Abstract
It has recently been recognized that the ability to use Fe(III) as a terminal electron acceptor is a highly conserved characteristic in hyperthermophilic microorganisms. This suggests that it may be possible to recover as-yet-uncultured hyperthermophiles in pure culture if Fe(III) is used as an electron acceptor. As part of a study of the microbial diversity of the Obsidian Pool area in Yellowstone National Park, Wyo., hot sediment samples were used as the inoculum for enrichment cultures in media containing hydrogen as the sole electron donor and poorly crystalline Fe(III) oxide as the electron acceptor. A pure culture was recovered on solidified, Fe(III) oxide medium. The isolate, designated FW-1a, is a hyperthermophilic anaerobe that grows exclusively by coupling hydrogen oxidation to the reduction of poorly crystalline Fe(III) oxide. Organic carbon is not required for growth. Magnetite is the end product of Fe(III) oxide reduction under the culture conditions evaluated. The cells are rod shaped, about 0.5 μm by 1.0 to 1.2 μm, and motile and have a single flagellum. Strain FW-1a grows at circumneutral pH, at freshwater salinities, and at temperatures of between 65 and 100°C with an optimum of 85 to 90°C. To our knowledge this is the highest temperature optimum of any organism in the Bacteria. Analysis of the 16S ribosomal DNA (rDNA) sequence of strain FW-1a places it within the Bacteria, most closely related to abundant but uncultured microorganisms whose 16S rDNA sequences have been previously recovered from Obsidian Pool and a terrestrial hot spring in Iceland. While previous studies inferred that the uncultured microorganisms with these 16S rDNA sequences were sulfate-reducing organisms, the physiology of the strain FW-1a, which does not reduce sulfate, indicates that these organisms are just as likely to be Fe(III) reducers. These results further demonstrate that Fe(III) may be helpful for recovering as-yet-uncultured microorganisms from hydrothermal environments and illustrate that caution must be used in inferring the physiological characteristics of at least some thermophilic microorganisms solely from 16S rDNA sequences. Based on both its 16S rDNA sequence and physiological characteristics, strain FW-1a represents a new genus among the Bacteria. The name Geothermobacterium ferrireducens gen. nov., sp. nov., is proposed (ATCC BAA-426).
The biogeochemistry of hydrothermal environments is poorly understood. One reason for this is the difficulty in assessing and intensively sampling such environments. Another reason is the steep physical-chemical gradients that are often present in these systems (24, 39-41). Yet another factor is the incomplete understanding of the physiological capabilities of the organisms inhabiting these environments. Analysis of 16S ribosomal DNA (rDNA) sequences in hydrothermal environments has typically revealed a wide diversity of Bacteria and Archaea, most of which have never been cultured (3, 11, 31). In some instances it may be possible to infer the physiological properties of these organisms from the physiology of closely related microorganisms that are available in pure culture. Often, however, sufficiently close relatives are not available, and thus any inferences on the physiological potential of the uncultured organisms do not have a high degree of confidence. Therefore, understanding of the biogeochemistry of hydrothermal environments could be improved if more of the microorganisms inhabiting these environments could be recovered in culture.
It has recently become apparent that one mechanism for recovering novel hyperthermophilic microorganisms in pure culture may be to use Fe(III) as the electron acceptor for enrichment and isolation (17). All hyperthermophilic Archaea and Bacteria that have been evaluated have the ability to reduce Fe(III), and many of these organisms can conserve energy to support growth from Fe(III) reduction (16, 17, 20, 36-38). Furthermore, the metabolic potential of some hyperthermophiles may be expanded when Fe(III) is provided as the electron acceptor. For example, Thermotoga maritima, which grows only fermentatively in the absence of Fe(III), can grow via Fe(III) respiration with hydrogen as the electron donor (38). In addition, Ferroglobus placidus, which was not previously known to oxidize organic compounds, has been found to oxidize acetate and a variety of aromatic compounds when Fe(III) is provided as the electron acceptor (36, 37). Isolation of microorganisms with Fe(III) may also be a good strategy for recovering organisms from hydrothermal environments, because Fe(III) may be an important electron acceptor in many of these environments (14, 15, 18).
Here we report on a microorganism isolated from Obsidian Pool in Yellowstone National Park which oxidizes hydrogen with Fe(III) serving as the electron acceptor. The 16S rDNA sequences most closely related to this isolate are sequences recovered from Obsidian Pool (11) and a hydrothermal spring in Iceland (33). These results further demonstrate that Fe(III) may be helpful for recovering as-yet-uncultured microorganisms from hydrothermal environments.
MATERIALS AND METHODS
Source of organism.
Geothermobacterium ferrireducens was isolated from hot sediment samples collected from Obsidian Pool in Yellowstone National Park, Wyo. Thermodesulfobacterium commune (DSM 2178) and Thermodesulfobacterium hveragerdense (DSM 12571) were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen in Braunschweig, Germany.
Enrichment and isolation.
A modified Hungate (12) technique was used throughout this study (4, 27). Gases were passed through a column of hot copper filings (350°C) to remove all traces of oxygen. All additions of solutions, transfers, and sampling of cultures (through thick butyl rubber-stoppered pressure tubes) were performed with disposable plastic syringes fitted with 23-gauge hypodermic needles that had been flushed with oxygen-free gas.
The freshwater enrichment medium contained the following components: poorly crystalline Fe(III) oxide (100 mM), MgCl2 · 6H2O (0.33 g liter−1), CaCl2 · 2H2O (0.33 g liter−1), KCl (0.33 g liter−1), KH2PO4 (0.33 g liter−1), NH4Cl (0.25 g liter−1), NaHCO3 (2.5 g liter−1), Na2SeO4 (100 μM), a vitamin solution (23) (10 ml liter−1), and a trace mineral solution (23) (10 ml liter−1). The final pH of the medium was about 7.0 (at room temperature, before autoclaving). The medium (10 ml) was dispensed into 26-ml anaerobic pressure tubes (Bellco Glass, Inc., Vineland, N.J.) and sparged with N2-CO2 (80:20, vol/vol) for 12 to 15 min to remove dissolved oxygen. The tubes were then sealed with thick butyl rubber stoppers and an aluminum crimp seal. The poorly crystalline Fe(III) oxide was synthesized by neutralizing a solution of FeCl3 as previously described (21).
After autoclaving, the medium in each tube was supplemented with 0.25 mM l-cysteine-HCl, 1.3 mM FeCl2 · 2H2O, and one of the following electron donors: 10 mM dl-lactate, 10 mM acetate, 10 mM pyruvate, 10 mM succinate, 10 mM fumarate, 10 mM formate, or H2-CO2 (80:20, vol/vol; 101 kPa). Electron donors were added from sterile anoxic stock solutions. No additional electron donor was added when H2 was tested as the electron donor. A 1.0-ml aliquot of the original sample was transferred into a tube containing 10 ml of the above-described freshwater medium with one of the electron donors and N2-CO2 (80:20, vol/vol; 101 kPa) or H2-CO2 (80:20, vol/vol; 101 kPa) as the gas phase.
A modification of the roll tube method (13) for cultivation of strict anaerobes by using serial dilution of enrichment culture on solid medium was employed to isolate single colonies (17). The solid medium was prepared by adding 2% Gelrite gellan gum (Sigma Chemical Co., St. Louis, Mo.) to anaerobically prepared Milli-Q H2O (1.0 g/50 ml in gassed and sealed 150-ml serum bottles) followed by autoclaving for 30 min. Immediately after being removed from the autoclave, the gellan gum was mixed well and placed in an 85 to 90°C water bath. A double-concentration freshwater medium was prepared with an additional 20 mM MgCl2 and 6 mM CaCl2, while the poorly crystalline Fe(III) oxide concentration was kept at 100 mM. While stirring, 3.5-ml aliquots of the double-concentration medium were anaerobically transferred into gassed-out (H2-CO2; 80:20, vol/vol) pressure tubes, which were then stoppered with thick butyl rubber stoppers, sealed (using aluminum crimp seals), and then autoclaved and placed in the same water bath as the molten gellan gum (at 85 to 90°C). The medium was then supplemented with l-cysteine-HCl (0.5 mM) and FeCl2 · 2H2O (2.6 mM). A dilution series (10−1 to 10−9) was established in nine of the tubes containing the double-concentrated medium. A 3.5-ml aliquot of 2% molten gellan gum was transferred into a pressure tube containing 3.5 ml of the double-concentrated freshwater medium, followed quickly by a 0.7-ml aliquot of the selected enrichment culture. After the contents of the tube were gently and thoroughly mixed (maintaining a temperature of between 85 to 90°C), 0.7 ml was quickly removed and transferred into the second pressure tube containing 3.5 ml of the double-concentration freshwater medium and mixed well. Using a tube spinner (Bellco Glass Inc.), the first pressure tube was then rolled at room temperature, taking care to ensure an even coating of the inner pressure tube wall. A 3.5-ml aliquot of 2% molten gellan gum was then transferred into the second tube, now containing 3.5 ml of the double-concentrated medium plus the 0.7-ml inoculum from the first tube. This was mixed as before, and the subsequent serial dilutions were performed in the same way. When complete, the serial dilutions were incubated vertically at 85°C.
Electron donor and acceptors.
To determine the ability of the isolate to use electron donors other than hydrogen, electron donors in the form of the sodium salt (when possible) were added individually from concentrated sterile and anoxic stock solutions to give a final concentration of 10 mM unless otherwise stated (Table 1). Hydrophobic long-chain fatty acid stocks (100 mM) were prepared in sterile anoxic water in sterile anaerobic tubes flushed with oxygen-free N2, which were then slowly heated to 80°C (until completely dissolved), and then 0.1-ml aliquots of these stocks were added to the growth medium to provide a final concentration of 1 mM. Substrate utilization was monitored by measuring growth and Fe(II) production over the incubation period at 85°C and under an N2-CO2 (80:20, vol/vol) atmosphere.
TABLE 1.
Substrate tests with strain FW-1a in pure culture at 85°C
| Category | Substrate (concna) |
|---|---|
| Electron donor | |
| Utilized | H2 (as H2-CO2; 80:20, vol/vol; 110 kPa) |
| Not utilized | Lactate (10), pyruvate (10), acetate (5), malate (10), succinate (10), peptone (0.1%), formate (10), fumarate (10), yeast extract (0.1%), glycerol (20), methanol (20)b, ethanol (20)b, citrate(10), alanine (11.2), histidine (6.4), proline (8.7), glycine (13.3), isoleucine (7.6), aspartic acid (7.5), glutamic acid (6.8), arginine (5.7), l-cysteine (8.3), serine (9.5), asparagine (5.7), palmitate (1), stearate (1), valerate (1-10), butyrate (5), propionate (5), catechol (1), phenol (0.25-0.50)b, toluene (1)b, benzoate (0.5-1.0)b, benzoic acid (0.5-1.0 mM)b, benzene (0.5-1.0 mM)b |
| Electron acceptor | |
| Utilized | Poorly crystalline Fe(III) (100) |
| Not utilized | SO42− (10), S2O32− (10), SO32− (10), S0 (2%, wt/vol), NO3− (10), NO2− (1.0-1.5), O2 (0.6-1.0% evaluated), MnO2 [poorly crystalline Mn(IV) (20)], 2,6-anthraquinone disulfonate (5), malate (10), fumarate (10-50), ferric pyrophosphate (10), ferric citrate (10-50), goethite (50), hematite (50), Co(III)-EDTA (200 μM), selenate (10), selenite (10), dimethyl sulfoxide (1-2) |
Concentrations are in millimolar unless otherwise indicated.
Tested at 70 to 75°C.
In experiments testing alternative electron acceptors (Table 1), poorly crystalline Fe(III) oxide was omitted from the medium and alternative electron acceptors were added to the sterile and anoxic basal medium from sterile anoxic stock solutions. Goethite (50 mM) (21) and hematite (50 mM) (21) were also used as the more crystalline forms of Fe(III) oxide and synthetic MnO2 (15 mM) (22) and were added from steam-sterilized anoxic stocks (steamed in a steam chamber for 1 h per day over a 3-day period) (1.0 mM) in order to further evaluate the potential for Fe(III) and Mn(IV) reduction. Fe(III)-citrate (20 to 50 mM) (23) and Fe(III)-pyrophosphate (3 g liter−1) (7) were provided as soluble forms of Fe(III). When noted, alternative electron acceptors (in the form of sodium salt when possible), such as nitrate (10 mM), nitrite (1.0 to 1.5 mM), sulfate (10 and 20 mM), thiosulfate (8 to 10 mM), sulfite (4 mM), dimethyl sulfoxide (1 to 2 mM), fumarate (10 and 20 mM), malate (10 mM), selenate (10 mM), selenite (10 mM), anthraquinone-2,6-disulfonate (5 mM), and Co(III)-EDTA (100 μM) (16), were added to the growth medium from sterile anoxic stock solutions in order to evaluate their potential reduction as alternative electron acceptors. O2 (0.5 to 1.0%, vol/vol) and colloidal S0 (20 g liter−1) (5) were also evaluated as potential electron acceptors.
Effect of temperature, salinity, and pH on growth and Fe(III) reduction.
The influence of temperature on growth rate was determined over a range of 58 to 102°C. The cultures were incubated in either temperature-controlled hot air incubators or water baths with a calibrated thermometer. The effect of NaCl on growth was determined by varying its concentration from 0 to 1% at 85°C.
The isolate was cultured in freshwater medium at neutral pH with 30 mM sodium bicarbonate as the buffering agent under an H2-CO2 (80:20, vol/vol; 101 kPa) atmosphere. The effect of pH on the growth of the isolate was not investigated. This is due to the inability of the new isolate to utilize electron acceptors other than Fe(III), which is abiotically reduced with MES (morpholinepropanesulfonic acid), PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], HEPES, and Tris buffers, particularly at hyperthermophilic temperatures.
Antibiotic susceptibility.
Sensitivity to chloramphenicol (100 μg ml−1), cycloheximide (100 μg ml−1), kanamycin (100 μg ml−1), neomycin sulfate (100 μg ml−1), novobiocin (100 μg ml−1), puromycin (100 μg ml−1), rifampin (100 μg ml−1), erythromycin (150 μg ml−1), tetracycline (150 μg ml−1), ampicillin (200 μg ml−1), penicillin G (200 μg ml−1), phosphomycin (200 μg ml−1), streptomycin (200 μg ml−1), vancomycin-HCl (200 μg ml−1), and trimethoprim (300 μg ml−1) (all from Sigma) was tested. Aliquots (1 ml) of an exponentially growing culture were anaerobically transferred into pressure tubes containing 10 ml of anaerobic freshwater medium containing poorly crystalline Fe(III) oxide and filter-sterilized antibiotic. The cultures were pressurized with H2-CO2 (80:20, vol/vol; 101 kPa), incubated at 75°C (in order to reduce the chance of thermal inactivation of these antibiotics), and routinely examined for cell growth and Fe(II) accumulation as a result of Fe(III) reduction.
Light and electron microscopy.
Cells were routinely examined with a Zeiss standard phase-contrast microscope (Oberkochen) with an oil immersion objective (100/1.25), and acridine orange-stained cells were observed by epifluorescence microscopy. Electron microscopy was carried out as previously described (19). Briefly, cells grown on poorly crystalline Fe(III) oxide were fixed for 1 h with glutaraldehyde (5%, vol/vol) in 0.1 M cacodylate buffer (pH 7.2), followed by brief washing with cacodylate buffer and postfixation with osmium tetroxide (1%, vol/vol) for 2 h. Samples were dehydrated through increasing concentrations of ethanol and embedded in epoxy resin, which was polymerized at 70°C for 8 h. Thin sections (70 nm) were stained with 5% uranyl acetate and 0.4% lead citrate and were examined with a JEOL 100S transmission electron microscope.
Analytical techniques.
Fe(III) reduction was monitored by measuring the accumulation of Fe(II) over time. Production of Fe(II) was measured with ferrozine (30) after a 2-h extraction at room temperature with anaerobic oxalate solution (28 g of ammonium oxalate and 15 g of oxalic acid liter−1) in the dark as previously described (30).
Cell numbers were determined by counting acridine orange-stained cells under oil immersion (×1,000) with an ocular grid and UV light (10). Cells were stained by anaerobically adding a 1-ml aliquot of the culture to 0.1 ml of glutaraldehyde (final concentration, 2.5%), waiting for 2 min, and then transferring the mixture to 5.9 ml of filter-sterilized oxalate solution to dissolve the poorly crystalline Fe(III) oxide. When the extraction was complete, a 2-ml aliquot of the sample was mixed with 3 drops (∼0.2 ml) of filtered acridine orange solution (final concentration, 0.01%) for 2 min. Finally the sample was filtered through a black Isopore membrane filter (0.2-μm pore diameter; Millipore) and examined under UV light.
DNA base composition.
Multiple attempts to grow strain FW-1a with an alternative electron acceptor (Table 1) in place of poorly crystalline Fe(III) oxide were unsuccessful. Therefore, DNA base composition analysis of the isolate could not be done.
Isolation of DNA and 16S rDNA phylogenetic analysis.
Cultures (10 ml) grown with poorly crystalline Fe(III) oxide as the electron acceptor were first treated with 30 ml of 300 mM filter-sterilized oxalate solution (30) in order to remove Fe(III), which inhibits Taq polymerase. Cells were collected by centrifugation, and genomic DNA was extracted as previously described in detail (32). Briefly, after the centrifugation, the resulting cell pellet was washed in 2 ml of 30 mM sodium bicarbonate buffer and resuspended in 0.5 ml of the same buffer. The resulting cell suspension was then subjected to five freeze-thaw cycles at −80 and 65°C and then extracted with phenol, phenol-chloroform-isoamyl alcohol (25:24:1), and chloroform-isoamyl alcohol (24:1).
Nearly the entire 16S rRNA gene of strain FW-1a was amplified by using primers 8 F (9) and 1525 R (1). PCR mixtures were prepared as previously described (28), with the following modifications: a total volume of 100 μl of reaction mixture contained 1.5 mM MgCl2, 2.5 U of AmpliTaq (Perkin-Elmer Cetus, Norwalk, Conn.), and 15% glycerol. Amplification was performed in a GeneAmp PCR System 2400 thermal cycler (Perkin-Elmer Cetus) with an initial denaturation step at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 30 s, which were then followed with a final extension at 72°C for 6 min. PCR products were prepared for sequencing with a QIAquick PCR purification kit (Qiagen Inc., Valencia, Calif.).
Complete bidirectional sequences were compared to the GenBank and Ribosomal Database Project databases by using the BLAST (2) and SMILARITY-RANK (26) algorithms. The secondary structure was verified prior to manual alignment with related 16S rDNA sequences obtained from GenBank and the Ribosomal Database Project by using the Genetic Computer Group (Madison, Wis.) sequence editor (Wisconsin package version 10). Aligned sequences were imported into PAUP 4.0b 4a (35), where phylogenetic trees were inferred. Branching order was determined and compared using character-based (maximum parsimony and maximum likelihood) and distance-based (using the HKY85 four-parameter, Jukes-Cantor, and Kimura two-parameter models) algorithms. Bootstrap analysis was performed using the distance-based HKY85 four-parameter model with 100 replicates.
The similarity matrix program (25), available on the Ribosomal Database Project II website, and ALIGN version 2.0 (29) were used to generate a similarity matrix with 1,200 alignment positions considered.
The GenBank and EMBL accession numbers for known sequences used in phylogenetic analyses are as follows: T. commune strain YSRA-1, L10662; T. hveragerdense, X96725; environmental clone SRI-93, AF255596; environmental clone SRI-27, AF255595; environmental clone OPT4, AF027093; environmental clone OPT53, AF027094; environmental clone OPS7, AF027095; environmental clone OPB45, AF027096; Thiobacillus ferrooxidans, AB039820; Deferribacter thermophilus, U75602; Desulfatotherma hydrogenophila, AF332514; Deinococcus radiodurans, Y11332; Geobacter metallireducens, L07834; Thermus thermophilus, TTRN16S; Fervidobacterium islandicum, FERRRDAAB; Thermotoga maritima, M21774; Desulfurobacterium thermolithotrophicum, DTBSA16SR; and Hydrogenothermus marinus, HMA292525.
Nucleotide sequence accession number.
The sequences determined in this study were deposited in GenBank under accession number AF411013.
RESULTS AND DISCUSSION
Enrichment and isolation.
Ten milliliters of freshwater medium containing 100 mM poorly crystalline Fe(III) as an electron acceptor and H2 (as H2-CO2; 80:20, vol/vol; 101 kPa) as the electron donor was inoculated with 1 ml of sample from Obsidian Pool, Yellowstone National Park. After 2 days at 80°C, the color of the poorly crystalline Fe(III) oxide changed from brick red-brown to a black magnetic mineral (probably magnetite). These enrichments were then used to inoculate (10%, vol/vol) 10 ml of the same medium in sets of five so that a member of each set could then be incubated at 75, 80, 85, 90, and 100°C. Only one of the original enrichments (designated FW-1a) was successfully transferred past the third transfer at 85°C. After the fifth successful transfer, serial dilutions (10−1 to 10−9) were made from this enrichment. The highest dilution (10−8) that reduced Fe(III) served as the inoculum for an additional series of dilutions. This procedure was then repeated a third time, and the highest positive dilution was selected for isolation of a pure culture.
Individual colonies were obtained in roll tube dilutions of hydrogen-Fe(III) oxide medium solidified with Gelrite. After 10 days at 85°C, single black colonies (0.5 to 1.0 mm) appeared in the more dilute tubes. Single colonies were picked from the highest dilutions and transferred into 2 ml of the freshwater medium containing 100 mM poorly crystalline Fe(III) oxide and H2 (as H2-CO2; 80:20, vol/vol; 101 kPa), supplemented with 0.25 mM cysteine and 1.3 mM FeCl2 · 4H2O, and were incubated at 85°C. Cultures with the highest rate of Fe(III) reduction were serially diluted into fresh medium, and then the highest positive dilution was serially diluted again. This process was repeated a total of three times, at which point the culture was considered to be pure. The culture, designated strain FW-1a, was morphologically uniform and contained only one 16S rDNA sequence.
Morphology.
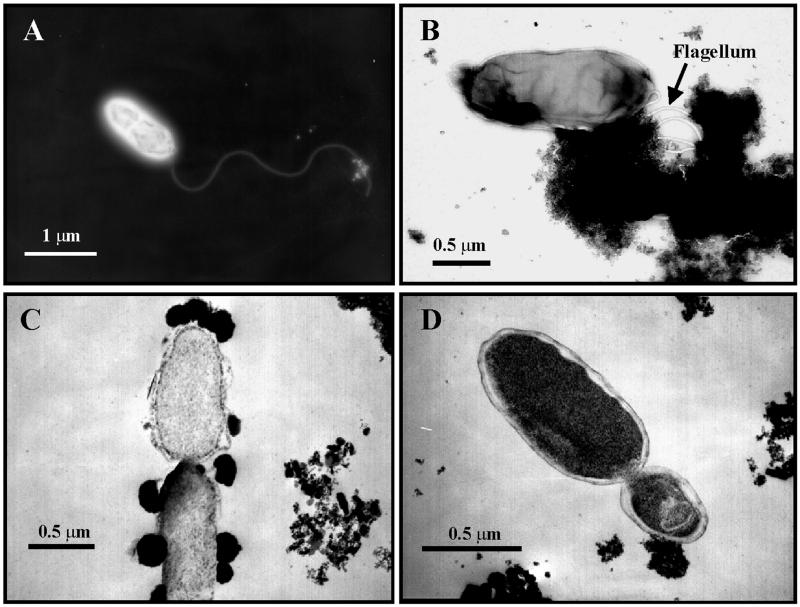
By epifluorescence microscopy, cells of strain FW-1a were rod shaped, ca. 1.0 to 1.2 μm in length and 0.5 μm in diameter, and usually observed as single cells or in pairs (not shown). The cells were highly motile, even at room temperature, when examined by phase-contrast microscopy. The electron micrograph of the cells exhibited monotrichous flagellation (a single flagellum about 12 nm thick and up to 8 μm long) (Fig. 1A and B). Several attempts to Gram stain the cells of strain FW-1a were unsuccessful, probably due to its exclusive growth on poorly crystalline Fe(III) oxide as the electron acceptor, which appears as dark deposits (electron-dense granules) on the cell surface (Fig. 1B and C) and interferes with the Gram staining reaction. However, electron micrographs from ultrathin sections of cells of strain FW-1a are typical of a gram-negative organism (Fig. 1D). Spore formation was not observed.
FIG. 1.
(A) Electron micrograph of strain FW-1a, showing a single flagellum. (B) Electron micrograph of strain FW-1a grown in freshwater medium with poorly crystalline Fe(III) oxide as the sole electron acceptor and H2 as the sole electron donor in early log phase. (C) Thin-section electron micrograph of strain FW-1a in freshwater medium with poorly crystalline Fe(III) oxide in mid-log phase; the electron-dense material attached to the cells surface appears to be poorly crystalline Fe(III) oxide, which is detached from the cells when it is reduced to magnetite. (D) Thin-section electron micrograph of strain FW-1a (in the same medium as in panels B and C) in late log phase, demonstrating a typical gram-negative bacterium.
Hydrogen oxidation coupled to Fe(III) oxide reduction.
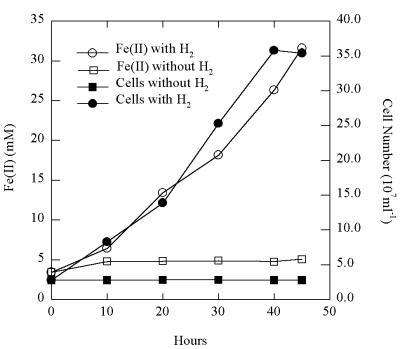
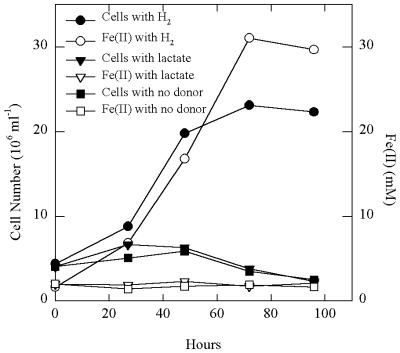
Strain FW-1a grew in defined medium at 85°C and under strictly anaerobic conditions, with H2 as the sole electron donor and poorly crystalline Fe(III) oxide as the sole electron acceptor, without the addition of an organic carbon source. In the presence of hydrogen, Fe(III) was reduced, as evidenced by an accumulation of Fe(II) in the medium over the incubation period (Fig. 2). There was no Fe(III) reduction or cell growth in the absence of added hydrogen. For each mole of H2 consumed, 1.97 ± 0.16 mol (mean ± standard deviation; n = 5) of Fe(II) was produced, which is in agreement with the expected stoichiometry according to the equation H2 + 2Fe(III) → 2H+ + 2Fe(II). This is only the second example of an Fe(III)-reducing microorganism capable of growing autotrophically on hydrogen (17).
FIG. 2.
Growth of strain FW-1a at 85°C with H2 as the electron donor and poorly crystalline Fe(III) oxide as the electron acceptor. The results are the means from duplicate cultures.
Other electron donors and acceptors.
Several attempts to grow strain FW-1a on a wide variety of electron donors and electron acceptors other than H2 and poorly crystalline Fe(III) oxide were unsuccessful (Table 1). Strain FW-1a was unable to reduce Fe(III) with a number of other organic electron donors, such as acetate, pyruvate, lactate, formate, fumarate, and aromatic compounds (Table 1). No growth was observed with complex organic compounds such as yeast extract, peptone, tryptone, or Casamino Acids as substrates (Table 1). None of the amino acids tested (Table 1) could serve as a sole electron donor for Fe(III) reduction. Growth on sugars (e.g., glucose, fructose, and ribose) could not be investigated because the sugars reacted abiotically with Fe(III) oxide at high temperatures.
A wide variety of commonly considered electron acceptors, including sulfate, thiosulfate, sulfite, S0, nitrate, nitrite, oxygen (0.6 to 1.0% evaluated), anthraquinone-2,6-disulfonate, Mn(IV), fumarate, Fe(III)-citrate, and Fe(III)-pyrophosphate did not support growth with hydrogen as the electron donor (Table 1).
Temperature, salt, and pH optima.
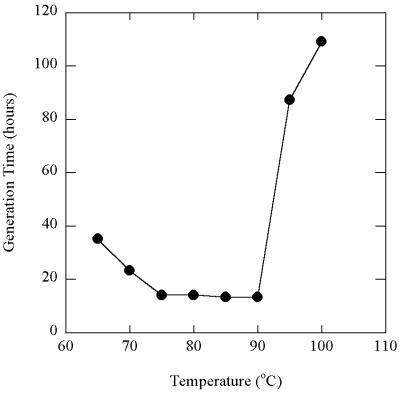
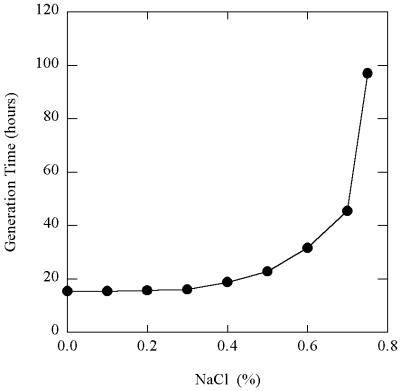
With hydrogen as the electron donor and Fe(III) oxide as the electron acceptor, FW-1a grew at between 65 and 100°C, with an optimum temperature at around 85 to 90°C (with a doubling time of about 14 to 15 h) (Fig. 3). The doubling time at 65°C was about 35 h, while the doubling time at 100°C was around 109 h. No growth was detected at 63°C or above 100°C. At 85°C strain FW-1a grew in medium containing 0 to 0.75% (wt/vol) NaCl, but growth was slow (doubling time, 97 h) at the highest concentration of NaCl, and no growth was detected with 0.8% (wt/vol) NaCl in the medium (Fig. 4).
FIG. 3.
Optimal growth temperature of isolate FW-1a during growth on H2 as the electron donor and poorly crystalline Fe(III) oxide as the electron acceptor. Doubling times were calculated from the slopes of the growth curves (not shown) at pH 7.0.
FIG. 4.
Effect of NaCl concentration on growth of isolate FW-1a at 85°C with H2 as the electron donor and poorly crystalline Fe(III) oxide as the electron acceptor. Doubling times were calculated from the slopes of the growth curves (not shown) at pH 7.0.
The effect of pH on the growth of strain FW-1a was not investigated due to the inability of the isolate to utilize electron acceptors other than Fe(III), which is abiotically reduced with MES, PIPES, HEPES, and Tris buffers, particularly at the optimum growth temperature.
Sensitivity to antibiotics.
Growth of strain FW-1a was inhibited by chloramphenicol (100 μg ml−1), puromycin (100 μg ml−1), rifampin (100 μg ml−1), erythromycin (150 μg ml−1), kanamycin (200 μg ml−1), phosphomycin (200 μg ml−1), vancomycin-HCl (200 μg ml−1), and trimethoprim (300 μg ml−1). The growth of strain FW-1a was not inhibited, however, by cycloheximide (100 μg ml−1), kanamycin (100 μg ml−1), neomycin sulfate (100 μg ml−1), novobiocin (100 μg ml−1), tetracycline (150 μg ml−1), ampicillin (200 μg ml−1), penicillin G (200 μg ml−1), and streptomycin (200 μg ml−1).
16S rDNA sequence analysis.
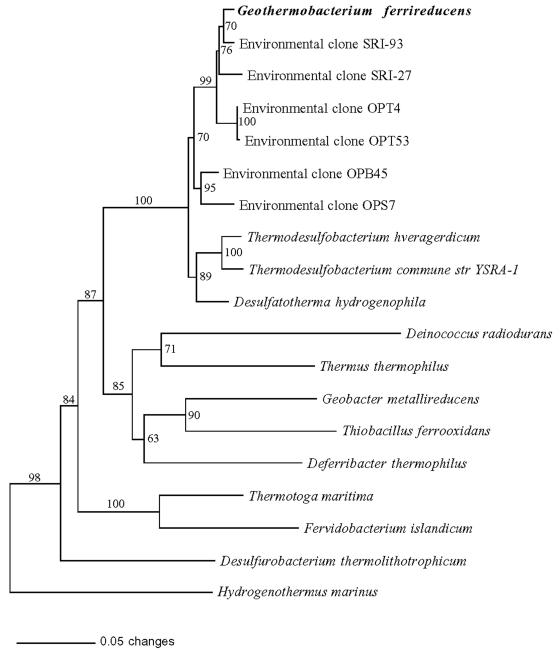
Phylogenetic analysis of the 16S rDNA sequence of strain FW-1a (GenBank accession number AF411013) indicated that its closest relatives are the environmental clones SRI-93 (33) (99.0% similar), SRI-27 (33) (96.2% similar), OPT4 (11) (98.1% similar), OPT53 (96.5% similar), OPB45 (96.8% similar), and OPS7 (97.6% similar) (11) (1,000 bases were considered in each case). The most closely related microorganisms available in culture are the sulfate-reducing microorganisms, T. commune strain YSRA-1 (42) (92% similar [1,200 bases considered]) and T. hveragerdense (34) (94.5% similar [1,200 bases considered]), which are members of Thermodesulfobacteriaceae (Fig. 5).
FIG. 5.
Phylogenetic tree generated by maximum-likelihood analysis of 16S rDNA sequences, showing the relationship of G. ferrireducens to previously described eubacterial sequences. Pyrococcus abyssi was used as the outgroup, and the branching order was identical when maximum-parsimony and distance-based algorithms were applied to aligned sequences in PAUP 4.0b 4a (35), with hypervariable regions masked (1,000 bases were considered).
Evaluation of ability of T. commune and T. hveragerdense to reduce Fe(III).
T. commune could be routinely transferred in medium with hydrogen as the sole electron donor and Fe(III) as the sole electron acceptor (Fig. 6). However, growth was much slower than that of strain FW-1a under the same conditions (doubling times of ≈ 20 versus ≈ 10 h). T. commune did not grow with lactate as the electron donor and Fe(III) as the electron acceptor. However, it grows with lactate and sulfate in the same medium. Multiple attempts to adapt T. hveragerdense to grow with poorly crystalline Fe(III) oxide as the electron acceptor with hydrogen and/or lactate as the electron donor were unsuccessful.
FIG. 6.
Growth of T. commune at 70°C with lactate as an electron donor and poorly crystalline Fe(III) oxide as an electron acceptor. The results are the means from duplicate cultures.
Ecological implications.
In two recent studies of terrestrial hot springs, 16S rDNA sequences closely related to strain FW-1a were found to be important components of the microbial community (8, 11). In both cases, it was inferred that the microorganisms with these sequences were sulfate-reducing microorganisms. Furthermore, in one instance (8) it was also inferred that the microorganisms had a heterotrophic metabolism. These inferences were valid based on the information available at that time, because the closest known relatives for these sequences were T. commune and T. hveragerdense, two thermophilic sulfate-reducing microorganisms (34, 42). However, all of the 16S rDNA sequences recovered from these hydrothermal environments are more closely related to the 16S rDNA sequence of strain FW-1a than to that of either T. commune or T. hveragerdense. This, coupled with the finding that strain FW-1a cannot reduce sulfate and is able to use only hydrogen as the electron donor, suggests that the microorganisms which were predicted to be sulfate reducers may in fact not have the capacity for sulfate reduction or for heterotrophic metabolism. Rather they may have been autotrophic hydrogen-oxidizing Fe(III)-reducing microorganisms, with a physiology similar to that of strain FW-1a.
It was suggested that in Obsidian Pool, the environment from which strain FW-1a was isolated, a source of hydrogen for the microbial community might be the reduction of water with Fe(II) (8). If so, this would produce not only hydrogen but also Fe(III), providing both the electron acceptor and the electron donor needed to support the growth of FW-1a and possibly the other organisms (8) which were previously thought to be sulfate-reducing microorganisms. It is also expected that Fe(III) will be available in this and other hydrothermal environments, as Fe(II)-rich waters contact oxygen which will abiotically oxidize Fe(II) to Fe(III) with the precipitation of Fe(III) oxide (6, 14, 15). Thus, the possibility that not only strain FW-1a but also the microorganisms associated with previously recovered 16S rDNA sequences from Obsidian Pool (8) may be growing via Fe(III) reduction is consistent with the geochemistry of this environment.
These results demonstrate the limitations to inferring physiology and likely biogeochemical reactions in hydrothermal environments based on analysis of 16S rDNA sequences. Ideally, the biogeochemical reactions in such environments should be measured directly, and the activity of different microbial populations should be assessed by monitoring the expression of genes related to the physiology of interest. However, the ability to infer physiology from 16S rDNA sequences will continue to improve as more organisms are recovered in pure culture and their physiology is characterized. Although it has been asserted that it is difficult or impossible to isolate most of the organisms from hydrothermal environments (8), the study reported here represents only the second (17) attempt to isolate a microorganism from a hydrothermal environment with Fe(III) oxide as the electron acceptor. Given the widespread ability of hyperthermophilic microorganisms to reduce Fe(III) and the fact that some of these organisms, such as FW-1a, can use only Fe(III) oxide as an electron acceptor (17), further attempts to recover more of the as-yet-uncultured organisms from hydrothermal environments with Fe(III) oxide as the electron acceptor seem warranted.
Comparison with Thermodesulfobacterium species.
The 16S rDNA sequence of strain FW-1a indicates that its closest known relatives are T. commune (92% similar) and T. hveragerdense (94.5% similar). Although analysis of its 16S rDNA sequence suggests that strain FW-1a is most closely related to Thermodesulfobacterium species, its metabolism is completely different. Unlike the Thermodesulfobacterium species, strain FW-1a is unable to use sulfate and thiosulfate as electron acceptors. Several attempts to grow strain FW-1a on a wide variety of commonly considered electron acceptors, including sulfate (10 to 20 mM), thiosulfate (10 mM), sulfite (2 to 4 mM), and S0 (20%, wt/vol), with H2 (as H2-CO2; 80:20%, vol/vol; 101 kPa), lactate (10 mM), pyruvate (10 mM), or a combination of H2-lactate, and H2-pyruvate as electron donors were unsuccessful. Strain FW-1a grows exclusively with poorly crystalline Fe(III) oxide as the electron acceptor. In contrast to T. commune and T. hveragerdense, which are heterotrophs, strain FW-1a is unable to grow with lactate and pyruvate as electron donors; it grows exclusively with hydrogen as the electron donor. The growth temperature ranges for T. commune and T. hveragerdense are between 45 and 85°C (with an optimum of 70°C) and between 55 and 74°C (with an optimum of between 70 and 74°C), respectively. Strain FW-1a, however, grows at between 65 and 100°C (optimum temperature, between 85 and 90°C). To our knowledge this is the highest optimum growth temperature for a bacterium.
On the basis of its unique metabolic properties, morphology, and 16S rDNA sequence, we conclude that strain FW-1a represents a new genus, and the name Geothermobacterium ferrireducens is proposed.
Description of Geothermobacterium gen. nov.
Geothermobacterium (Ge.o.thermo. bacterium. Gr. n. geo, the earth; Gr. n. thermos, heat; Gr. n. bakterion, a small rod; N.L. masc.n. Geothermobacterium, a small rod from hot earth). Cells are rod shaped, 0.5 by 1.0 to 1.2 μm, occurring singly and in pairs, highly motile (even at room temperature), by means of a monotrichous flagellum. Cell wall structure typical of a gram-negative bacterium. Strictly anaerobic chemoautotroph, which conserves energy to support growth, by coupling the oxidation of hydrogen to the reduction of poorly crystalline Fe(III) oxide. Grows exclusively with hydrogen as the sole electron donor and poorly crystalline Fe(III) oxide as the sole electron acceptor. The genus Geothermobacterium is in the family Thermodesulfobacteriaceae within the division δ-Proteobacteria. Habitat, hot springs (Obsidian Pool area) in Yellowstone National Park.
Description of Geothermobacterium ferrireducens sp. nov.
Geothermobacterium ferrireducens (fer.ri.re.du′cens. L.n. ferrum, iron; L. part. adj. reducens, converting to a different state; N.L. adj. ferrireducens, reducing iron). Gram-negative rods, 0.5 by 1.0 to 1.2 μm, occurring singly and in pairs, motile (even at room temperature) by means of a monotrichous flagellum. Strictly anaerobic autotroph, grows exclusively with hydrogen as the sole electron donor and poorly crystalline Fe(III) oxide as the sole electron acceptor. No growth with sulfate, thiosulfate, sulfite, sulfur, nitrate, fumarate, ferric citrate, or ferric pyrophosphate as electron acceptor with hydrogen, dl-lactate, pyruvate, formate, fumarate, yeast extract, peptone, amino acids, aromatic compounds, short- and long-chain fatty acids, or carbohydrates. Growth optimum between 85 and 90°C, at near-neutral pH (pH 6.8 to 7.0). No growth at 58 or 102°C. Can tolerate NaCl concentration of up to 0.75%, with an optimum at 0.0 to 0.05%. Growth inhibited by 0.8% NaCl. Growth also inhibited by chloramphenicol (100 μg ml−1), puromycin (100 μg ml−1), rifampin (100 μg ml−1), erythromycin (150 μg ml−1), kanamycin (200 μg ml−1), phosphomycin (200 μg ml−1), vancomycin-HCl (200 μg ml−1), and trimethoprim (300 μg ml−1). Isolated from hot sediment samples from Obsidian Pool, Yellowstone National Park (Wyoming, United States).
The strain has been deposited in the American Type Culture Collection (ATCC BAA-426).
Acknowledgments
We thank Lucy Ru-Sin Yin for support with electron microscopy.
This research was supported by grants to D.R.L. (MCB-0085365) from the LExEn program and A.L.R. (OCE-9996160) from the National Science Foundation.
REFERENCES
- 1.Achenbach, L., and C. Woese. 1995. 16S and 23S rRNA-like primers, p. 201-203. In F. T. Robb, A. R. Place, K. R. Sowers, H. J. Schreier, S. DasSarma, and E. M. Fleischmann (ed.), Archaea: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: reevaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blumentals, I. I., M. Itoh, G. J. Olson, and R. M. Kelly. 1990. Role of polysulfides in reduction of elemental sulfur by the hyperthermophilic archaebacterium Pyrococcus furiosus. Appl. Environ. Microbiol. 56:1255-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brock, T. D., C. S., S. Petersen, and J. L. Mosser. 1976. Biogeochemistry and bacteriology of ferrous iron oxidation in geothermal habitats. Geochim. Cosmochim. Acta 40:493-500. [Google Scholar]
- 7.Caccavo, F., D. J. Lonergan, D. R. Lovley, M. Davis, J. F. Stolz, and M. J. McInerney. 1994. Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60:3752-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dojka, M. A., J. Kirk Harris, and N. R. Pace. 2000. Expanding the known diversity and environmental distribution of an uncultured phylogenetic division of bacteria. Appl. Environ. Microbiol. 66:1617-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eden, P. E., T. M. Schmidt, R. P. Blakemore, and N. R. Pace. 1991. Phylogenetic analysis of Aquaspirillum magnetotacticum using polymerase chain reaction-amplified 16S rRNA-specific DNA. Int. J. Syst. Bacteriol. 41:324-325. [DOI] [PubMed] [Google Scholar]
- 10.Hobbie, J. E., R. J. Daley, and S. Jasper. 1977. Use of Nuclepore filters for counting bacteria by fluorescence microscopy. Appl. Environ. Microbiol. 33:1225-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hungate, R. E. 1950. The anaerobic mesophilic cellulytic bacteria. Bacteriol. Rev. 14:1-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hungate, R. E. 1969. A roll tube method for cultivation of strict anaerobes. Methods Microbiol. 3B:117-132. [Google Scholar]
- 14.Jannasch, H. W. 1995. Microbial interactions with hydrothermal fluids. Seafloor hyrothermal systems: physical, chemical, biological, and geological interactions. Geophys. Monogr. 91:273-296. [Google Scholar]
- 15.Karl, D. M. 1995. Ecology of free-living hydrothermal vent microbial communities, p. 35-124. In D. M. Karl (ed.), The microbiology of deep-sea hydrothermal vents. CRC Press, New York, N.Y.
- 16.Kashefi, K., and D. R. Lovley. 2000. Reduction of Fe(III), Mn(IV), and toxic metals at 100°C by Pyrobaculum islandicum. Appl. Environ. Microbiol. 66:1050-1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kashefi, K., J. M. Tor, D. E. Holmes, C. Gaw Van Praagh, A.-L. Reysenbach, and D. R. Lovley. Geoglobus ahangari, gen. nov., sp. nov., a novel hyperthermophilic archaeon capable of oxidizing organic acids and growing autotrophically on hydrogen with Fe(III) serving as the sole electron acceptor. Int. J. Syst. Bacteriol., in press. [DOI] [PubMed]
- 18.Lovley, D. R. 2000. Fe(III) and Mn(IV) reduction, p. 3-30. In D. R. Lovley (ed.), Environmental microbe-metal interactions. ASM Press, Washington, D.C.
- 19.Lovley, D. R., S. J. Giovannoni, D. C. White, J. E. Champine, E. J. P. Phillips, Y. A. Gorby, and S. Goodwin. 1993. Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159:336-344. [DOI] [PubMed] [Google Scholar]
- 20.Lovley, D. R., K. Kashefi, M. Vargas, J. M. Tor, and E. L. Blunt-Harris. 2000. Reduction of humic substances and Fe(III) by hyperthermophilic microorganisms. Chem. Geol. 169:289-298. [Google Scholar]
- 21.Lovley, D. R., and E. J. P. Phillips. 1986. Availability of ferric iron for microbial reduction in bottom sediments of the freshwater tidal Potomac river. Appl. Environ. Microbiol. 52:751-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lovley, D. R., and E. J. P. Phillips. 1988. Manganese inhibition of microbial iron reduction in anaerobic sediments. Geomicrobiol. J. 6:145-155. [Google Scholar]
- 23.Lovley, D. R., and E. J. P. Phillips. 1988. Novel mode of microbial energy metabolism: organic carbon oxidation coupled to dissimilatory reduction of iron or manganese. Appl. Environ. Microbiol. 54:1472-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luther, G. W., III, T. F. Rozan, M. Taillefert, D. B. Nuzzio, C. Di Meo, T. M. Shank, and R. A. Lutz. 2001. Chemical speciation drives hyrothermal vent ecology. Nature 410:813-816. [DOI] [PubMed] [Google Scholar]
- 25.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, J. M. Stredwick, G. M. Garrity, B. Li, G. J. Olsen, S. Pramanik, T. M. Schmidt, and J. M. Tiedje. 2000. The RDP (Ribosomal Database Project) continues. Nucleic Acids Res. 28:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maidak, B. L., G. J. Olsen, N. Larsen, R. Overbeek, M. J. McCaughey, and C. R. Woese. 1997. The ribosomal database project. Nucleic Acids Res. 25:109-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, T. L., and M. J. Wolin. 1974. A serum bottle modification of the Hungate technique for cultivating obligate anaerobes. Appl. Microbiol. 27:985-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray, A. E., J. T. Hollinbaugh, and C. Orrego. 1996. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl. Environ. Microbiol. 62:2676-2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Myers, E. W., and W. Miller. 1988. Optimal alignments in linear-space. Comput. Appl. Biol. Sci. 4:11-17. [DOI] [PubMed] [Google Scholar]
- 30.Phillips, E. J. P., and D. R. Lovley. 1987. Determination of Fe(III) and Fe(II) in oxalate extracts of sediment. Soil Sci. Soc. Am. J. 51:938-941. [Google Scholar]
- 31.Reysenbach, A.-L., M. Ehringer, and K. L. Hershberger. 2000. Microbial diversity at 83°C in calcite spring, Yellowstone National Park: another environment where the Aquificales and “Korarchaeota” coexist. Extremophiles 4:61-67. [DOI] [PubMed] [Google Scholar]
- 32.Rooney-Varga, J. N., R. T. Anderson, J. L. Fraga, D. Ringelberg, and D. R. Lovley. 1999. Microbial communities associated with anaerobic benzene mineralization in a petroleum-contaminated aquifer. Appl. Environ. Microbiol. 65:3056-3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skirnisdottir, S., G. O. Hreggvidsson, S. Hjorleifsdottir, and V. T. Marteinsson. 2000. Influence of sulfide and temperature on species composition and community structure of hot spring microbial mats. Appl. Environ. Microbiol. 66:2835-2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sonne-Hansen, J., and B. K. Ahring. 1999. Thermodesulfobacterium hveragerdense sp. nov., and Thermodesulfovibrio islandicus sp. nov., two thermophilic sulfate reducing bacteria isolated from Icelandic hot spring. Syst. Appl. Microbiol. 22:559-564. [DOI] [PubMed] [Google Scholar]
- 35.Swofford, D. L. 1998. PAUP*. Phylogenetic Analysis Using Parsimony (* and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 36.Tor, J. M., K. Kashefi, and D. R. Lovley. 2001. Acetate oxidation coupled to Fe(III) reduction in hyperthermophilic microorganisms. Appl. Environ. Microbiol. 67:1363-1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tor, J. M., and D. R. Lovley. 2001. Anaerobic oxidation of benzoate by hyperthermophilic Archaea. Environ. Microbiol. 3:281-287.11359514 [Google Scholar]
- 38.Vargas, M., K. Kashefi, E. L. Blunt-Harris, and D. R. Lovley. 1998. Microbiological evidence for Fe(III) reduction on early Earth. Nature 395:65-67. [DOI] [PubMed] [Google Scholar]
- 39.Xu, Y., M. A. A. Schoonen, D. K. Nordstrom, K. M. Cunningham, and J. W. Ball. 1998. Sulfur geochemistry of hydrothermal waters in Yellowstone National Park. I. The origin of thiosulfate in hot spring waters. Geochim. Cosmochim. Acta 62:3729-3743. [Google Scholar]
- 40.Zehnder, A. J. B., and W. Stumm. 1988. Geochemistry and biogeochemistry of anaerobic habitats, p. 1-38. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. John Wiley & Sons, New York, N.Y.
- 41.Zeikus, J. G., A. Ben-Bassat, and P. W. Hegge. 1980. Microbiology of methanogenesis in thermal volcanic environments. J. Bacteriol. 143:432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeikus, J. G., M. A. Dawson, T. E. Thompson, K. Ingvorsen, and E. C. Hatchikian. 1983. Microbial ecology of volcanic sulphidogenesis: isolation and characterization of Thermodesulfobacterium commune gen. nov. and sp. nov. J. Gen. Microbiol. 129:1159-1169. [Google Scholar]