGAAGCGATTTA-3′ introduces an NcoI site (underlined) around the authentic ATG codon and is located between positions 10960 and 10979 of the cyclomaltodextrinase gene (GenBank accession no. AB015670). The reverse primer 5′-TTCGGTACCTTAAATCACCTTTATAACACC-3′ introduces a KpnI site (underlined) and is located between positions 9217 and 9236 of the gene. These primers generate mutations at the second codon from TTA to GTA and at the stop codon from TAG to TAA. The amplified fragment was digested with NcoI and KpnI and then introduced into the NcoI-KpnI site in pKK388-1 (Clontech, Palo Alto, Calif.) to obtain the expression plasmid pNPR63.
E. coli TG-1 {supE hsdΔ5 thi Δ(lac-proAB)/F′ [traD36 proAB+ lacIq lacZΔM15]} carrying pNPR63 was grown in Terrific broth (Gibco BRL) containing 50 μg of ampicillin per ml at 37°C until the late log phase. The inducer isopropyl-β-d-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.1 mM, and the culture was incubated for 20 h at 15°C. Cells were harvested and washed with 10 mM sodium phosphate buffer (pH 7.5) (buffer A) and disrupted by sonication in buffer A at 4°C. Cell debris was removed from the crude extract by centrifugation, and the enzyme was precipitated by adding solid (NH4)2SO4 to 80% saturation. The precipitate was redissolved and dialyzed against buffer A. The dialysate was loaded onto a Q-Sepharose Fast Flow column (Pharmacia) equilibrated with buffer A. After the column was washed with buffer A containing 0.2 M NaCl, the enzyme was eluted with buffer A containing 0.4 M NaCl. The enzyme was dialyzed against buffer A, loaded onto a Resource Q column (Pharmacia) equilibrated with buffer A, and eluted with a linear gradient of 0.2 to 0.4 M NaCl in buffer A. The active fractions were pooled, dialyzed against buffer A, and stored at 4°C. The optimum activity of the cyclomaltodextrinase was observed at pH 6.5 and 35 to 40°C.
Expression of the maltogenic amylase gene in E. coli.
The maltogenic amylase gene was amplified by PCR from chromosomal DNA of B. licheniformis ATCC27811. The forward primer 5′-TGGCATATGATCGAATTAGCAGCGATAC-3′ introduces an NdeI site (underlined) upstream of the start codon and is located between positions 292 and 313 of the maltogenic amylase gene (GenBank accession no. X67133). The reverse primer 5′-CTTGAATTCTTAACAGAATTTAGACCGC-3′ introduces an EcoRI site (underlined), changes the stop codon from the authentic TAG to TAA, and is located between positions 2010 and 2027 of the gene. The amplified fragment was digested with NdeI and EcoRI and then introduced into the NdeI-EcoRI site in pGEX-Nde2 to obtain the expression plasmid pNPR422. The expression vector pGEX-Nde2 is a derivative of pGEX-5X-3 (Amersham) and was constructed by introducing four nucleotides into the unique XbaI site of pGEX-Nde (30).
E. coli TG-1 carrying pNPR422 was grown in Terrific broth containing 50 μg of ampicillin per ml at 37°C until the late log phase. IPTG was then added to a final concentration of 0.1 mM, and the culture was incubated for 20 h at 15°C. Cells were harvested and washed with buffer A and disrupted by sonication in buffer A at 4°C. Cell debris was removed from the crude extract by centrifugation, and the enzyme was precipitated by adding solid (NH4)2SO4 to 70% saturation. The precipitate was redissolved and dialyzed against buffer A. The dialysate was loaded onto a Q-Sepharose Fast Flow column equilibrated with buffer A. After the column was washed with buffer A containing 0.4 M NaCl, the enzyme was eluted with buffer A containing 1 M NaCl. The enzyme was dialyzed against buffer A and stored at 4°C.
Expression of the B. megaterium neopullulanase gene in B. subtilis.
Yebra et al. (33) reported an extracellular neopullulanase from B. polymyxa CECT155. The enzyme gene was cloned and expressed in Saccharomyces cerevisiae (34). Since we could not obtain B. polymyxa CECT155, we used B. megaterium NCIMB11568 as a source for the gene of an extracellular neopullulanase. The B. polymyxa and B. megaterium neopullulanases are 63% identical at the amino acid sequence level (20). Both enzymes have a typical signal sequence for secretion (20, 34), which is not found in B. stearothermophilus neopullulanase (12). Therefore, the B. megaterium neopullulanase gene was likely to encode an extracellular neopullulanase. This was confirmed after characterization of the B. megaterium neopullulanase as described below. The B. megaterium neopullulanase gene was amplified by PCR from chromosomal DNA of B. megaterium NCIMB11568. The forward primer 5′-TCCACTAGTAAAGGGAAAAAATGGACAGC-3′ introduces an SpeI site (underlined) upstream of the second codon of the B. megaterium neopullulanase gene nucleotide sequence (GenBank accession no. X07261) and is located between bases 110 and 129. The reverse primer 5′-TTCGGATCCTCCGTGGGTTCATC-3′ creates a BamHI site and is located between bases 1687 and 1700. The amplified gene was digested with SpeI and BamHI and then introduced into the SpeI-BamHI site in the E. coli-Bacillus shuttle vector pWH1520 (MoBiTec GmbH) to obtain the expression plasmid pNPR78. This plasmid encodes a fusion protein of the four N-terminal amino acids of the XylA protein of pWH1520 and B. megaterium neopullulanase.
B. subtilis ANA1 (arg-15 hsdR hsdM ΔaprA3 amyE npr) carrying pNPR78 was grown at 37°C in L broth (1% Tryptone [Difco], 0.5% yeast extract [Difco], 0.5% NaCl [pH 7.3]) containing 20 μg of tetracycline per ml. At the mid log phase, xylose was added to a final concentration of 0.5% to induce expression of the B. megaterium neopullulanase gene. After 22 h, the culture supernatant was collected and the enzyme was precipitated by adding solid (NH4)2SO4 to 80% saturation. The precipitate was redissolved and dialyzed against buffer A. The dialysate was loaded onto a Q-Sepharose Fast Flow column equilibrated with buffer A. The active fraction was not absorbed by the column. The enzyme solution was again passed through the column, concentrated using polyethylene glycol (molecular mass, 20 kDa), and stored at 4°C.
TLC.
Thin-layer chromatography (TLC) was carried out in the ascending mode on a silica gel plate (Merck, Darmstadt, Germany) with a solvent system of acetonitrile-water (8:2, vol/vol) and developed three times. Sugar spots were visualized by spraying with H2SO4-methanol (1:1, vol/vol) and then heating at 130°C.
Gel permeation chromatography.
The degradation of amylose and amylopectin was analyzed by gel permeation chromatography using connected Superose 6 (10 by 300 mm; Pharmacia) and Superdex 30 (10 by 300 mm; Pharmacia) columns. Elution was carried out at room temperature with 100 mM NaCl at a flow rate of 1 ml/min. Eluted carbohydrates were detected with a refractive index (RI) detector (RID-6A; Shimadzu, Kyoto, Japan).
Measurement of amylose or amylopectin contents during neopullulanase action on starch.
Amylose was fractionated from the reaction mixture by a modification (4) of the method of Lansky et al. (18) and purified by recrystallization from aqueous 10% 1-butanol. Amylopectin was fractionated by precipitation by adding a sevenfold volume of ethanol after removing the amylose fraction. The low-molecular-mass products, which contain maltose as a main component, remained in the supernatant after the precipitation of amylopectin. The amylose and amylopectin fractions were dissolved in 90% dimethyl sulfoxide (DMSO). The total carbohydrate contents of these three fractions were measured by the phenol-sulfuric acid method (2). The degradation ratio was calculated from the contents of the reducing sugar as maltose per total carbohydrate. The content of reducing sugar was measured by the Somogyi-Nelson method (26).
Measurement of the molecular mass of each fractionated product from amylose or amylopectin.
The molecular mass was determined by high-performance liquid chromatography with a multiangle-laser light-scattering photometer (MALLS) (DAWN DSP; Wyatt Technology Co., Ltd., Santa Barbara, Calif.) and a differential RI detector (RI-71; Showa Denko, Tokyo, Japan). The columns were Shodex OH-pak SB-806 M HQ (6.0 by 300 mm; Showa Denko) with an OH-pak SB-G precolumn. Elution was carried out at 40°C with 0.1 M NaNO3 at a flow rate of 1.0 ml/min. Samples (5 mg/ml; 25 μl) dissolved in the eluent were injected into the column after filtration through a 0.22-μm-pore-size membrane (Millex GS; Millipore). Light scattering and RI signals were transferred to the computer to calculate the weight-average molecular mass according to the instruction manual (Wyatt Technology Co.) for the DAWN DSP.
Analysis of the chain length of amylopectin.
Amylopectin and amylopectin reacted with neopullulanase were debranched with isoamylase at 40°C for 16 h as described previously (29). The products were analyzed by high-performance anion-exchange chromatography (HPAEC) with a DX-300 (Dionex Corp., Sunnyvale, Calif.) gradient chromatography system. The chromatographic conditions were as follows: column, CarboPac PA-100 (4 by 250 mm; Dionex Corp.); detector, pulsed amperometric detector; meter scale, 1 μC; temperature, ambient. Elution was performed using 100 mM NaOH solution containing the following gradient of 1 M sodium acetate: 0% (vol/vol) at 0 min, 10% (vol/vol) at 12 min, 20% (vol/vol) at 32 min, 20% (vol/vol) at 37 min, and 80% (vol/vol) at 57 min (flow rate, 1 ml/min).
RESULTS
Degradation of amylose or amylopectin with B. stearothermophilus neopullulanase and other amylolytic enzymes.
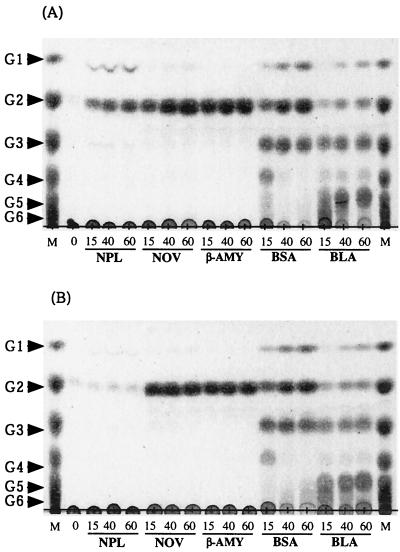
The 1% amylose and 1% amylopectin (both from potato starch) solutions were prepared from the 10% substrates solutions in 90% DMSO by dilution with deionized water. The diluted solution was used immediately for the enzyme reaction. The substrate solution (300 μl) was individually mixed with enzyme solutions (300 μl each) that contained equal amounts of activity (12 U) toward potato amylose. Bacterial liquefying α-amylase, bacterial saccharifying α-amylase, and β-amylase were solubilized in 200 mM sodium acetate (pH 5.5); Novamyl was solubilized in 200 mM sodium acetate buffer (pH 5.0); and B. stearothermophilus neopullulanase was solubilized in 200 mM sodium MES-Na buffer (pH 6.0). After an appropriate incubation period at 50°C, 20-μl samples were collected, and the reaction was stopped by heat treatment at 100°C for 5 min. Three microliters of each sample was applied to TLC (Fig. 1). Novamyl, β-amylase, bacterial liquefying α-amylase, and bacterial saccharifying α-amylase efficiently hydrolyzed both amylose and amylopectin and produced the same products from each. Although B. stearothermophilus neopullulanase efficiently hydrolyzed amylose to produce maltose and a small amount of glucose, it scarcely hydrolyzed amylopectin (Fig. 1). These results indicate that the specificity of B. stearothermophilus neopullulanase toward amylose and amylopectin was clearly different from those of other amylolytic enzymes.
FIG. 1.
Thin-layer chromatograms of reaction products of amylolytic enzymes from amylose (A) or amylopectin (B). Lane M, standard maltooligosaccharides; G1, G2, G3, G4, G5, and G6, glucose, maltose, maltotriose, maltotetraose, maltopentaose, and maltohexaose, respectively. NPL, NOV, β-AMY, BSA, and BLA, B. stearothermophilus neopullulanase, Novamyl, β-amylase, bacterial saccharifying α-amylase, and bacterial liquefying α-amylase, respectively. The reaction times (minutes) are indicated below the lanes.
Confirmation of the selective hydrolysis of amylose by B. stearothermophilus neopullulanase.
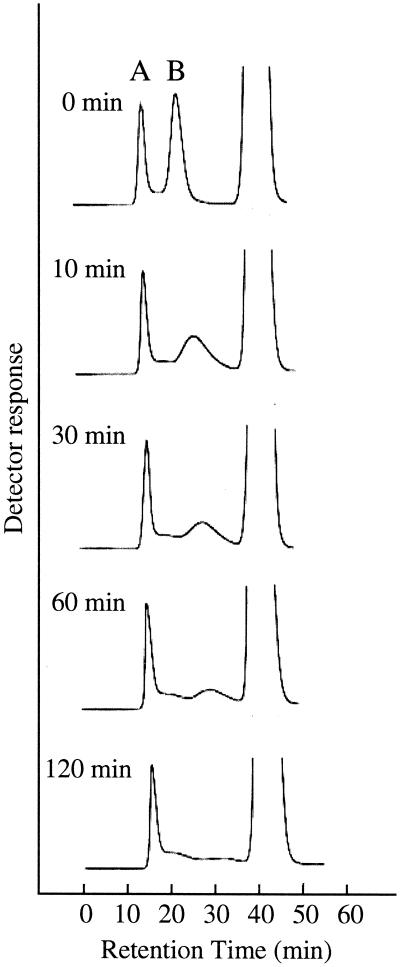
Synthetic amylose (amylose AS-70) or amylopectin (from potato starch) solutions were prepared from the 10% substrate solutions in 90% DMSO by dilution with deionized water. The diluted solution was used immediately for enzyme treatment. A reaction mixture (1.5 ml) consisting of 0.5% synthetic amylose, 0.5% potato amylopectin, and B. stearothermophilus neopullulanase (0.6 U) in 100 mM Mes-Na buffer (pH 6.0) was incubated at 50°C. After incubation for 0, 10, 30, 60, and 120 min, 270-μl samples were collected, and the reaction was terminated by heat treatment at 100°C for 5 min. After removal of the precipitate by centrifugation, the supernatant (250 μl) was subjected to gel permeation chromatography. Since synthetic amylose with an average molecular mass of 70 kDa and potato amylopectin were used as the substrates, these two components were clearly fractionated by column chromatography, as shown in Fig. 2. Although the peak of amylose gradually disappeared upon treatment with B. stearothermophilus neopullulanase, the peak of amylopectin was not affected even after 120 min. This result indicates that B. stearothermophilus neopullulanase selectively hydrolyzes amylose even in a mixture of amylose and amylopectin.
FIG. 2.
Time course of the reaction of B. stearothermophilus neopullulanase on a mixture of amylopectin (peak A) and amylose (peak B). The elution profile was analyzed by gel permeation chromatography.
Action of B. stearothermophilus neopullulanase on starch.
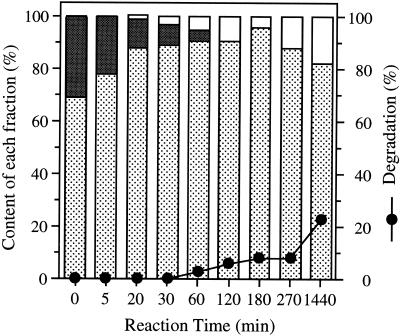
Potato starch, consisting of about 20% amylose and 80% amylopectin, was solubilized by heating and used as a substrate for B. stearothermophilus neopullulanase. A reaction mixture (10 ml) consisting of 1.0% potato starch and B. stearothermophilus neopullulanase (40 U) in 100 mM MES-Na buffer (pH 6.0) was incubated at 50°C. After incubation for 0, 5, 10, 20, 30, 60, 120, 180, 270, and 1,440 min, 1-ml samples were collected, and the reaction was terminated by heat treatment at 100°C for 5 min. After removal of the precipitate by centrifugation, the supernatant was used to analyze of the contents of amylose, amylopectin, and low-molecular-mass fractions. These three fractions were obtained by using 1-butanol as described in Materials and Methods. Figure 3 shows the contents of these fractions during the action of B. stearothermophilus neopullulanase on potato starch. The amylose fraction gradually decreased with enzyme treatment, while the amylopectin fraction was not significantly affected during the reaction period. The low-molecular-mass fraction increased in response to the decrease in the amylose fraction. The degradation rate was calculated from the amount of reducing sugar as maltose per total carbohydrate, and it was assumed that the low-molecular-mass fraction was mainly maltose and was produced only from the amylose fraction (Fig. 3). The results indicated that B. stearothermophilus neopullulanase selectively hydrolyzed amylose even in native starch.
FIG. 3.
Time courses of the contents of amylose, amylopectin, and low-molecular-mass fractions during the reaction of B. stearothermophilus neopullulanase on potato starch. The contents of amylopectin (stippled bars), amylose (gray bars), and low-molecular-mass (white bars) fractions are shown against the amount of total carbohydrate. The degradation ratio in each reaction period is shown as a percentage of reducing sugar as maltose per total carbohydrate.
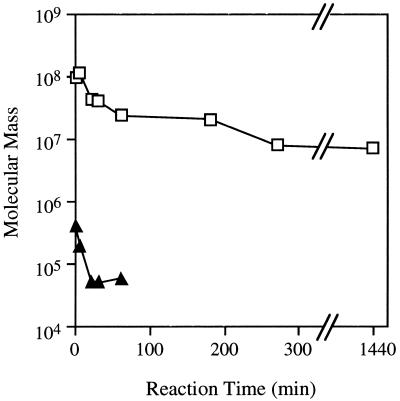
The time courses of the changes in the molecular masses of the amylose and amylopectin fractions during the B. stearothermophilus neopullulanase reaction were analyzed by high-performance liquid chromatography with MALLS and RI (Fig. 4). The molecular mass of amylose rapidly decreased below 105 Da within 120 min, and the amylose fraction was not observed after 120 min (Fig. 3). Therefore, it is most likely that the amylose was completely hydrolyzed to maltooligosaccharides, mainly maltose. The molecular mass of the native amylopectin from potato was greater than 108 Da. Although the molecular mass of potato amylopectin slightly decreased to approximately 107 Da, it was not reduced further by B. stearothermophilus neopullulanase (Fig. 4). Even when an excess amount of the enzyme (400 U, 10 times the amount used in the case described above) was used for the same reaction period (1,440 min), the same phenomenon was observed. Similar results were observed in experiments using starches from other sources, such as tapioca, maize, rice, and wheat.
FIG. 4.
Time courses of the molecular masses of the amylopectin (□) and amylose (•) fractions during the reaction of B. stearothermophilus neopullulanase on potato starch. The molecular mass was measured by high-performance liquid chromatography with the MALLS system.
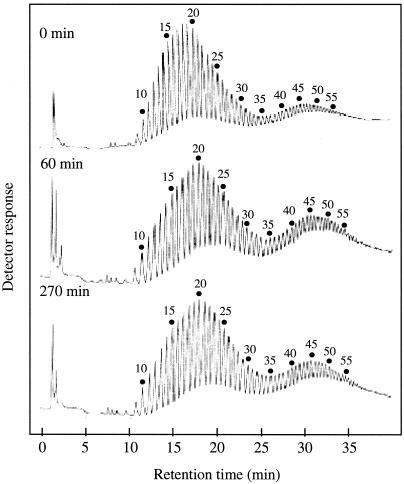
The change in the chain length distribution of amylopectin after treatment with B. stearothermophilus neopullulanase was investigated by HPAEC (Fig. 5). There was little difference in chain length distribution between the neopullulanase-treated amylopectin and unreacted amylopectin.
FIG. 5.
Chain length distribution of amylopectin after treatment with B. stearothermophilus neopullulanase. A reaction mixture (1.5 ml) consisting of 0.5% potato amylopectin and B. stearothermophilus neopullulanase (0.6 U) in 100 mM Mes-Na buffer (pH 6.0) was incubated at 50°C. After incubation for 0, 60, and 270 min, 270-μl samples were collected, and the reaction was terminated by heat treatment at 100°C for 5 min. The products were completely debranched by isoamylase as described previously (29). The solutions of the debranched products (25 μl; 100 μg each) were analyzed by HPAEC. The numbers with dots are degrees of polymerization.
Comparison of the substrate specificities of B. stearothermophilus neopullulanase and other amylolytic enzymes.
The initial velocities of the reactions of B. stearothermophilus neopullulanase and other amylolytic enzymes with various substrates were measured quantitatively (Table 1). Reaction mixtures (200 μl) containing 0.5% substrates and enzyme were incubated at 40°C (cyclomaltodextrinase, B. licheniformis maltogenic amylase, and B. megaterium neopullulanase) or 50°C (B. stearothermophilus neopullulanase, bacterial saccharifying α-amylase, bacterial liquefying α-amylase, Novamyl, and β-amylase) for 10 min. Potato amylose, synthetic amylose, maltooligosaccharides, amylopectin, waxy maize starch, highly branched cyclic dextrin, potato starch, cyclomaltodextrins, and pullulan were used as the substrate for the enzyme reaction. The same buffer as in the enzyme assay was used for each enzyme. The same amount of each enzyme (0.76 U) was used, based on potato amylose-hydrolyzing activity.
TABLE 1.
Comparison of specificities of various amylolytic enzymes
| Substrate | Initial velocitya
|
|||||||
|---|---|---|---|---|---|---|---|---|
| NPL | CDase | BLMA | BMNP | BSA | BLA | NOV | β-AMY | |
| Potato amylose | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 | 100.0 |
| Amylose AS-1000 | 114.3 | 122.9 | 140.6 | 106.6 | 116.5 | 95.5 | 66.1 | 14.9 |
| Amylose AS-320 | 106.1 | 130.2 | 130.4 | 96.6 | 101.9 | 82.0 | 64.6 | 26.4 |
| Amylose AS-110 | 119.8 | 148.6 | 154.6 | 89.1 | 106.6 | 85.3 | 65.0 | 65.8 |
| Amylose AS-70 | 133.1 | 155.4 | 153.0 | 89.8 | 112.1 | 90.1 | 65.4 | 98.0 |
| Amylose EX-III | 161.7 | 212.0 | 232.6 | 85.1 | 107.9 | 85.8 | 89.8 | 138.6 |
| Amylose EX-I | 194.2 | 265.6 | 261.7 | 60.3 | 66.6 | 56.5 | 80.5 | 143.6 |
| Maltoheptaose | 117.1 | 173.1 | 291.0 | 23.8 | 19.6 | 44.7 | 67.1 | 38.0 |
| Maltohexaose | 115.3 | 141.5 | 254.3 | 23.5 | 22.2 | 13.9 | 64.3 | 42.6 |
| Maltopentaose | 107.0 | 178.1 | 246.1 | 13.6 | 13.3 | 8.2 | 61.1 | 29.6 |
| Maltotetraose | 67.4 | 87.9 | 171.4 | 14.6 | 8.6 | 11.2 | 35.2 | 22.5 |
| Maltotriose | 24.9 | 69.9 | 46.7 | NDb | ND | ND | 22.6 | ND |
| Potato amylopectin | ND | ND | ND | 74.9 | 83.5 | 76.4 | 115.9 | 85.9 |
| Waxy maize starch | ND | ND | ND | 61.9 | 76.5 | 78.6 | 129.6 | 114.2 |
| Higly branched cyclic dextrin | 14.9 | ND | ND | 57.7 | 74.2 | 70.0 | 127.1 | 98.5 |
| Potato starch | 31.9 | 10.8 | 26.0 | 59.5 | 77.2 | 66.5 | 92.5 | 62.4 |
| α-Cyclomaltodextrin | 413.7 | 400.5 | 568.9 | ND | ND | ND | ND | ND |
| β-Cyclomaltodextrin | 401.8 | 481.6 | 449.8 | 18.2 | ND | ND | ND | ND |
| γ-Cyclomaltodextrin | 379.9 | 596.7 | 526.4 | 53.8 | 55.3 | ND | ND | ND |
| Pullulan | 59.0 | 7.0 | ND | 9.5 | ND | ND | ND | ND |
The initial velocity is indicated with respect to that for potato amylose, which was defined as 100. NPL, B. stearothermophilus neopullulanase; CDase, cyclomaltodextrinase; BLMA, B. licheniformis maltogenic amylase; BMNP, B. megaterium neopullulanase; BSA, bacterial saccharifing α-amylase; BLA, bacterial liquefying α-amylase; NOV, Novamyl; β-AMY, soybean β-amylase.
ND, enzyme activity was not detectable (the initial velocity was less than 1/100 of that for potato amylose).
The substrate specificity of B. stearothermophilus neopullulanase was similar to those of cyclomaltodextrinase and B. licheniformis maltogenic amylase but was quite different from those of the other amylolytic enzymes (Table 1). B. stearothermophilus neopullulanase, cyclomaltodextrinase, and B. licheniformis maltogenic amylase efficiently hydrolyzed amylose, maltooligosaccharides, and cyclomaltodextrins but scarcely hydrolyzed amylopectin. On the other hand, the other amylolytic enzymes did not exhibit significantly different specificities toward amylose and amylopectin. B. megaterium neopullulanase was secreted in the culture broth of B. subtilis ANA-1 carrying pNPR78 (with the B. megaterium neopullulanase gene) as described in Materials and Methods. Since B. megaterium neopullulanase hydrolyzed pullulan (Table 1) to produce panose (data not shown), it could be classified as a neopullulanase (33). However, B. megaterium neopullulanase did not exhibit a significantly different specificity toward amylose and amylopectin.
DISCUSSION
We found a unique macromolecule recognition by the neopullulanase from B. stearothermophilus TRS40. Although B. stearothermophilus neopullulanase hydrolyzed amylose to produce maltose and a small amount of glucose (Fig. 1), it scarcely hydrolyzed amylopectin (Fig. 1 and 2). Furthermore, it selectively hydrolyzed amylose when starch was used as the substrate (Fig. 3). While the molecular mass of amylose decreased sharply below 105 Da, that of amylopectin decreased by only one order of magnitude, from approximately 108 to 107 Da (Fig. 4). There was little difference in the chain length distribution between the neopullulanase-treated amylopectin and unreacted amylopectin (Fig. 5). Other amylolytic enzymes such as α-amylase and β-amylase did not exhibit this distinct substrate specificity (Fig. 1; Table 1).
Amylose and cyclomaltodextrins were good substrates for B. stearothermophilus neopullulanase (Table 1). It is speculated that the helical structure of amylose and the pseudohelical structure of cyclomaltodextrins might fit into the active center of the enzyme. Amylose is a relatively small, linear molecule of 5 × 105 to 1 × 106 Da, and amylopectin is a much larger (1 × 107- to 1 × 108-Da), branched molecule with a large spherical shape. Based on the observation of a polymodal distribution of chain lengths in amylopectin, a cluster model has been suggested (3). Based on the chain lengths (3), the packing model of chains (7), and the size of clusters derived from electron microscopic analysis (32), the molecular mass of each cluster is calculated as being on the order of 105 Da (28). However, the amylopectin molecule is organized at a further large level of structure that has been called a blocklet (11). The structure of amylopectin at the level higher than the cluster has not been fully understood. Since the degradation of amylopectin completely halted at a molecular mass of approximately 107 Da, a structure susceptible to the action by B. stearothermophilus neopullulanase may exist at an interval of every several tens of clusters in amylopectin. Indeed, B. stearothermophilus neopullulanase did not hydrolyze waxy maize starch, which is composed only of amylopectin with a molecular mass of approximately 107 Da (data not shown), or highly branched cyclic dextrin, which is composed of a cluster structure with a molecular mass of approximately 5 × 105 Da (28) (Table 1).
This unique macromolecule recognition by B. stearothermophilus neopullulanase was also observed in the reactions of cyclomaltodextrinase and B. licheniformis maltogenic amylase (Table 1). These three enzymes exhibit 40 to 60% amino acid sequence identity (24). There is presently great interest in the structure-function relationships of these enzymes (19). Although B. megaterium neopullulanase hydrolyzed pullulan to produce panose, it did not exhibit distinct substrate specificity toward amylose and amylopectin (Table 1). Since neopullulanase from B. stearothermophilus TRS40 and that from B. megaterium NCIMB11568 have less than 30% sequence identity, B. megaterium neopullulanase and the neopullulanase from B. polymyxa CECT155 (33) can be classified into different categories. In this context, B. stearothermophilus neopullulanase, cyclomaltodextrinase, and B. licheniformis maltogenic amylase are intracellular enzymes, while B. megaterium neopullulanase and B. polymyxa neopullulanase are extracellular enzymes (10). The problem of identification and the physiological roles of the enzymes in the α-amylase family have been discussed previously (19, 24).
A partially degraded starch, dextrin, of various sizes is widely used in the food and chemical industries. A new type of dextrin, highly branched cyclic dextrin, with a molecular mass of approximately 5 × 105 Da was recently produced in our laboratory (28). We also produced a new dextrin with a molecular mass of approximately 107 Da which was 20 times larger than highly branched cyclic dextrin, using B. stearothermophilus neopullulanase. Some characteristics of this new dextrin have been investigated. The viscosity of the dextrin paste was much lower than that of gelatinized intact starch paste. The new dextrin solution also has a low propensity for retrogradation. B. stearothermophilus neopullulanase may also be used to produce low-amylose or amylose-free starch. Investigation of the industrial application of the unique properties of this enzyme is now in progress.
Acknowledgments
We thank Y. Takeda, Kagoshima University, for valuable discussions. We also appreciate the helpful advice of S. Okada.
REFERENCES
- 1.Dauter, Z., M. Dauter, A. M. Brozozowski, S. Christensen, T. V. Borchert, L. Beier, K. S. Wilson, and G. J. Davies. 1999. X-ray structure of Novamyl, the five-domain “maltogenic” α-amylase from Bacillus stearothermophilus: maltose and acabose complex at 1.7 Å resolution. Biochemistry 38:8385-8392. [DOI] [PubMed] [Google Scholar]
- 2.Dubois, M., K. A. Gilles, J. K. Hamilton, P. A. Robers, and F. Smith. 1956. Colorimetric method for determination of sugars and related substances. Anal. Chem. 28:350-356. [Google Scholar]
- 3.Hizukuri, S. 1986. Polymodal distribution of the chain lengths of amylopectin, and its significance. Carbohydr. Res. 147:342-347. [Google Scholar]
- 4.Hizukuri, H., Y. Takeda, M. Yasuda, and A. Suzuki. 1981. Multi-branched nature of amylose and the action of debranched enzymes. Carbohydr. Res. 94:205-213. [Google Scholar]
- 5.Igarashi, K., K. Ara, K. Saeki, K. Ozaki, S. Kawai, and S. Ito. 1992. Nucleotide sequence of the gene that encodes a neopullulanase from alkalophilic Bacillus. Biosci. Biotechnol. Biochem. 56:514-516. [DOI] [PubMed] [Google Scholar]
- 6.Imanaka, T., and T. Kuriki. 1989. Pattern of action of Bacillus stearothermophilus neopullulanase on pullulan. J. Bacteriol. 171:369-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imberty, A., H. Chanzy, S. Perez, A. Buleon, and V. Tran. 1988. The double-helical nature of the crystalline part of A-starch. J. Mol. Biol. 201:365-378. [DOI] [PubMed] [Google Scholar]
- 8.Kato, K., T. Sugimoto, A. Amemura, and T. Harada. 1975. A Pseudomonas intracellular amylase with high activity on maltodextrin and cyclodextrin. Biochim. Biophys. Acta 391:96-108. [DOI] [PubMed] [Google Scholar]
- 9.Kim, I.-C., J.-H. Cha, J.-R. Kim, S.-Y. Jang, B.-C. Seo, T.-K. Cheong, D. S. Lee, Y. D. Choi, and K.-H. Park. 1992. Catalytic properties of the cloned amylase from Bacillus licheniformis. J. Biol. Chem. 267:22108-22114. [PubMed] [Google Scholar]
- 10.Kim, J.-S., S.-S. Cha, H.-J. Kim, T.-J. Kim, N.-C. Ha, S.-T. Oh, H.-S. Cho, M.-J. Cho, M.-J. Kim, H.-S. Lee, J.-W. Kim, K. Y. Choi, K.-H. Park, and B.-H. Oh. 1999. Crystal structure of a maltogenic amylase provides insights into a catalytic versatility. J. Biol. Chem. 274:26279-26286. [DOI] [PubMed] [Google Scholar]
- 11.Kossmann, J., and J. Lloyd. 2000. Understanding and influencing starch biochemistry. Crit. Rev. Plant Sci. 19:171-226. [PubMed] [Google Scholar]
- 12.Kuriki, T., and T. Imanaka. 1989. Nucleotide sequence of the neopullulanase gene from Bacillus stearothermophilus. J. Gen. Microbiol. 135:1521-1528. [DOI] [PubMed] [Google Scholar]
- 13.Kuriki, T., and T. Imanaka. 1999. The concept of the α-amylase family: structural similarity and common catalytic mechanism. J. Biosci. Bioeng. 87:557-565. [DOI] [PubMed] [Google Scholar]
- 14.Kuriki, T., H. Kaneko, M. Yanase, H. Takata, J. Shimada, S. Handa, T. Takada, H. Umeyama, and S. Okada. 1996. Controlling substrate preference and transglycosylation activity of neopullulanase by manipulating steric constraint and hydrophobicity in active center. J. Biol. Chem. 271:17321-17329. [DOI] [PubMed] [Google Scholar]
- 15.Kuriki, T., S. Okada, and T. Imanaka. 1988. New type of pullulanase from Bacillus stearothermophilus and molecular cloning and expression of the gene in Bacillus subtilis. J. Bacteriol. 170:1554-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuriki, T., D. C. Stewart, and J. Preiss. 1997. Construction of chimeric enzymes out of maize endosperm branching enzymes I and II: activity and properties. J. Biol. Chem. 272:28999-29004. [DOI] [PubMed] [Google Scholar]
- 17.Kuriki, T., H. Takata, S. Okada, and T. Imanaka. 1991. Analysis of active center of Bacillus stearothermophilus neopullulanase. J. Bacteriol. 173:6147-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lansky, S., M. Kooi, T. J. Schoch, and J. Thomas. 1949. Properties of the fractions and linear subfractions from various starches. J. Am. Chem. Soc. 71:4066-4075. [Google Scholar]
- 19.MacGregor, E. A., S. Janecek, and B. Svensson. 2001. Relationship of sequence and structure to specificity in the α-amylase family of enzymes. Biochim. Biophys. Acta 1546:1-20. [DOI] [PubMed] [Google Scholar]
- 20.Metz, R. J., L. N. Allen, T. M. Cao, and N. W. Zeman. 1988. Nucleotide sequence of an amylase from Bacillus megaterium. Nucleic Acids Res. 16:5203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohdan, K., T. Kuriki, H. Kaneko, J. Shimada, T. Takada, Z. Fujimoto, H. Mizuno, and S. Okada. 1999. Characteristics of two forms of α-amylase and structural implication. Appl. Environ. Microbiol. 65:4652-4658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohdan, K., T. Kuriki, H. Takata, H. Kaneko, and S. Okada. 2000. Introduction of raw starch-binding domains into Bacillus subtilis α-amylase by fusion with the starch-binding domains of Bacillus cyclomaltodextrin glucanotransferase. Appl. Environ. Microbiol. 66:3058-3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohdan, K., T. Kuriki, H. Takata, and S. Okada. 2000. Cloning of the cyclodextrin glucanotransferase gene from alkalophilic Bacillus sp. A2-5a and analysis of the raw starch-binding domain. Appl. Microbiol. Biotechnol. 53:430-434. [DOI] [PubMed] [Google Scholar]
- 24.Park, K.-H., T.-J. Kim, T.-K. Cheong, J.-W. Kim, B.-H. Oh, and B. Svensson. 2000. Structure specificity and function of cyclomaltodextrinase, a multispecific enzyme of the α-amylase family. Biochim. Biophys. Acta 1478:165-185. [DOI] [PubMed] [Google Scholar]
- 25.Smith, K. A., and A. A. Salyers. 1991. Characterization of a neopullulanase and an α-glucosidase from Bacteroides thetaiotaomicron 95-1. J. Bacteriol. 173:2962-2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Somogyi, M. 1952. Notes on sugar determination. J. Biol. Chem. 195:19-23. [PubMed] [Google Scholar]
- 27.Takata, H., T. Kuriki, S. Okada, Y. Takesada, M. Iizuka, N. Minamiura, and T. Imanaka. 1992. Action of neopullulanase: neopullulanase catalyzes both hydrolysis and transglycosylation at α-(1→4) and α-(1→6)-glucosidic linkages. J. Biol. Chem. 267:18447-18452. [PubMed] [Google Scholar]
- 28.Takata, H., T. Takaha, H. Nakamura, K. Fujii, S. Okada, M. Takagi, and T. Imanaka. 1997. Production and some properties of a dextrin with narrow size distribution by the cyclization reaction of branching enzyme. J. Ferment. Bioeng. 84:119-123. [Google Scholar]
- 29.Takata, H., T. Takaha, S. Okada, S. Hizukuri, M. Takagi, and T. Imanaka. 1996. Structure of the cyclic glucan produced from amylopectin by Bacillus stearothermophilus branching enzyme. Carbohydr. Res. 295:91-101. [DOI] [PubMed] [Google Scholar]
- 30.Terada, Y., K. Fujii, T. Takaha, and S. Okada. 1999. Thermus aquaticus ATCC 33923 amylomaltase gene cloning and expression and enzyme characterization: production of cycloamylose. Appl. Environ. Microbiol. 65:910-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tonozuka, T., M. Ohtsuka, S. Mogi, H. Sakai, T. Ohta, and Y. Sakano. 1993. A neopullulanase-type α-amylase gene from Thermoactinomyces vulgaris R-47. Biosci. Biotechnol. Biochem. 57:395-401. [DOI] [PubMed] [Google Scholar]
- 32.Yamaguchi, M., K. Kainuma, and D. French. 1979. Electron microscopic observation of waxy maize starch. J. Ultrastruct. Res. 69:249-261. [DOI] [PubMed] [Google Scholar]
- 33.Yebra, M. J., J. Arroyo, P. Sanz, and J. A. Priet. 1997. Characterization of novel neopullulanase from Bacillus polymyxa. Appl. Biochem. Biotechnol. 68:113-120. [Google Scholar]
- 34.Yebra, M. J., A. Blasco, and P. Sanz. 1999. Expression and secretion of Bacillus polymyxa neopullulanase in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 170:41-49. [DOI] [PubMed] [Google Scholar]