Abstract
By proteomic analysis we found a 21-kDa protein (P21) from Acidithiobacillus ferrooxidans ATCC 19859 whose synthesis was greatly increased by growth of the bacteria in pyrite, thiosulfate, elemental sulfur, CuS, and ZnS and was almost completely repressed by growth in ferrous iron. After we determined the N-terminal amino acid sequence of P21, we used the available preliminary genomic sequence of A. ferrooxidans ATCC 23270 to isolate the DNA region containing the p21 gene. The nucleotide sequence of this DNA fragment contained a putative open reading frame (ORF) coding for a 23-kDa protein. This difference in size was due to the presence of a putative signal peptide in the ORF coding for P21. When p21 was cloned and overexpressed in Escherichia coli, the signal peptide was removed, resulting in a mature protein with a molecular mass of 21 kDa and a calculated isoelectric point of 9.18. P21 exhibited 27% identity and 42% similarity to the Deinococcus radiodurans thiosulfate-sulfur transferase (rhodanese; EC 2.8.1.1) and similar values in relation to other rhodaneses, conserving structural domains and an active site with a cysteine, both characteristic of this family of proteins. However, the purified recombinant P21 protein did not show rhodanese activity. Unlike cytoplasmic rhodaneses, P21 was located in the periphery of A. ferrooxidans cells, as determined by immunocytochemical analysis, and was regulated depending on the oxidizable substrate. The genomic context around gene p21 contained other ORFs corresponding to proteins such as thioredoxins and sulfate-thiosulfate binding proteins, clearly suggesting the involvement of P21 in inorganic sulfur metabolism in A. ferrooxidans.
Acidithiobacillus ferrooxidans is a chemolithoautotrophic bacterium that obtains its energy from the oxidation of ferrous iron, elemental sulfur, or partially oxidized sulfur compounds (5, 14, 19, 24, 35, 38). The ability of these and other microorganisms present in their habitat to solubilize metal sulfides is successfully applied in biomining operations (5). Two mechanisms have been proposed for the dissolution of metal sulfides: the direct one and the indirect one (see reference 28). In the direct mechanism, the bacteria directly oxidize the mineral by biological means, without any requirement for ferric or ferrous ions (37).
Recent evidence favors the indirect mechanism of sulfide dissolution (8, 28-30) by which metal sulfides are degraded by a chemical attack of Fe(III) ions and/or protons on the crystal lattice (28, 29). The primary Fe(III) ions are supplied by the bacterial exopolymers (28), where they are complexed to glucuronic acid residues. The mechanism of degradation is determined by the mineral structure. According to the proposed model, the disulfides pyrite (FeS2), molybdenite (MoS2), and tungstenite (WS2) are degraded, generating thiosulfate as the main intermediate (30). Iron(III) ions are exclusively the oxidizing agents for the dissolution. Thiosulfate would be consequently degraded in a cyclic process to sulfate, with elemental sulfur being a side product. This explains why only Fe(II) ion-oxidizing bacteria are capable of oxidizing these metal sulfides (29).
Metal sulfides such as sphalerite (ZnS), galena (PbS), covelite (CuS), and chalcopyrite (CuFeS2), among others, are degradable by iron(III) ion and proton attack. As a consequence, polysulfides and elemental sulfur are the main intermediates. In this case, thiosulfate is only a by-product of further degradation steps (29).
Because thiosulfate is the key compound in the oxidation of the sulfur moiety of pyrite, the mechanism has recently been defined as the thiosulfate mechanism. All reactions comprising this mechanism have been shown to occur on a purely chemical basis (30). However, sulfur compounds oxidizing enzymes such as the tetrathionate hydrolase of A. ferrooxidans, Acidithiobacillus thiooxidans, or Thiobacillus acidophilus may be involved (7, 9, 10, 15, 35). In addition, enzymes for thiosulfate or sulfite oxidation from A. ferrooxidans or A. thiooxidans may succesfully compete with the chemical reactions with iron(III) ions as an oxidizing agent (30). A rhodanese activity has been previously described in A. ferrooxidans (36). This enzyme is a thiosulfate sulfur-transferase which breaks the S-S bond present in thiosulfate, generating sulfur and sulfite. Other enzymes may also participate in the thiosulfate mechanism, such as the thiosulfate-oxidizing enzyme of A. ferrooxidans (33).
By using two-dimensional (2-D) polyacrylamide gel electrophoresis (PAGE), we have previously studied the global changes in gene expression of A. ferrooxidans when the microorganism is grown on ferrous iron or sulfur and have identified some of these proteins (12, 23). To further define some of the components involved in the oxidation of sulfur and metal sulfides, we analyzed here by means of 2-D PAGE the proteins synthesized by the bacterium when grown on Fe(II) ions, elemental sulfur, pyrite, thiosulfate, ZnS, CuS, and PbS. Our results indicate that the synthesis of an exported rhodanese-like P21 protein was greatly induced when the microorganisms were grown in FeS2 and other reduced inorganic sulfur forms. Although yet to be demonstrated, this protein may be an important part of the indirect thiosulfate mechanism of dissolution of minerals and/or sulfur metabolism in A. ferrooxidans.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
A. ferrooxidans ATCC 19859 was grown in ferrous iron-containing modified 9K medium as described previously (2, 39), and growth on elemental sulfur was done with sulfur prills (3). Pyrite (40% [wt/wt] Fe, found to pass a 75-μm-pore-size sieve) was kindly given to us by P. Norris. ZnS and CuS were purchased from Aldrich and contained 28 and 125 ppm of trace iron, respectively. Growth of cells in these substrates was done in modified 9K medium (pH 2.5), replacing ferrous iron with 1% (wt/vol) of the corresponding metal sulfide. Growth of A. ferrooxidans in thiosulfate was done in DSMZ medium 71 containing 20 mM thiosulfate and the following components (in g liter−1): KH2PO4, 3.0; MgSO4 · 7H2O, 0.5; (NH4)2SO4, 3.0; and CaCl2 · 2H2O, 0.25. The pH was kept at ca. 4.6 by addition of 1 M NaOH. Washed cells (1010) resuspended in 10 ml of the corresponding medium were labeled in the presence of 0.1 mCi of [35S]methionine (specific activity, 1,087 Ci/mmol) for 40 h. Escherichia coli BL21(DE3) containing plasmid pGZ105 with the glpE insert coding for the E. coli rhodanese (26) was a kind gift of T. Larson. E. coli strains BL21(DE3) and derivatives were grown in Luria-Bertani medium (27).
2-D NEPHGE, SDS-PAGE, and autoradiography.
Total cell proteins were separated by 2-D nonequilibrium pH PAGE (2-D NEPHGE) (21), and this was performed as described before for A. ferrooxidans (31) with ampholites (pH 3 to 10) from Bio-Rad. The cell samples (4 mg [wet weight] of unlabeled cells or 500,000 cpm contained in cells labeled by growth in [35S]methionine) were resuspended in 80 μl of sonication buffer (10 mM Tris-HCl [pH 7.4], 5 mM MgCl2, and 50 μg of pancreatic RNase per ml), sonicated, and treated with DNase (final concentration, 50 μg/ml). The mixture was then lyophilized and dissolved in lysis buffer as described previously (2). Sodium dodecyl sulfate (SDS)-PAGE (18) consisted of 12.5% or 5 to 20% polyacrylamide gradients that were stained with Coomassie blue. Autoradiography was done as described previously (2).
Microsequencing of proteins extracted from 2-D gels.
Proteins of interest were recovered from Coomassie blue-stained and heat-dried 2-D gels by excising the protein spots. After rehydration and concentration of the spots by SDS-PAGE, the proteins were electroblotted onto a polyvinylidene difluoride (PVDF) membrane (20) and subjected to microsequencing by the Laboratoire de Microséquençage des Protéines of the Institut Pasteur.
Primers and PCR conditions.
The oligonucleotide primers were purchased from Genset Corporation. Taq and Pwo polymerases were from Promega and Roche, respectively, and were used according to the manufacturer's recommendations. The oligonucleotide primer sequences were deduced from the open reading frames (ORFs) found in the available almost finished DNA genomic sequence of A. ferrooxidans ATCC 23270 (http://www.tigr.org). These primers were: P21NH2-NdeI (5′-GGAGAATCATATGTTCAAGCGCCTAGCA-3′), P21 M-NdeI (5′TGGCCTCCCATATGGACAGTGGTAACCAAG-3′), and P21CTER-XhoIHT (5′-TCCAAACCTCGAGGATGCGCCTGCGGGTTGTC-3′).
To amplify the p21 gene, we used a two-step HotPCR protocol: 2 min at 95°C, followed by 25 cycles at 95°C for 30 s and 72°C for 1 min, and finally 4 min at 72°C.
DNA manipulations.
Restriction enzyme digestions and T4 DNA ligase reactions were performed according to the manufacturer's recommendations. Recombinant DNA techniques were carried out according to standard laboratory procedures (27). The dideoxy chain termination method was employed to sequence DNA by using [γ-33P]ATP and the dsDNA Cycle Sequencing System from Gibco-BRL. The DNA sequences were compiled and analyzed with the University of Wisconsin GCG Package (version 9.1; Genetics Computer Group, Madison, Wis.).
p21 gene cloning and expression.
We used pGEM-T (Promega) and the pET System (Novagen). The p21 gene with signal peptide was obtained by PCR with P21NH2-NdeI and P21CTER-XhoIHT primers corresponding to the N-terminal and C-terminal end sequences of P21 and containing NdeI and XhoI restriction sites, respectively. The p21 gene without leader peptide was cloned with the P21 M-NdeI and P21 XhoIHT primers. We used Pwo polymerase (Roche) and a low number of amplification cycles to decrease the number of sequence errors. The DNA fragments separated by electrophoresis in 1% agarose gels were recovered, purified with Wizard PCR Prep (Promega), and ligated to pGEM-T vector (Promega). The ligation products were used to transform E. coli JM109. The positive clones were analyzed by using colony PCR. Pure plasmids with inserts were obtained by using the Wizard Plus Minipreps DNA purification system (Promega). The DNA fragments were ligated to pET21b(+) vector (Novagen), both previously digested with NdeI and XhoI. The ligation products (pPR21H and pPR21MH vectors with or without the coding sequence for the leader peptide, respectively) were used to transform E. coli BL21(DE3). The recombinant clones were selected on Luria-Bertani solid medium supplemented with ampicillin (100 μg/ml). The induction-expression analysis was done in the presence or absence of 1 mM IPTG, added when the cultures reached an optical density at 600 nm (OD600) of 0.6. Expression of the recombinant P21 (rP21) with or without signal peptide was analyzed by SDS-PAGE of total cell extracts.
Purification of rP21.
rP21 was purified under denaturing conditions as follows. A 300-ml culture of BL21(DE3) transformed with pPR21H or pPR21MH vectors was grown to an OD600 of 0.6 and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Cells were harvested by centrifugation, and the pellet was resuspended in 30 ml of 1× binding buffer containing 5 mM imidazole, 0.5 M NaCl, and 20 mM Tris-HCl (pH 7.9). Cell disruption was performed by sonication (six times during 30 s). After centrifugation (at 20,000 × g for 15 min), the pellet was resuspended in 5 ml of 1× binding buffer containing 6 M urea and incubated for 1 h in an ice bath. The sample was centrifuged (39,000 × g for 20 min), and the supernatant, previously filtered through a 0.45-μm-pore-size Millipore filter, was applied to a column containing 1 ml of His-nitrilotriacetic acid resin. rP21 was eluted with 10 ml of Elute buffer containing 300 mM imidazole, 0.25 mM NaCl, 10 mM Tris-HCl (pH 7.9), and 6 M urea. The collected fractions (0.5 ml) were analyzed by SDS-PAGE. Finally, rP21 containing fractions, which were essentially free from other proteins, were pooled and renatured by removal of the urea in five sequential dialysis steps with 67 mM phosphate (pH 5) buffer containing 6, 2, 1, and 0.5 M urea and no urea. All dialyses were performed in the presence of 50 mM thiosulfate and 200 mM β-mercaptoethanol.
Preparation of antibodies against rP21.
The antiserum against rP21 was made by immunizing intraperitoneally a BALB/c mouse with ca. 50 μg of rP21 (this corresponded to one 2-D gel spot from the respective gel in each immunization). To prepare the samples for immunization, the gel piece containing the rP21 spot was loaded in one well of a slab gel prepared with a meltable synthetic electrophoresis matrix (ProtoPrep; National Diagnostics) and was allowed to rehydrate for 1 h in 1× sample buffer diluted with 50 mM H3BO3-0.1% SDS (pH 8). After electrophoresis the gel was stained with Coomassie blue, and the concentrated rP21 band was excised and washed four times with distilled water. The slice containing rP21 was weighed, 1 volume (assuming 1 mg = 1 μl) of ProtoPrep Dissolution Reagent was added, and the mixture was incubated for 1 h at 80°C in a water bath. The melted viscous ProtoPrep mixture was then directly mixed with 1 volume of Freund complete adjuvant (Gibco-BRL) and vortexed for 1 h to produce an injectable emulsion with a final volume of 800 μl. Immunization was done four times at 1-week intervals. One week after the last injection, the blood was collected from the mouse, and the serum was obtained by centrifugation.
Western immunoblotting.
The proteins synthesized by A. ferrooxidans when grown in different oxidizable substrates were separated by SDS-PAGE and electrotransferred to a PVDF membrane as already described (13). For the antigen-antibody reaction, the membrane containing the transferred proteins was treated with the antiserum against rP21 as the primary antibody (1:2,500 dilution) and monoclonal anti-mouse antibodies conjugated with peroxidase (Amersham) as the secondary antibodies (1:5,000 dilution). The specificity of the mouse anti-rP21 serum was tested with both the preimmune and the immune sera (1:4,000 dilution) against pure rP21 and total proteins from A. ferrooxidans and E. coli BL21(DE3). No cross-reaction was observed with E. coli proteins (results not shown), while only one reacting band was detected in A. ferrooxidans samples and no band was observed with the preimmune serum, indicating that the serum was monospecific. A colorimetric method was used to develop Western blots as recommended by Promega. The relative intensities of the P21 protein bands in Western blots were determined by using a scanner and an image analysis program (www.scioncorp.com).
Immunocytochemistry with thin sections and cell fractionation.
Cells of A. ferrooxidans grown in sulfur and ferrous iron were harvested by centrifugation (10 min, 6,000 × g, 4°C) and washed in water (pH 1.9), and the pellets were fixed with 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.0) for 2 h at 4°C. After being washed, the cells were dehydrated with an ethanol series and then infiltrated with 1:1 Unicryl-ethanol and Unicryl pure (Energy Beam Sciences, Inc.) for 4 h and overnight, respectively. The samples were then placed in gelatin capsules and polymerized for 72 h at 45°C. Thin sections were cut with a Sorval MT-2B Ultramicrotome and collected onto nickel grids.
Ultrathin sections were blocked overnight with 1% bovine serum albumin (BSA) in 20 mM Tris-HCl buffer (pH 7.6) containing 137 mM NaCl and 0.05% Tween 20 (TBST). Sections were washed five times with TBST and incubated with anti-rP21 antibody at a dilution of 1:50 in TBST-1% BSA for 2 h at room temperature. Sections were washed five times in a similar manner (10 min each time) and then incubated with goat anti-mouse immunoglobulin G conjugated with 10-nm gold particles (Sigma) at a dilution of 1:10 for 1 h in TBST-1% BSA to allow the detection of primary antibody binding. Thereafter, the sections were washed five times with TBST and three times with water. Finally, the sections were stained with uranyl acetate and viewed at 80 kV in a transmission electron microscope (Phillips Tecnai 12).
To locate P21, we fractionated the A. ferrooxidans cells into outer-membrane, soluble, and inner-membrane fractions. The outer membrane was obtained as described earlier (13). After this fractionation to obtain the outer membrane, we defined the soluble fraction as the supernatant obtained after the first centrifugation at 100,000 × g for 2 h to pellet the total membrane fraction. The inner- membrane fraction was considered to be the last supernatant obtained after treatment of the total membrane fraction in the presence of 2% sodium laurylsarcosinate and pelleting of the final outer-membrane preparation (13).
Determination of rhodanese activity.
Rhodanese (thiosulfate-cyanide sulfurtransferase; EC 2.8.1.1) activity was assayed with crude enzyme extracts or with the purified recombinant protein rP21. As a comparison, we used the recombinant rhodanese GlpE from E. coli (26). The assay was done at pH 7.5 to 8.5 essentially as described by Singleton and Smith (34) and Gardner and Rawlings (11).
Sequence analysis.
Identity-similarity searching in databases was done by using the BLASTP program (1) from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov) and from the unfinished A. ferrooxidans ATCC 23270 genome site (http://www.tigr.org). Multiple alignments were performed with CLUSTALW 1.8 (http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html) and edited by using BOXSHADE 3.21 (http://www.isrec.isb-sib.ch:8080/software/BOX_form.html.). Molecular masses and isoelectric points of ORFs were obtained by using Protparam (http://www.expasy.ch/cgi-bin/protparam). The possible presence of transmembrane domains in the analyzed ORFs was analyzed by using TMPRED (http://www.ch.embnet.org/software/TMPRED). The putative functions and predicted subcellular locations of the proteins coded by the different ORFs analyzed were obtained by using BLAST (http://www.ncbi.nlm.nih.gov/BLAST).
Nucleotide sequence accession number.
The nucleotide sequence of the p21 gene is available in the EMBL database under accession no. AJ31223.
RESULTS
Pattern of global protein synthesis of A. ferrooxidans grown with different oxidizable substrates.
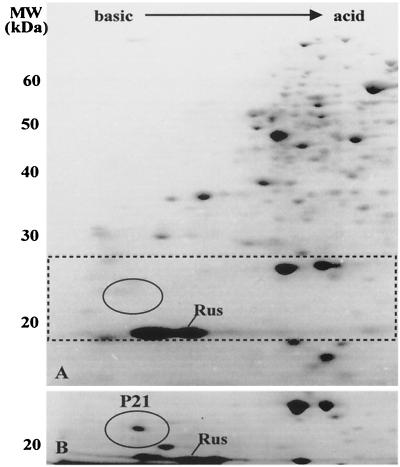
To characterize some of the proteins synthesized by A. ferrooxidans when grown in different oxidizable substrates, we employed 2-D NEPHGE analysis of the total cell proteins stained with Coomassie blue as shown in Fig. 1. We have previously observed that the general pattern of protein synthesis for ferrous-iron-grown A. ferrooxidans showed several differences compared with that observed for the sulfur-grown bacterium (23). The differences involved both the induction (or derepression) and the repression of the synthesis of numerous polypeptides. Based on this previous analysis, we have chosen to study in greater detail protein P21, indicated by the window in the gels of Fig. 1. It is clear that P21 was entirely absent when A. ferrooxidans was grown in ferrous iron (gel A) and was greatly induced when the cells grew in pyrite (gel B). Rusticyanin (Rus), a protein known to be induced by growth of the microorganism in ferrous iron, was present in both conditions, although in larger amounts in cells grown in ferrous iron.
FIG. 1.
Induction of the expression of protein P21 by growth of A. ferrooxidans on pyrite. Total proteins from cells grown in ferrous iron (A) or in pyrite (B) were separated by nonequilibrium pH 2-D PAGE with a pH gradient between 3.0 (right side of the gel) and 10.0 (left side of the gel), and the proteins were stained with Coomassie blue. In panel A, the window indicated by dotted lines contains an oval showing the region where P21 migrates. In panel B, only the portion of the gel where P21 migrates is shown. Numbers to the left of the gels indicate the molecular mass standards in kilodaltons. Rus, the migrating position of Rus.
The isolated p21 gene shows similarity to rhodaneses.
Protein P21 was isolated from 2-D gels and subjected to Edman microsequencing, yielding the following amino acid sequence: DDSGNQAAQQVLNARMEKFFADQKPYG. Using this sequence, we performed a tBLASTN analysis of the available preliminary genome sequence of A. ferrooxidans ATCC 23270 (http://www.tigr.org) and identified one ORF of 645 bp. On the basis of this information, we cloned and sequenced the p21 gene from A. ferrooxidans ATCC 19859. The result of this sequence can be found in the database under accession no. AJ31223.
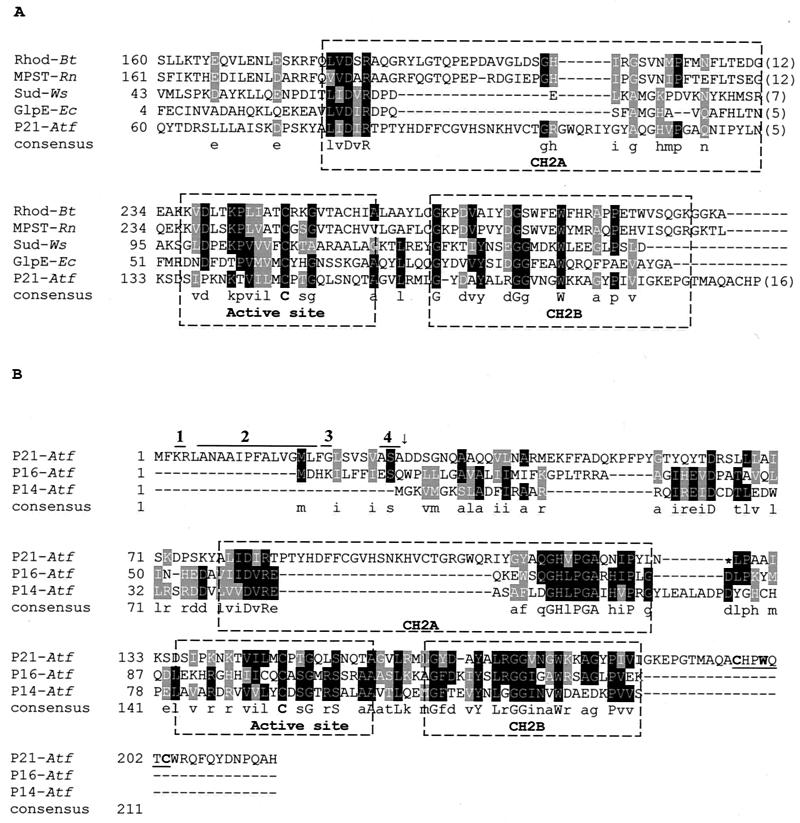
The BLASTP analysis of the amino acid sequence of P21 indicated that this protein had 27% identity and 42% similarity to the sulfurtransferase (rhodanese) from Deinococcus radiodurans and comparable values with those of many other rhodaneses. The active site of the rhodanese and two characteristically conserved motifs (CH2A and CH2B) were also present in all of the proteins compared (Fig. 2A). Interestingly, most described bacterial rhodaneses have an isoelectric point of 4 to 5 and various molecular masses (12,000 to 35,000 kDa). The A. ferrooxidans P21 rhodanese-like protein has an isoelectric point of 9.18 (without the signal peptide), which is similar to other very basic proteins present in the acidic periplasm of this bacterium, such as Rus. The p21 gene encoded a polypeptide of 215 amino acids with a putative signal peptide (Fig. 2B), suggesting that P21 is exported, possibly to the periplasm of A. ferrooxidans. The amino-terminal sequence contains all of the features for the gram-negative signal peptide sequence: positively charged residues at the N terminus, a stretch of hydrophobic residues, a glycyl residue at the end of this stretch that would allow the formation of turns, and a consensus cleavage sequence (Ala/Gly/Ser)-X-(Ala/Gly/Ser), a motif characteristic of those proteins transported via the Sec system (25).
FIG. 2.
(A) Partial sequence alignment of the P21 protein of A. ferrooxidans with those of representative sulfurtransferases. The active site and two conserved structural motifs, designated CH2A and CH2B, are labeled. A consensus is indicated in which the active site presents a cysteine conserved in all of the sequences shown. Rhod-Bt, bovine liver rhodanese; MPST-Rn, rat mercaptopyruvate sulfurtransferase; Sud-Ws, sulfide dehydrogenase from W. succinogenes; GlpE-Ec, E. coli rhodanese-like protein; P21-Atf, putative periplasmic rhodanese-like protein P21 from A. ferrooxidans. (B) Sequence alignment of A. ferrooxidans proteins with similarity to rhodaneses. The P21 protein sequence contains a 28-amino-acid leader peptide. Within this leader peptide, indicated by the numbered lines above, are the following: the positively charged residues at the N terminus (line 1), a stretch of hydrophobic residues (line 2), a glycyl residue at the end of this stretch (line 3), and a consensus cleavage sequence (Ala/Gly/Ser)-X-(Ala/Gly/Ser) (line 4). The arrow indicates the site where the cleavage of the leader peptide would occur. P14-Atf, putative cytoplasmic rhodanese-like from A. ferrooxidans; P16-Atf, putative cytoplasmic rhodanase-like from A. ferrooxidans. The underlined sequence in P21-Atf indicates a putative cysteine motif known to bind iron-sulfur clusters. Accession numbers: Sud, X81642; Rhod-Bt, M58561; MPST-Rn, D50564; GlpE-Ec, M96795; P21-Atf, AJ312238; and P14-Atf and P16-Atf, derived from the genome sequence of A. ferrooxidans 23270.
Upon analyzing the unfinished A. ferrooxidans genome sequence, we found that not only was the p21 gene present but so, too, were at least two other small sequences with rhodanese-like similarities: P14 and P16 (Fig. 2B). These two putative ORFs did not present signal peptides, and the corresponding putative proteins had isoelectric points of 4.8 (P14) and 9.3 (P16). It is also interesting that only P21 contained at its C-terminal end a cysteine motif, Cys-XX-Trp-XX-Cys (underlined in Fig. 2B), known to bind iron-sulfur clusters (4).
Genomic analysis of the P21 region.
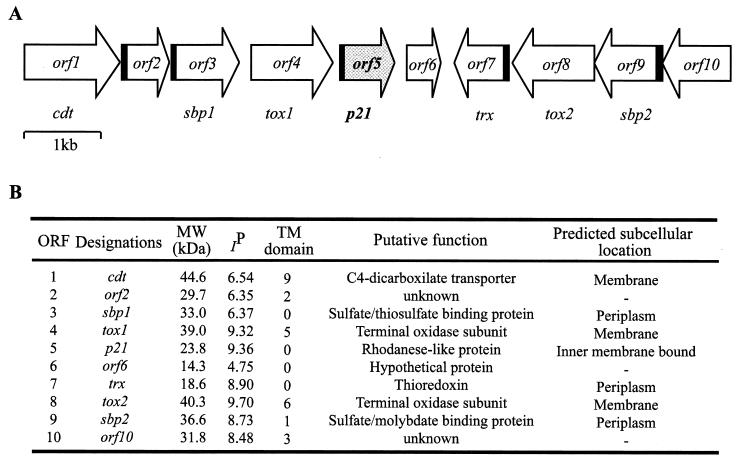
When the regions surrounding gene p21 were analyzed on the incomplete genomic sequence of A. ferrooxidans ATCC 23270, several putative ORFs related to sulfur metabolism were deduced. Upstream of P21 a terminal oxidase subunit (ORF4) and a sulfate-thiosulfate binding protein (ORF3) were located (Fig. 3A). These putative genes together with ORF1, ORF2, and ORF6 apparently form a cluster with the same orientation. On the other hand, a putative gene with high similarity to a periplasmic thioredoxin (ORF7), together with ORF8, ORF9, and ORF10, was oriented in a divergent way from the P21 cluster. The ORFs coding for P14 and P16 were located in different contigs of the incomplete genome from A. ferrooxidans ATCC 23270 (contigs 7920 and 7913, respectively) and with entirely different neighboring putative genes (not shown).
FIG. 3.
(A) Schematic map of the contig region containing the putative gene cluster context around gene p21 from A. ferrooxidans. Coding regions containing putative signal peptides for the Sec system are indicated in black. (B) Some properties of the putative ORFs present in this region. The programs used to determine the properties of the listed gene products are indicated in Materials and Methods.
In a separate study, we also isolated a 2,991-bp chromosomal DNA fragment from A. ferrooxidans ATCC 19859 that was cloned and sequenced. The nucleotide sequences for the p21 gene with its signal peptide and the ORFs corresponding to thioredoxin and the terminal oxidase subunit present downstream and upstream of it, respectively, were the same as those obtained from the ATCC 23270 genomic data (results not shown), demonstrating the great similaritiy between these putative genes in the two A. ferrooxidans strains.
Analysis of the expression of P21 in A. ferrooxidans grown under different conditions.
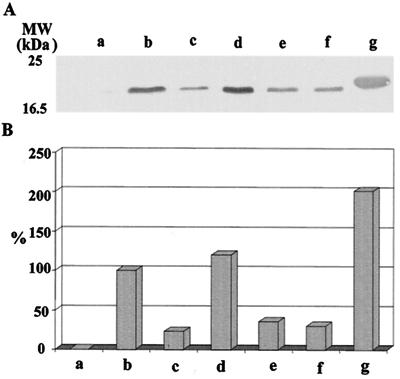
The synthesis of the rhodanese-like P21 appeared to be regulated by the presence of iron in the growth medium, with the synthesis being strongly inhibited by an increasing Fe(II) concentration. Table 1 shows the changes observed in proteins P21 and Rus when A. ferrooxidans grown in elemental sulfur was supplemented with increasing ferrous iron concentrations. A clear decrease in the levels of P21 synthesized and an increase in the levels of Rus synthesis were observed. This suggests a functional role for P21 in sulfur metabolism and the regulation of the expression of P21 by the ferrous ion concentration present in the growth medium. When cells were grown in ferrous iron, elemental sulfur, thiosulfate, and different metal sulfides and their total cell proteins were separated by SDS-PAGE, followed by Western blotting with antiserum against rP21, the results shown in Fig. 4 were obtained. Clearly, there was a great induction of P21 in cells grown in pyrite (lanes d) and an almost entire lack of P21 in cells grown in ferrous iron (lanes a), in agreement with the results shown in Table 1. Cells grown in the presence of elemental sulfur (lanes b) showed lower induced levels of P21 compared with those of cells grown in FeS2 but greater levels compared with those of microorganisms grown in thiosulfate (lanes c), CuS (lanes e), or ZnS (lanes f) (Fig. 4).
TABLE 1.
Effect of ferrous iron concentration on the expression of protein P21a
| Fe(II) (mM) | Relative level of protein (%)
|
|
|---|---|---|
| P21 | Rus | |
| 0.5 | 100.0 | 100.0 |
| 5 | 44.4 | 260.8 |
| 10 | 2.7 | 391.3 |
A. ferrooxidans cells growing in sulfur medium to which the indicated ferrous iron concentrations and [35S]methionine were added. After 30 h of incubation under these conditions, total cell proteins were separated by 2-D PAGE and subjected to autoradiography, and the intensities of the spots corresponding to the rhodanese-like protein (P21) and Rus were determined by using an image analysis program. The positions of these two spots were identified previously by their coordinates in the gels and by N-terminal amino acid sequence analysis of the isolated spots (23).
FIG. 4.
Western blotting analysis of P21 in A. ferrooxidans grown under different conditions. (A) Equivalent amounts of total cell proteins from A. ferrooxidans grown in ferrous iron (lane a), elemental sulfur (lane b), thiosulfate (lane c), pyrite (lane d), copper sulfide (lane e), zinc sulfide (lane f), and a sample of pure recombinant P21 (2 μg) (lane g) were separated by SDS-PAGE and were then transferred to a PVDF membrane, followed by reaction with antiserum against rP21 and colorimetric development as described in Materials and Methods. (B) Quantification of the bands in panel A by using the Scion image-processing program. The values shown are relative to those obtained in sulfur grown cells taken as 100%.
In vivo expression and processing of P21 from A. ferrooxidans in E. coli.
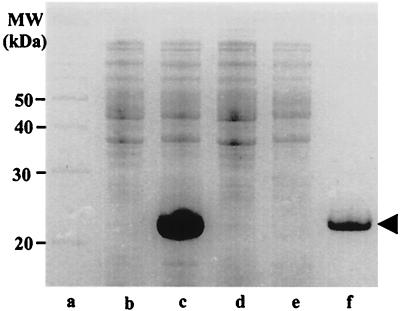
The expression vectors pPR21H and pPR21MH containing the ORF corresponding to P21, with or without leader peptide, respectively, were used to transform E. coli BL21(DE3), and the overexpression of the protein was studied in the presence or absence of IPTG as shown in Fig. 5. The start codon downstream from the T7 promoter is overlapped by an NdeI site (CATATG), and the genes to be cloned for overexpression in this system are usually amplified by PCR with a primer to introduce an NdeI site at the start of the coding sequence. The p21 genes contained in the resulting plasmids pPR21H and pPR21MH were highly expressed. About 2 h after the addition of IPTG to a mid-log-phase culture of E. coli BL21(DE3) carrying pPR21H, the protein P21 synthesized represented ca. 40% of the total protein present in crude extracts (Fig. 5).
FIG. 5.
In vivo overexpression of A. ferrooxidans p21 gene in E. coli. The pET21b(+) plasmid containing the p21 gene insert (lanes b and c) or the vector without the insert (lanes d and e) was used to transform E. coli BL21(DE3). All of the strains were grown for 2 h in the presence (lanes c and e) or in the absence (lanes b and d) of 1 mM IPTG added at the half-logarithmic phase of growth. The total cell proteins (lanes b, c, d, and e), molecular mass markers (lane a), and rP21 purified by affinity chromatography (lane f) were separated by SDS-PAGE and stained with Coomassie blue. The arrowhead indicates the migrating position of protein P21.
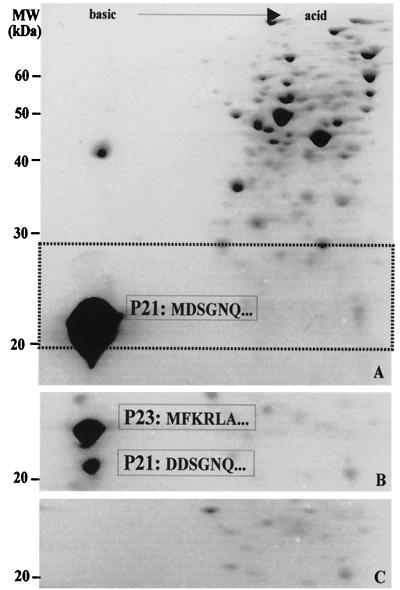
To obtain information about the processing of the rhodanese-like P21 protein in E. coli BL21(DE3), total cell proteins from this strain were subjected to 2-D PAGE analysis. The overexpressed P21 from a plasmid containing the p21 gene without signal peptide is seen in Fig. 6A. When the p21 gene with signal peptide was overexpressed in E. coli BL21(DE3), as shown in Fig. 6B, two spots with apparent masses of 23 and 21 kDa were obtained. The migrations of these spots were in agreement with the predicted sizes and isoelectric points obtained for each of these proteins before and after processing to remove the leader peptide of P23. This was confirmed by determining the N-terminal sequence of each of the spots being analyzed and suggests that the E. coli Sec machinery recognizes the A. ferrooxidans secretion signals.
FIG. 6.
The signal peptide of A. ferrooxidans P21 is processed when the p21 gene is expressed in E. coli. The pET21b(+) plasmid containing the p21 gene insert without the signal peptide (A), the vector with the signal peptide (B), or the same plasmid without the insert (C) was used to transform E. coli strain BL21(DE3). All of the strains were grown for 2 h in the presence of 1 mM IPTG added at the half-logarithmic phase of growth. The total cell proteins were then separated by 2-D NEPHGE and stained with Coomassie blue. Numbers to the left indicate molecular mass markers. In gel A, the dotted horizontal lines indicate the area where P21 migrates. Only this area is shown in gels B and C. The N-terminal amino acid sequences of the processed (P21) and nonprocessed (P23) forms of P21 are indicated. The sequence of P21 in gel A starts with a different amino acid due to a change of an aspartic acid residue for a methionine during the cloning procedure to eliminate the signal peptide sequence.
Measuring rhodanese activity of P21 and crude extracts from A. ferrooxidans.
The ability of rP21 to transfer sulfane sulfur from thiosulfate to cyanide was tested (11, 34) in crude extracts of E. coli overexpressing rP21 or with rP21 purified by affinity chromatography. However, no thiosulfate-cyanide sulfurtransferase activity was detected under these conditions (results not shown). This lack of activity could be due to the presence of a His tag in P21. However, this was not the case, since we detected no rhodanese activity in cell extracts from E. coli expressing P21 with no His tag (not shown). When we measured rhodanese activity in crude extracts from A. ferrooxidans ATCC 19859 grown in sulfur, ferrous iron, or pyrite, we obtained values in the range of those reported by Tabita et al. (0.250 μmol of SCN/min/mg of protein), the differences between iron- and sulfur-grown cells being only ca. 25% (data not shown). This result is clearly different from that expected if the levels of P21 synthesized in ferrous iron- or sulfur-grown cells were responsible of the observed rhodanese activity.
Localization of P21 in A. ferrooxidans by using immunocytochemistry and cell fractionation.
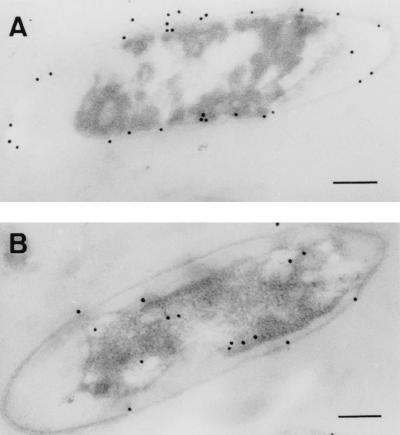
We used the monospecific antibodies recognizing P21 to localize this protein in A. ferrooxidans cells. Figure 7 shows electron micrographs of cells grown in sulfur and ferrous iron. Gold particles attached to the secondary antibodies recognizing the primary antibodies against P21 were concentrated at the cell periphery in sulfur-grown cells (Fig. 7A), suggesting that the protein is associated with the cell wall envelope of A. ferrooxidans. On the other hand, cells grown on ferrous iron (Fig. 7B) showed reduced affinity for the antibody, with low levels of randomly distributed colloidal gold particles. These results are in agreement with the expression of P21 under these two conditions (Fig. 4; Table 1). The antibodies against P21, being monospecific, did not recognize the putative proteins P14 and P16 encoded by the other A. ferrooxidans ORFs in spite of their similarity to rhodanese (results not shown). The presence of a signal peptide and the absence of transmembrane domains in P21 strongly suggest that this protein could be located in the periplasmic space of A. ferrooxidans. By the use of osmotic shock of A. ferrooxidans we obtained a periplasmic fraction which gave no reaction with anti-P21 antibodies by Western analysis. On the other hand, we determined by cell fractionation that P21 was present in both soluble and inner-membrane fractions and not in the outer membrane (results not shown).
FIG. 7.
Localization of P21 in A. ferrooxidans cells grown with different oxidizable substrates. Cells were grown in medium containing either elemental sulfur (A) or ferrous iron (B). Ultrathin sections of both kinds of cells were then subjected to immunocytochemistry with colloidal gold particles and were observed by electron microscopy. Bar, 100 nm.
DISCUSSION
The existence in A. ferrooxidans ATCC 19859 of an exported protein, P21, similar to a thiosulfate-sulfur transferase and which is regulated depending on the oxidizable substrate is very interesting considering the proposal that the oxidation of pyrite generates thiosulfate as one of the main intermediates (30). On the other hand, it has been shown that A. ferrooxidans generates thiosulfate when grown in a medium containing elemental sulfur (32). This could explain why P21 was induced when cells were grown in elemental sulfur. The soluble thiosulfate generated from sulfur would allow the growth of cells not attached to the sulfur particles. If P21 is involved in thiosulfate metabolism, one should expect an increased expression of the protein when the cells are grown in pyrite, thiosulfate, or sulfur, as we have observed. The lack of repression of P21 synthesis by growth in pyrite compared with that obtained by growth in ferrous iron was unexpected. However, during pyrite attack, much smaller amounts of free ferrous iron are probably present and, as we have shown, the levels of P21 drastically decreased at higher concentrations of ferrous iron in the growth medium (Table 1).
Tabita et al. (36) reported the partial purification of a rhodanese from A. ferrooxidans having an optimal transferase activity in the 7.5 to 9.0 pH range. Recently, Gardner and Rawlings (11) detected thiosulfate-sulfur transferase activity in whole cells and crude extracts from A. ferrooxidans, A. thiooxidans, and Acidithiobacillus caldus, whereas this activity was absent from Leptospirillum ferrooxidans, since this microorganism is only capable of oxidizing ferrous iron or the iron contained in pyrite but not its sulfur moiety (29, 30). These results support the idea of rhodanese being involved during in vivo sulfur oxidation. Nevertheless, in the studies of Gardner and Rawlings (11), the rhodanese activity levels in A. ferrooxidans were the same when cells were grown either in ferrous iron or in sulfur. We also determined this activity in crude cell extracts from A. ferrooxidans ATCC 19859 grown in the two substrates and found similar results (data not shown). Therefore, this rhodanese activity does not behave like our reported P21, whose levels were clearly different under the aforementioned conditions. Rhodaneses have been reported as constitutive (6, 9). Therefore, the rhodanese activity measured by Gardner and Rawlings most likely corresponded to the cytoplasmic rhodanese. The regulated protein P21 that we describe here could be anchored on the periplasmic side of the cytoplasmic membrane of A. ferrooxidans and have a different role during sulfur metabolism.
The comparative analysis of the amino acid sequence of P21 with several known thiosulfate-sulfur transferases whose activity has been demonstrated in vitro showed a significant similarity (42%). P21 also contained the highly conserved structural domains CH2A, CH2B, and a catalytic site with a Cys-119, typical of thiosulfate-sulfur transferases (Fig. 2). The putative proteins P14 and P16 also showed rhodanese-like conserved regions, but they did not contain signal peptides on their ORFs, suggesting their cytoplasmic localization, and therefore may correspond to the previously described rhodanese activity from A. ferrooxidans (36). However, proteins P14 and P16 may also correspond to other bacterial proteins related to stress responses. The existence of an extensive family of such proteins containing rhodanese-like domains is well documented (16).
The lack of activity detection for P21 in vitro in our assays may be due to the need for an expected periplasmic acidic optimum pH for the protein. The pH (7.5 to 9.0) conditions for the standard rhodanese activity may not be appropriate to measure the putative rhodanese activity of P21. On the other hand, lowering the pH to 3.0 to 4.0 did not allow us to use cyanide as the sulfur acceptor. Alternatively, the lack of rhodanase activity of P21 may be due to the need for additional polypeptides required for it to be active, since it occurs with the thiosulfate-oxidizing complex from P. versutus (9) and the mitochondrial rhodanese (22).
The studies of a small rhodanese-like protein from Wolinella succinogenes showed that it acts as a periplasmic sulfide dehydrogenase and uses the same catalytic cysteine involved in anion transferase and hydrolase activity (17). This suggests a possible redox function for rhodanese-like proteins similar to that of the thioredoxin proteins. This idea is supported by the presence on the C-terminal end of P21 of a putative cysteine motif involved in the binding of iron-sulfur clusters in electron transport complexes (4). It is also possible that P21 from A. ferrooxidans is a dithiol-disulfide redox activity analogous to the one in W. succinogenes. Nevertheless, it is known that rhodanese-like proteins could show several alternative catalytic activities, among them: (i) detoxification of toxic compounds, such as arsenate and cyanide, by either transferring anions or reducing them and (ii) a chaperone activity to allow efficient assembly of iron-sulfur complex-containing proteins (16).
Due to the lack of an appropriate workable genetic system to perform functional genomics with A. ferrooxidans, at this point it is not possible to assign a definitive role to P21 in sulfur metabolism in A. ferrooxidans. However, given its proximity to genes encoding the sulfate-thiosulfate transport system (Fig. 3), its specific and high level of accumulation in pyrite, sulfur, thiosulfate, and other sulfides, its possible localization bound to the inner membrane but facing the periplasmic space, and its similarity to rhodaneses, the protein is probably involved in the oxidation or in the acquisition of sulfur compounds.
Acknowledgments
This work was supported in part by grants from FONDECYT (1000967), ICGEB (project CRP/CHI00-04, contract 01/001) and ICM-P99-031-F. P.R. was the recipient of a DAAD Ph.D. scholarship.
We thank P. Varela for some initial experiments on this work. We acknowledge Maria-Rosa Bono for assistance with the preparation of the anti-P21 serum. Preliminary sequence data for the A. ferrooxidans strain 23270 was obtained from The Institute for Genomic Research (http://www.tigr.org).
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. L. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaro, A. M., D. Chamorro, M. Seeger, R. Arredondo, I. Peirano, and C. A. Jerez. 1991. Effect of external pH perturbations on in vivo protein synthesis by the acidophilic bacterium Thiobacillus ferrooxidans. J. Bacteriol. 173:910-915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arredondo, R., A. García, and C. A. Jerez. 1994. The partial removal of lipopolysaccharide from Thiobacillus ferrooxidans affects its attachment to solids. Appl. Environ. Microbiol. 60:2846-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berks, B. C., S. J. Ferguson, J. W. B. Moir, and D. J. Richardson. 1995. Enzymes and associated electron transport systems that catalyze the respiratory reduction of nitrogen oxides and oxyanions. Biochim. Biophys. Acta 1232:97-173. [DOI] [PubMed] [Google Scholar]
- 5.Brierley, C. L. 1978. Bacterial leaching. Crit. Rev. Microbiol. 6:207-262. [DOI] [PubMed] [Google Scholar]
- 6.Chandra, T. S., and C. G. Friedrich. 1986. Tn5-induced mutations affecting sulfur-oxidizing ability (Sox) of Thiosphaera pantotropha. J. Bacteriol. 166:446-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Jong, G. A. H., W. Hazeu, P. Bos, and G. Kuenen. 1997. Polythionate degradation by tetrathionate hydrolase of Thiobacillus ferrooxidans. Microbiology 143:499-504. [DOI] [PubMed] [Google Scholar]
- 8.Fowler, T. A., and F. K. Crundwell. 1998. Leaching of zinc sulfide by Thiobacillus ferrooxidans: experiments with a controlled redox potential indicate no direct bacterial mechanism. Appl. Environ. Microbiol. 64:3570-3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedrich, C. G. 1998. Physiology and genetics of sulfur-oxidizing bacteria. Adv. Microb. Physiol. 39:235-289. [DOI] [PubMed] [Google Scholar]
- 10.Friedrich, C. G., D. Rother, F. Bardischewsky, A. Quentmeier, and J. Fischer. 2001. Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl. Environ. Microbiol. 67:2873-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardner, M. N., and D. E. Rawlings. 2000. Production of rhodanese by bacteria present in bio-oxidation plants to recover gold from arsenopyrite concentrates. J. Appl. Microbiol. 89:185-190. [DOI] [PubMed] [Google Scholar]
- 12.Guiliani, N., and C. A. Jerez. 1999. Protein genes from Thiobacillus ferrooxidans that change their expression by growth under different energy sources, p. 79-87. In R. Amils and A. Ballester (ed.), Biohydrometallurgy and the environment toward the mining of the 21st century, part B. Elsevier, Amsterdam, The Netherlands.
- 13.Guiliani, N., and C. A. Jerez. 2000. Molecular cloning, sequencing, and expression of omp-40, the gene coding for the major outer membrane protein from the acidophilic bacterium Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 66:2318-2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison, A. P., Jr. 1984. The acidophilic thiobacilli and other acidophilic bacteria that share their habitat. Annu. Rev. Microbiol. 38:265-292. [DOI] [PubMed] [Google Scholar]
- 15.Kelly, D. P., J. K. Shergill, W.-P. Lu, and A. P. Wood. 1997. Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Leeuwenhoek 71:95-107. [DOI] [PubMed] [Google Scholar]
- 16.Koonin, E. V., L. Aravind, and M. Y. Galperin. 2000. A comparative-genomic view of the microbial stress response, p. 417-444. In G. Storz and R. Hengge-Aronis (ed.), Bacterial stress responses. ASM Press, Washington, D.C.
- 17.Kreis-Kleinschmidt, V., F. Fahrenholz, E. Kojro, and A. Kröger. 1995. Periplasmic sulphide dehydrogenase (Sud) from Wolinella succinogenes: isolation, nucleotide sequence of the sud gene and its expression in Escherichia coli. Eur. J. Biochem. 227:137-142. [DOI] [PubMed] [Google Scholar]
- 18.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 19.Lundgren, D. G. 1980. Ore leaching by bacteria. Annu. Rev. Microbiol. 34:263-283. [DOI] [PubMed] [Google Scholar]
- 20.Matsudaira, P. T. 1989. A practical guide to protein and peptide purification for microsequencing. Academic Press, Inc., New York, N.Y.
- 21.O'Farrell, P. Z., H. M. Goodman, and P. H. O'Farrell. 1977. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell 12:1133-1141. [DOI] [PubMed] [Google Scholar]
- 22.Ogata, K., and M. Volini. 1990. Mitochondrial rhodanese: membrane bound and complexed activity. J. Biol. Chem. 265:8087-8093. [PubMed] [Google Scholar]
- 23.Osorio, G., P. Varela, R. Arredondo, M. Seeger, A. M. Amaro, and C. A. Jerez. 1993. Changes in global expression of Thiobacillus ferrooxidans when grown on elementary sulphur, p. 565-575. In A. E. Torma, M. L. Apel, and C. L. Brierley (ed.), Biohydrometallurgical technologies, vol 2. TMS Press, Warrendale, Pa. [Google Scholar]
- 24.Pronk, J. T., R. Meulenberg, W. Hazeu, P. Bos, and J. G. Kuenen. 1990. Oxidation of reduced inorganic sulphur compounds by acidophilic thiobacilli. FEMS Microbiol. Rev. 75:293-306. [Google Scholar]
- 25.Pugsley, A. 1993. The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ray, W. K., G. Zeng, M. B. Potters, A. M. Mansuri, and T. J. Larson. 2000. Characterization of a 12-kilodalton rhodanese encoded by glpE of Escherichia coli and its interaction with thioredoxin. J. Bacteriol. 182:2277-2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York, N.Y.
- 28.Sand, W., T. Gehrke, R. Hallmann, and A. Schippers. 1995. Sulfur chemistry, biofilm, and the (in)direct attack mechanism—a critical evaluation of bacterial leaching. Appl. Microbiol. Biotechnol. 43:961-966. [Google Scholar]
- 29.Sand, W., T. Gehrke, P. G. Jozsa, and A. Schippers. 2001. (Bio)chemistry of bacterial leaching—direct vs. indirect bioleaching. Hydrometallurgy 59:159-175. [Google Scholar]
- 30.Schippers, A., and W. Sand. 1999. Bacterial leaching of metal sulfides proceeds by two indirect mechanisms via thiosulfate or via polysulfides and sulfur. Appl. Environ. Microbiol. 65:319-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seeger, M., G. Osorio, and C. A. Jerez. 1996. Phosphorylation of GroEL, DnaK, and other proteins from Thiobacillus ferrooxidans grown under different conditions. FEMS Microbiol. Lett. 138:129-134. [DOI] [PubMed] [Google Scholar]
- 32.Shihari, S. R. Bhavaraju, J. M. Modak, R. Kumar, and K. S. Gandhi. 1993. Dissolution of sulfur particles by Thiobacillus ferrooxidans: substrate for unattached cells. Biotechnol. Bioeng. 41:612-616. [DOI] [PubMed] [Google Scholar]
- 33.Silver, M., and D. G. Lundgren. 1968. The thiosulfate-oxidizing enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 46:1215-1220. [DOI] [PubMed] [Google Scholar]
- 34.Singleton, D. R., and D. W. Smith. 1988. Improved assay for rhodanese in Thiobacillus spp. Appl. Environ. Microbiol. 54:2866-2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Suzuki, I. 1999. Oxidation of inorganic sulfur compounds: chemical and enzymatic reactions. Can. J. Microbiol. 45:97-105. [Google Scholar]
- 36.Tabita, R., M. Silver, and D. G. Lundgren. 1969. The rhodanese enzyme of Ferrobacillus ferrooxidans (Thiobacillus ferrooxidans). Can. J. Biochem. 47:1141-1145. [DOI] [PubMed] [Google Scholar]
- 37.Tributsch, H. 1999. Direct versus indirect bioleaching, p. 51-60. In R. Amils and A. Ballester (ed.), Biohydrometallurgy and the environment toward the mining of the 21st century, part A. Elsevier, Amsterdam, The Netherlands.
- 38.Tuovinen, O. 1990. Biological fundamentals of mineral leaching processes, p. 55-77. In H. L. Ehrlich and C. L. Brierley (ed.), Microbial mineral recovery. McGraw-Hill Book Co., New York, N.Y.
- 39.Varela, P., G. Levican, F. Rivera, and C. A. Jerez. 1998. An immunological strategy to monitor in situ the phosphate starvation state in Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 64:4990-4993. [DOI] [PMC free article] [PubMed] [Google Scholar]