Abstract
For industrial applications in animal feed, a phytase of interest must be optimally active in the pH range prevalent in the digestive tract. Therefore, the present investigation describes approaches to rationally engineer the pH activity profiles of Aspergillus fumigatus and consensus phytases. Decreasing the negative surface charge of the A. fumigatus Q27L phytase mutant by glycinamidylation of the surface carboxy groups (of Asp and Glu residues) lowered the pH optimum by ca. 0.5 unit but also resulted in 70 to 75% inactivation of the enzyme. Alternatively, detailed inspection of amino acid sequence alignments and of experimentally determined or homology modeled three-dimensional structures led to the identification of active-site amino acids that were considered to correlate with the activity maxima at low pH of A. niger NRRL 3135 phytase, A. niger pH 2.5 acid phosphatase, and Peniophora lycii phytase. Site-directed mutagenesis confirmed that, in A. fumigatus wild-type phytase, replacement of Gly-277 and Tyr-282 with the corresponding residues of A. niger phytase (Lys and His, respectively) gives rise to a second pH optimum at 2.8 to 3.4. In addition, the K68A single mutation (in both A. fumigatus and consensus phytase backbones), as well as the S140Y D141G double mutation (in A. fumigatus phytase backbones), decreased the pH optima with phytic acid as substrate by 0.5 to 1.0 unit, with either no change or even a slight increase in maximum specific activity. These findings significantly extend our tools for rationally designing an optimal phytase for a given purpose.
Active sites typically contain ionizable groups (Arg, Lys, His, Glu, and Asp) that are involved in substrate or product binding and/or catalysis and that determine the pH activity profile of an enzyme (2). Both for physiological constraints and for industrial applications, it is crucial that an enzyme works properly at the appropriate pH value(s). However, no truly reliable methods for modifying the pH activity profile of an enzyme are yet available. In order to understand more clearly how nature masters catalysis at physiological pH values and to rationally tailor enzymes for industrial use at a predefined pH value, it is important to gain more experimental experience on the pH optimum engineering of enzymes.
Different strategies can be chosen to modify the pH activity profile of an enzyme. (i) The first is the replacement of ionizable groups that are directly involved in substrate or product binding and/or catalysis by nonionizable ones or by amino acids with different charge or pK values (i.e., “A residues” [13, 23]). (ii) The second is the replacement of residues that are in direct contact with A residues by forming hydrogen bonds and/or salt bridges. Substitution of such residues may disturb the hydrogen-bonding network in the active site or alter the electronic environment of A residues (1, 11, 19, 27). The effects on the pH activity profile caused by this type of mutations are particularly difficult to predict. (iii) The third is the alteration of longer-range (indirect) charge-charge interactions by modification of the surface charge of the enzyme. This can be achieved by either (nonselective) chemical modification of surface residues (16, 20, 22, 26) or by selective, site-directed modification of surface charge (10, 11, 17, 18). Making the surface more negatively charged raises the pKa values of ionizable groups (and, thus, the pH optimum) in proteins. Conversely, making the surface more positively charged lowers the pKa of ionizable groups. The latter effects are maximal at low ionic strength and almost vanish at an ionic strength of 1 M (10).
To mention just a few successful examples, Rashid and Siddiqui (16) managed—by chemical modification of surface carboxy groups with either glycinamide or ethylenediamine (in the presence of 1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide)—to decrease the pH optimum of Aspergillus niger β-glucosidase from pH 4.5 to 5.0 to pH 2.5, with only a slight decrease or even an increase in Vmax/Km. In their pioneering work on the serine protease subtilisin, Fersht and coworkers showed that single changes of surface charge at a distance of 12.2 to 24.4 Å from His-64 (acting as a general base in catalysis) change the pKa of the latter by ±(0.11 to 0.40) unit; in addition, these authors showed that the effects of single changes were nearly additive in multiple mutants (10, 17, 18). In Aspergillus awamori glucoamylase, Ser-411 forms a hydrogen bond with Glu-400, the catalytic base. Disruption of this hydrogen bond by site-directed mutagenesis of Ser-411 to either Ala or Cys had only minor effects on kcat but increased the pH optima with maltose or maltoheptaose as substrates by 0.8 to 0.9 unit (1). Catalysis in alcohol dehydrogenase is effected by a proton release system involving a zinc atom, a water molecule, a Ser/Thr residue, and a His residue. Site-directed mutagenesis of the respective His-43 in Bacillus stearothermophilus alcohol dehydrogenase to Arg increased the pH optimum from 7.8 to 9.0 and also increased the specific activity with ethanol as a substrate (19). In the histone acetyltransferase GCN5 from yeast, Glu-173 seems to be an essential catalytic residue, acting as a general base catalyst by deprotonating ɛ-amino groups of histone Lys residues in order to allow direct nucleophilic attack of bound acetyl coenzyme A by these Lys residues. In the E173Q mutant, Gln-173 no longer acts as a base catalyst, thereby shifting the pH activity profile by ca. 1.5 units toward alkaline pH values (23). Finally, in family 11 xylanases, the pH optimum was found to correlate well with the nature of a residue adjacent to the acid or base catalyst of the reaction. In xylanases that function optimally under acidic conditions, this residue is Asp, whereas it is Asn in xylanases that function under more-alkaline conditions. In full agreement with this correlation, but in contrast to electrostatic considerations discussed above, substitution of Asn-35 with Asp decreased the pH optimum of Bacillus circulans xylanase from 5.7 to 4.6 (3).
Phytases (myo-inositol hexakisphosphate phosphohydrolases; EC 3.1.3.8 and 3.1.3.26) are found primarily in microorganisms and plants and catalyze the hydrolysis of the phosphoester bonds of phytic acid (myo-inositol hexakisphosphate), the major storage form of phosphorus in plant seeds, to liberate inorganic phosphate. Since monogastric animals (pigs and poultry) virtually lack phytase activity in their digestive tract, phytic acid phosphorus is metabolically unavailable to these animals, necessitating supplementation of the feed with inorganic phosphate. The latter, however, increases the phosphorus burden in the manure, causing environmental problems by eutrophication of surface waters. These problems can be circumvented by supplementation of the feed with a recombinantly produced phytase that has a pH activity profile ideally suited for maximal activity in the digestive tract of either pigs or poultry. We describe here our attempts to engineer the pH activity profiles of fungal and consensus phytases by both chemical modification and site-directed mutagenesis.
MATERIALS AND METHODS
Source, overproduction, and purification of fungal and consensus phytases.
Wild-type, synthetic (consensus), and mutant phytases (obtained by site-directed mutagenesis) were expressed in either Saccharomyces cerevisiae or Hansenula polymorpha, followed by purification to apparent homogeneity, according to procedures described previously (7, 25).
Chemical modification of Aspergillus fumigatus Q27L phytase.
Glycinamidylation of A. fumigatus Q27L phytase was largely based on the procedure used by Siddiqui et al. (20) for pH optimum engineering of Arthrobacter d-xylose isomerase. To a 1-ml sample of 0.5 mg of A. fumigatus Q27L phytase/ml in 50 mM KH2PO4-K2HPO4 (pH 5.7) containing 1.27 M glycinamide and 10 mM myo-inositol hexakissulfate, three 19-mg aliquots of EDC (1-ethyl-3-[3-dimethylaminopropyl]-carbodiimide) were added at 90-min intervals. After a total incubation time at room temperature of 5.5 h, the sample was dialyzed against 50 mM Tris-HCl (pH 7.5) overnight at 4°C. Subsequently, the sample was split into two equal aliquots, and one aliquot was incubated with 1 M hydroxylamine for 5.25 h at 35°C. The pH activity profile for both preparations (with or without hydroxylamine) was determined as described previously (30).
Other methods.
Homology modeling of fungal phytase structures was based on the crystal structure of A. niger NRRL 3135 phytase (4) and performed according to the method of Tomschy et al. (25). For the determination of pH activity profiles as described previously (30), purified enzymes were diluted in 10 mM sodium acetate (pH 5.0). Incubations at 37°C were started by mixing aliquots of the diluted protein with an equal volume of 1% phytic acid (∼10 mM) in a series of different buffers: 0.4 M glycine, pH 2.5 (HCl); 0.4 M acetate, pH 3.0, 3.5, 4.0, 4.5, 5.0, or 5.5 (NaOH); 0.4 M imidazole, pH 6.0, 6.5, or 7.0 (HCl); and 0.4 M Tris, pH 7.5, 8.0, 8.5, or 9.0 (HCl). Substrate specificity was determined at 37°C and pH 5.0 as described previously (30) with 5 mM concentrations of either phytic acid, p-nitrophenyl phosphate, or one of several other phosphate compounds as a substrate. One unit of phytase (or acid phosphatase, glucose-6-phosphatase, etc.) activity catalyzes the liberation of 1 μmol of inorganic phosphate per min. Protein concentrations were calculated from the optical density at 280 nm by using theoretical absorption values calculated from the known protein sequences with the DNA* Software (DNASTAR, Inc.). One A at 280 nm corresponds to 0.94 mg of A. fumigatus phytase and 0.95 mg of consensus phytase/ml. All amino acid numberings used here correspond to the numbering of mature A. niger phytase (29). Unless stated otherwise, all data presented are given as means ± the standard deviation (SD) of at least three measurements; in many cases, the SDs were smaller than the data symbols.
RESULTS AND DISCUSSION
Chemical modification of A. fumigatus Q27L phytase.
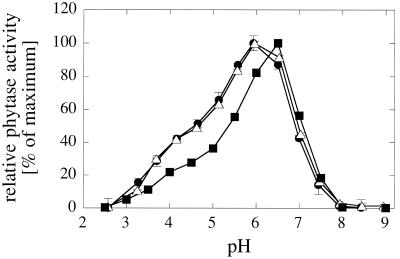
A. fumigatus wild-type phytase displays a series of favorable properties such as, for example, a broad pH optimum (see Fig. 2), broad substrate specificity (30), the capacity to refold properly after heat denaturation (28), and a high potency to increase the bioavailability of phytic acid phosphorus in animal experiments (21). Unfavorably, its specific activity is rather low (26.5 U/mg of protein at pH 5.0 [30]). The Q27L mutation increases the specific activity of A. fumigatus phytase mainly around pH 6.5 (up to 230 U/mg of protein), while the increase is much smaller in the pH range 2 to 5 that seems most relevant for maximal performance of the enzyme in animals (25).
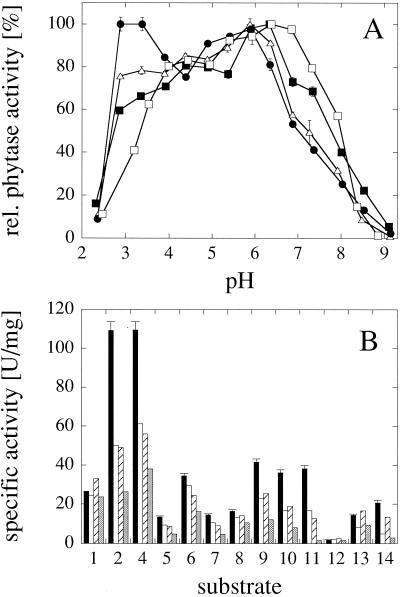
FIG. 2.
pH optimum engineering of A. fumigatus phytase based on a comparison with A. niger NRRL 3135 phytase. (A) pH activity profiles of A. fumigatus wild-type phytase (□), A. fumigatus G277K phytase (▪), A. fumigatus Y282H phytase (▵) and A. fumigatus G277K Y282H phytase (•). (B) Specific activities of A. fumigatus wild-type phytase (black bars), A. fumigatus G277K phytase (white bars), A. fumigatus Y282H phytase (hatched bars), and A. fumigatus A205E phytase (gray bars) at pH 5.0 with 5 mM concentrations of a series of phosphate compounds: 1, phytic acid; 2, p-nitrophenyl phosphate; 3, phenyl phosphate; 4, fructose-1,6-bisphosphate; 5, fructose-6-phosphate; 6, glucose-6-phosphate; 7, ribose-5-phosphate; 8, α-glycerophosphate; 9, β-glycerophosphate; 10, 3-phosphoglycerate; 11, phosphoenolpyruvate; 12, AMP; 13, ADP; 14, ATP. While the data for the wild-type enzyme represent the means ± SD of three measurements, the data for the mutants (B) represent single measurements only.
As a first step toward shifting the pH optimum of A. fumigatus Q27L phytase to lower pH values, the charge of the surface carboxylate groups was neutralized by chemical modification with glycinamide plus EDC. Lowering the negative surface charge should decrease the pKa of ionizable groups and therefore lower the pH optimum of the enzyme. As can be seen in Fig. 1, glycinamidylation of A. fumigatus Q27L phytase in fact decreased the pH optimum from 6.5 to 6.0, independently of whether the chemically modified enzyme was subsequently exposed to hydroxylamine treatment or not. Hydroxylamine was used to regenerate Tyr residues that potentially might also have been modified by glycinamide-EDC (see reference 20). With or without hydroxylamine treatment, and independently of whether the substrate analogue myo-inositol hexakissulfate was included in the reaction mix to protect the active site from modification, glycinamidylation of A. fumigatus Q27L phytase was associated with a 70 to 75% loss of activity (measured at pH 5.0 [results not shown]). Because of this shortcoming, and based on the fact that chemical modification of phytase would not be economically feasible on an industrial scale, we focused our efforts on site-directed mutagenesis approaches to engineer the pH activity profiles of fungal and consensus phytases.
FIG. 1.
Effect of chemical modification on the pH activity profile of A. fumigatus Q27L phytase. The surface carboxy groups of A. fumigatus Q27L phytase were glycinamidylated as described in Materials and Methods. Subsequently, one aliquot of the modified enzyme was treated with hydroxylamine in order to regenerate modified Tyr residues, while the other aliquot was not. Chemical modification, independently of hydroxylamine treatment, caused 70 to 75% inactivation of the enzyme (measured at pH 5.0). Symbols: ▪, nonmodified; •, modified and hydroxylamine treated; ▵, modified but not exposed to hydroxylamine treatment.
General approach.
Over the last few years, we have reported the cloning, sequencing, and biochemical, as well as biophysical, characterization of a series of homologous fungal phytases (12, 14, 15, 28-30). In addition, we have determined the crystal structure of A. niger phytase at a 2.5-Å resolution (4). Inspection of this three-dimensional (3D) structure, comparison with other high-Mr histidine acid phosphatases, and site-directed mutagenesis of residues in the presumed active site (8, 25, 25) yielded strong evidence for a highly positively charged cleft at the interface between the smaller α-domain and the larger α/β-domain of the molecule as the substrate binding (active) site.
The fungal phytases characterized thus far display largely different catalytic properties in terms of specific activity, pH optima, and substrate specificity. Through a comparison of the catalytic properties and amino acid sequences, together with the 3D structures of the enzymes (obtained by homology modelling from the crystal structure of A. niger phytase), we tried to identify divergent residues in the active site of phytase that might correlate with a given property of interest. As a proof of concept, among 12 divergent residues between two wild-type A. niger phytases, only three were found to be located in (residue 274) or close (residues 66 and 269) to the presumed active site. Site-directed mutagenesis confirmed that the R274Q mutation fully accounts for the differences in catalytic properties (e.g., a threefold difference in specific activity at pH 5.0) between these two wild-type A. niger phytases (24). Moreover, comparison of the catalytic properties and the amino acid sequences of A. niger and Aspergillus terreus phytase, together with site-directed mutagenesis, allowed the identification of another residue (residue 27; see also above) that has a strong impact on the specific activity of fungal phytases (25). We now sought to apply the same principle to the pH optimum engineering of fungal and consensus phytases.
In regard to increased activity at acidic pH values, three phytases or acid phosphatases seemed particularly attractive as benchmarks for comparison: A. niger phytase, A. niger pH 2.5 acid phosphatase, and Peniophora lycii phytase. A. niger NRRL 3135 phytase is the only known wild-type phytase with two distinct pH optima at pH 2.5 and pH 5.0 to 5.5, displaying specific activities at the two pH optima of ca. 60 and 100 U/mg of protein, respectively (see reference 30). Peniophora lycii phytase has a pH optimum of 4.0 to 4.5, at which it displays an unusually high specific activity of ∼2,000 U/mg of protein (6). Finally, A. niger pH 2.5 acid phosphatase—although displaying (much) higher activity with classical acid phosphatase substrates such as phenyl phosphate or p-nitrophenyl phosphate and despite almost no phytase activity at pH 5.0—is homologous in amino acid sequence to the fungal phytases and displays considerable phytase activity at pH 2.5 (77 U/mg of protein [30]).
pH optimum engineering of A. fumigatus phytase based on a comparison with A. niger NRRL 3135 phytase.
A. fumigatus phytase has a rather broad pH optimum, displaying ≥80% of the maximal specific activity between pH 4.0 and 7.3 (Fig. 2). On the other hand, A. niger NRRL 3135 phytase has two pH optima at pH 2.5 and pH 5.0 to 5.5, at which it displays considerably higher specific activity than A. fumigatus phytase (60 to 100 versus 26.5 U/mg of protein [30]). Inspection of the phytase amino acid sequence alignment (Fig. 3) and of the 3D structure (4) led to the hypothesis that Lys-277, His-282, and/or Glu-205 may be responsible for the second pH optimum of A. niger NRRL 3135 phytase at pH 2.5. In fact, the G277K and Y282H mutations both increased the activity of A. fumigatus phytase at pH 2.8 to 3.4, and their effect was nearly additive (Fig. 2A). On the other hand, both mutations decreased the relative activity at pH values of >6.3. Below pH 2.8, the activity decreased precipitously, which may be due to acid-induced denaturation of the protein (not shown). Compared to G277K and Y282H, the A205E mutation—either alone or when combined with the former two mutations—had only a minor influence on the pH activity profile of A. fumigatus phytase (D. Kostrewa, L. Pasamontes, A. Tomschy, A. van Loon, K. Vogel, and M. Wyss, [European Patent Application 0-897-010]).
FIG. 3.
Amino acid sequence alignment of fungal phytases or acid phosphatases around the mutated residues (indicated in boldface). Afum, A. fumigatus ATCC 13073 phytase; Anid, A. nidulans phytase; Ater, A. terreus 9A1 phytase; Anig, A. niger NRRL 3135 phytase; Mthe, Myceliophthora thermophila phytase; Plyc, Peniophora lycii phytase; Agro, Agrocybe pediades phytase; A2.5, A. niger pH 2.5 acid phosphatase; con1, consensus phytase-1. The amino acids thought to correlate with the low-pH phytase activity of the respective proteins and tested for pH optimum engineering of A. fumigatus and consensus phytases are boxed.
Although the G277K and Y282H mutations gave rise to a second pH optimum for A. fumigatus phytase in the acidic range, all three mutations—G277K, Y282H, and A205E—had only a minor influence, if any, on the specific phytase activity at pH 5.0 (Fig. 2B), thus showing that these residues do not account for the considerably higher specific activity of A. niger NRRL 3135 phytase. Much to the contrary, all three mutations, when analyzed separately, considerably decreased the activity of A. fumigatus phytase with p-nitrophenyl phosphate or fructose-1,6-bisphosphate as a substrate and also affected the specific activities with other phosphate compounds. When the individual mutations were combined (A205E + G277K, G277K + Y282H, and A205E + G277K + Y282H), the specific activities with individual phosphate compounds remained on the level of the single mutants or were even further decreased (Kostrewa et al., Patent Application).
pH optimum engineering of A. fumigatus and consensus phytases based on a comparison with Peniophora lycii phytase.
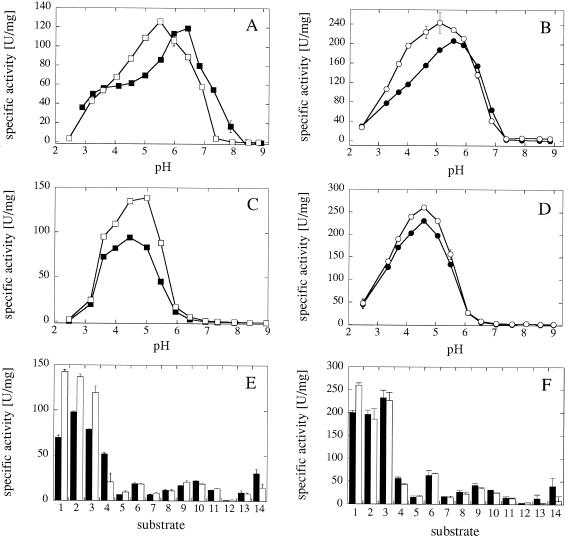
Peniophora lycii phytase has a relatively low pH optimum of 4.0 to 4.5, at which it displays an unusually high specific activity of ∼2,000 U/mg of protein (6). Inspection of the amino acid sequence and of the homology-modeled 3D structure suggested that Ala-68 (Lys in A. fumigatus phytase) of Peniophora lycii phytase might be a residue contributing to these particular catalytic properties. Remarkably, A. niger pH 2.5 acid phosphatase, also displaying phytase activity at low pH, has an Ala at this position as well (Fig. 3). The K68A mutation in fact decreased the pH optimum of the A. fumigatus Q27T variant (which displays a considerably higher specific activity than the A. fumigatus wild-type enzyme [Fig. 2 and 4] [see reference 25]) with phytic acid as a substrate by roughly 1 pH unit (Fig. 4A) while having a relatively minor effect on the pH optimum with p-nitrophenyl phosphate as substrate (Fig. 4C). A similar shift in pH optimum with the K68A mutation and with phytic acid as a substrate was also seen for the A. fumigatus Q27I variant and for several consensus phytases (consensus phytase-1 Q27T, consensus phytase-1 thermo[8] Q27T, and consensus phytase-10 thermo[3] Q27T; Fig. 4B and data not shown; for a detailed description of the different consensus phytases, see references 7 and 9 and M. Lehmann, C. Loch, A. Middendorf, D. Studer, S. F. Lassen, L. Pasamontes, A. P. G. M. van Loon, and M. Wyss, unpublished data). The consensus phytases are based on amino acid sequence alignments of several homologous ascomycete or ascomycete plus basidiomycete phytases where, at every position of the alignment, the “consensus amino acid” was calculated by an appropriate computer program. Surprisingly, these consensus phytases revealed considerably (by at least 16°C) increased intrinsic thermostability relative to all parent phytases.
FIG. 4.
pH optimum engineering of A. fumigatus and consensus phytase based on a comparison with Peniophora lycii phytase. (A and B) pH activity profiles with phytic acid as substrate; (C and D) pH activity profiles with p-nitrophenyl phosphate as substrate; (E and F) specific activities at pH 5.0 with 5 mM concentrations of a series of phosphate compounds (see legend to Fig. 2). (A, C, and E) A. fumigatus Q27T phytase (▪) and A. fumigatus Q27T K68A phytase (□); (B, D, and F) consensus phytase-10 thermo[3] Q27T (•) and consensus phytase-10 thermo[3] Q27T K68A (○).
In regard to substrate specificity (Fig. 4E and F), the K68A mutation primarily affected the specific activities with the bulkiest and most negatively charged substrates (phytic acid and ATP). Only in selected variants (e.g., A. fumigatus Q27T phytase; Fig. 4E) were the specific activities with p-nitrophenyl phosphate, phenyl phosphate, and fructose-1,6-bisphosphate also affected to some extent.
pH optimum engineering of A. fumigatus phytase based on a comparison with A. niger pH 2.5 acid phosphatase.
A. niger pH 2.5 acid phosphatase, although it has almost no phytase activity at pH 5.0, displays considerable phytase activity at pH 2.5 (77 U/mg of protein). Since residue 27 was shown previously to have a significant effect on the specific activity of A. fumigatus and consensus phytases (25; Lehmann, unpublished), this and the neighboring residue (i.e., residues 27 and 28) of A. fumigatus phytase were replaced by the respective A. niger pH 2.5 acid phosphatase residues (Gly and Asn, respectively; Fig. 3). In addition, inspection of the amino acid sequence alignment and of the crystal structure of A. niger pH 2.5 acid phosphatase (5) suggested that Tyr-140 and Gly-141 of the latter might correlate with its phytase activity at low pH.
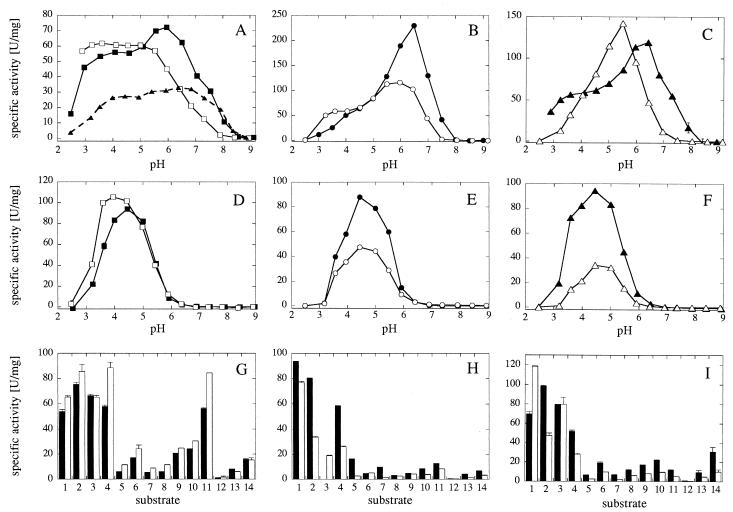
As can be seen in Fig. 5A, the Q27G mutation caused a general twofold increase in specific activity of A. fumigatus phytase, as well as a shift of the entire pH activity profile by 0.5 unit toward more acidic pH values (25). In the A. fumigatus Q27G background, the Y28N mutation eliminated the local phytase activity peak at ca. pH 6, while at a pH <5.0 the mutation may have increased the specific activity slightly (Fig. 5A). In the A. fumigatus Q27L background, the Y28N mutation also decreased the specific phytase activity at a pH of >5.0 and caused an increase in specific activity between pH 3.2 and 3.6 (Fig. 5B). In contrast, the Y28N mutation had relatively minor effects on the pH optima with p-nitrophenyl phosphate as a substrate (Fig. 5D and E). With regard to substrate specificity (Fig. 5G and H), the high specific activity of the Q27G and Q27G Y28N variants with phosphoenolpyruvate is surprising and therefore particularly interesting. A satisfactory explanation for this finding has not yet been found.
FIG. 5.
pH optimum engineering of A. fumigatus phytase variants based on a comparison with A. niger pH 2.5 acid phosphatase. (A to C) pH activity profiles with phytic acid as substrate; (D to F) pH activity profiles with p-nitrophenyl phosphate as substrate; (G to I) specific activities with 5 mM concentrations of a series of phosphate compounds (see legend to Fig. 2). (A, D, and G) A. fumigatus Q27G phytase (▪), A. fumigatus Q27G Y28N phytase (□), and A. fumigatus wild-type phytase (▴); (B, E, and H) A. fumigatus Q27L phytase (•) and A. fumigatus Q27L Y28N phytase (○); (C, F, and I) A. fumigatus Q27T phytase (▴) and A. fumigatus Q27T S140Y D141G phytase (▵). In panel H, the specific activity of A. fumigatus Q27L phytase with phenyl phosphate (substrate 3) has not been determined.
As can be seen in Fig. 5C, the S140Y D141G double mutation decreased the pH optimum of A. fumigatus Q27T phytase by roughly 1 pH unit and slightly increased the maximum specific activity. In contrast, this double mutation had no effect on the pH optimum, but considerably decreased the specific activity with p-nitrophenyl phosphate as a substrate (Fig. 5F). Very similar effects for the S140Y D141G double mutation were also observed when introduced into the A. fumigatus Q27T S66D and A. fumigatus Q27L phytase variants (not shown).
Conclusions.
A. niger NRRL 3135 phytase, A. niger pH 2.5 acid phosphatase, and Peniophora lycii phytase all have pH optima for phytase activity in the low-pH range (2.5 to 4.5). Inspection of their experimentally determined or homology-modeled 3D structures, together with a detailed comparison of their amino acid sequences with those of homologous fungal phytases, allowed the identification of active-site residues that (might) correlate with high phytase activity at low pH. Introduction of these favorable residues into the backbones of either A. fumigatus phytase, consensus phytase, or variants thereof—all with an activity optimum at higher pH values—confirmed that the Q27G, Y28N, K68A, S140Y D141G, G277K, and Y282H mutations (i) increase the specific phytase activity at a pH of <4.0, (ii) give rise to an additional (second) pH optimum at pH 2.8 to 3.4, or (iii) decrease the pH optimum by 0.5 to 1.0 unit with either no decrease or even a slight increase in maximal specific phytase activity.
Some of the mutations (e.g., Y28N and K68A) were shown to exert very similar effects on the pH activity profiles when introduced into different backbones (e.g., in either A. fumigatus or consensus phytase showing 80% amino acid sequence identity). Therefore, these mutations may be generally applicable for the pH optimum engineering of fungal, consensus, or even more distantly related phytases and acid phosphatases. It remains to be established, however, whether the effects of these mutations, when combined, are additive or not. In addition, we miss detailed mechanistic explanations as to how these mutations affect substrate or product binding and/or catalysis to bring about the observed changes in pH optima and substrate specificity. Given that phytic acid is highly negatively charged at a physiological pH, that a considerable number of positively charged amino acids are located in or close to the active site of the phytase molecule, and that many titratable groups might therefore contribute to the shape of the pH activity profile, this seems to be a very complicated and time-consuming task. This is even more so the case since our knowledge on the rate-limiting step(s) of the reaction is very limited. Very recently, we obtained preliminary evidence that, in A. niger T213 and A. fumigatus wild-type phytase, product (myo-inositol pentakisphosphate) release may be the rate-limiting step of the reaction and that product release (and thus specific activity) can be increased significantly by site-directed mutagenesis of Arg-274 to Gln (24) and of Gln-27 to Leu, Thr, or Ile, respectively (25). Clearly, these investigations need to be extended to other (fungal and consensus) phytases.
Acknowledgments
We thank Roland Rémy and Nicole van Loon for technical assistance and Claus C. Fuglsang for valuable discussions.
REFERENCES
- 1.Fang, T.-Y., and C. Ford. 1998. Protein engineering of Aspergillus awamori glucoamylase to increase its pH optimum. Protein Eng. 11:383-388. [DOI] [PubMed] [Google Scholar]
- 2.Fersht, A. 1985. Enzyme structure and mechanism, 2nd ed. W. H. Freeman and Company, New York, N.Y.
- 3.Joshi, M. D., G. Sidhu, I. Pot, G. D. Brayer, S. G. Withers, and L. P. McIntosh. 2000. Hydrogen bonding and catalysis: a novel explanation for how a single amino acid substitution can change the pH optimum of a glycosidase. J. Mol. Biol. 299:255-279. [DOI] [PubMed] [Google Scholar]
- 4.Kostrewa, D., F. Grüninger-Leitch, A. D'Arcy, C. Broger, D. Mitchell, and A. P. G. M. van Loon. 1997. Crystal structure of phytase from Aspergillus ficuum at 2.5 Å resolution. Nat. Struct. Biol. 4:185-190. [DOI] [PubMed] [Google Scholar]
- 5.Kostrewa, D., M. Wyss, A. D'Arcy, and A. P. G. M. van Loon. 1999. Crystal structure of Aspergillus niger pH 2.5 acid phosphatase at 2.4 Å resolution. J. Mol. Biol. 288:965-974. [DOI] [PubMed] [Google Scholar]
- 6.Lassen, S. F., J. Breinholt, P. R. Ostergaard, R. Brugger, A. Bischoff, M. Wyss, and C. C. Fuglsang. 2001. Expression, gene cloning, and characterization of five novel phytases from four basidiomycete fungi: Peniophora lycii, Agrocybe pediades, a Ceriporia sp., and Trametes pubescens. Appl. Environ. Microbiol. 67:4701-4707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lehmann, M., D. Kostrewa, M. Wyss, R. Brugger, A. D'Arcy, L. Pasamontes, and A. P. G. M. van Loon. 2000. From DNA sequence to improved functionality: using protein sequence comparisons to rapidly design a thermostable consensus phytase. Protein Eng. 13:49-57. [DOI] [PubMed] [Google Scholar]
- 8.Lehmann, M., R. Lopez-Ulibarri, C. Loch, C. Viarouge, M. Wyss, and A. P. G. M. van Loon. 2000. Exchanging the active site between phytases for altering the functional properties of the enzyme. Protein Sci. 9:1866-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehmann, M., L. Pasamontes, S. F. Lassen, and M. Wyss. 2000. The consensus concept for thermostability engineering of proteins. Biochim. Biophys. Acta 1543:408-415. [DOI] [PubMed] [Google Scholar]
- 10.Loewenthal, R., J. Sancho, T. Reinikainen, and A. R. Fersht. 1993. Long-range surface charge-charge interactions in proteins. Comparison of experimental results with calculations from a theoretical method. J. Mol. Biol. 232:574-583. [DOI] [PubMed] [Google Scholar]
- 11.Mantafounis, D., and J. Pitts. 1990. Protein engineering of chymosin: modification of the optimum pH of enzyme catalysis. Protein Eng. 3:605-609. [DOI] [PubMed] [Google Scholar]
- 12.Mitchell, D. B., K. Vogel, B. J. Weimann, L. Pasamontes, and A. P. G. M. van Loon. 1997. The phytase subfamily of histidine acid phosphatases: isolation of genes for two novel phytases from the fungi Aspergillus terreus and Myceliophthora thermophila. Microbiology 143:245-252. [DOI] [PubMed] [Google Scholar]
- 13.Myers, M. A., M. J. Healy, and J. G. Oakeshott. 1993. Effects of the residue adjacent to the reactive serine on the substrate interactions of Drosophila esterase 6. Biochem. Genet. 31:259-278. [PubMed] [Google Scholar]
- 14.Pasamontes, L., M. Haiker, M. Wyss, M. Tessier, and A. P. G. M. van Loon. 1997. Gene cloning, purification, and characterization of a heat-stable phytase from the fungus Aspergillus fumigatus. Appl. Environ. Microbiol. 63:1696-1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasamontes, L., M. Haiker, M. Henriquez-Huecas, D. B. Mitchell, and A. P. G. M. van Loon. 1997. Cloning of the phytases from Emericella nidulans and the thermophilic fungus Talaromyces thermophilus. Biochim. Biophys. Acta 1353:217-223. [DOI] [PubMed] [Google Scholar]
- 16.Rashid, M. H., and K. S. Siddiqui. 1998. Carboxy-group modification: high-temperature activation of charge-neutralized and charge-reversed β-glucosidases from Aspergillus niger. Biotechnol. Appl. Biochem. 27:231-237. [PubMed] [Google Scholar]
- 17.Russell, A. J., and A. R. Fersht. 1987. Rational modification of enzyme catalysis by engineering surface charge. Nature 328:496-500. [DOI] [PubMed] [Google Scholar]
- 18.Russell, A. J., P. G. Thomas, and A. R. Fersht. 1987. Electrostatic effects on modification of charged groups in the active site cleft of subtilisin by protein engineering. J. Mol. Biol. 193:803-813. [DOI] [PubMed] [Google Scholar]
- 19.Sakoda, H., and T. Imanaka. 1992. Cloning and sequencing of the gene coding for alcohol dehydrogenase of Bacillus stearothermophilus and rational shift of the optimum pH. J. Bacteriol. 174:1397-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddiqui, K. S., T. Loviny-Anderton, M. Rangarajan, and B. S. Hartley. 1993. Arthrobacter d-xylose isomerase: chemical modification of carboxy groups and protein engineering of pH optimum. Biochem. J. 296:685-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simões Nunes, C., and P. Guggenbuhl. 1998. Comparative effects of Aspergillus fumigatus and A. niger phytases on phosphorus and calcium digestibilities and phosphorus faecal excretion in the growing pig. J. Anim. Feed Sci. 7:177-180. [Google Scholar]
- 22.Spomer, W. E., and J. F. Wootton. 1971. The hydrolysis of α-N-benzoyl-l-argininamide catalyzed by trypsin and acetyltrypsin. Dependence on pH. Biochim. Biophys. Acta 235:164-171. [DOI] [PubMed] [Google Scholar]
- 23.Tanner, K. G., R. C. Trievel, M.-H. Kuo, R. M. Howard, S. L. Berger, C. D. Allis, R. Marmorstein, and J. M. Denu. 1999. Catalytic mechanism and function of invariant glutamic acid 173 from the histone acetyltransferase GCN5 transcriptional coactivator. J. Biol. Chem. 274:18157-18160. [DOI] [PubMed] [Google Scholar]
- 24.Tomschy, A., M. Wyss, D. Kostrewa, K. Vogel, M. Tessier, S. Höfer, H. Bürgin, A. Kronenberger, R. Rémy, A. P. G. M. van Loon, and L. Pasamontes. 2000. Active site residue 297 of Aspergillus niger phytase critically affects the catalytic properties. FEBS Lett. 472:169-172. [DOI] [PubMed] [Google Scholar]
- 25.Tomschy, A., M. Tessier, M. Wyss, R. Brugger, C. Broger, L. Schnoebelen, A. P. G. M. van Loon, and L. Pasamontes. 2000. Optimization of the catalytic properties of Aspergillus fumigatus phytase based on the three-dimensional structure. Protein Sci. 9:1304-1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valenzuela, P., and M. L. Bender. 1971. Kinetic properties of succinylated and ethylenediamine-amidated δ-chymotrypsins. Biochim. Biophys. Acta 250:538-548. [DOI] [PubMed] [Google Scholar]
- 27.Wind, R. D., J. C. M. Uitdehaag, R. M. Buitelaar, B. W. Dijkstra, and L. Dijkhuizen. 1998. Engineering of cyclodextrin product specificity and pH optima of the thermostable cyclodextrin glycosyltransferase from Thermoanaerobacterium thermosulfurigenes EM1. J. Biol. Chem. 273:5771-5779. [DOI] [PubMed] [Google Scholar]
- 28.Wyss, M., L. Pasamontes, R. Rémy, J. Kohler, E. Kusznir, M. Gadient, F. Müller, and A. P. G. M. van Loon. 1998. Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger pH 2.5 acid phosphatase. Appl. Environ. Microbiol. 64:4446-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wyss, M., L. Pasamontes, A. Friedlein, R. Rémy, M. Tessier, A. Kronenberger, A. Middendorf, M. Lehmann, L. Schnoebelen, U. Röthlisberger, E. Kusznir, G. Wahl, F. Müller, H.-W. Lahm, K. Vogel, and A. P. G. M. van Loon. 1999. Biophysical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): molecular size, glycosylation pattern, and engineering of proteolytic resistance. Appl. Environ. Microbiol. 65:359-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wyss, M., R. Brugger, A. Kronenberger, R. Rémy, R. Fimbel, G. Oesterhelt, M. Lehmann, and A. P. G. M. van Loon. 1999. Biochemical characterization of fungal phytases (myo-inositol hexakisphosphate phosphohydrolases): catalytic properties. Appl. Environ. Microbiol. 65:367-373. [DOI] [PMC free article] [PubMed] [Google Scholar]