Abstract
A strain of Geobacillus caldoxylosilyticus from central heating system water could utilize a number of organophosphonates as the sole phosphorus source for growth at 60°C. During growth on glyphosate, aminomethylphosphonate release to the medium was observed, and in cell extracts, a glyphosate oxidoreductase-type activity, producing stoichiometric amounts of aminomethylphosphonate and glyoxylate from glyphosate, was detectable.
Organophosphonates, characterized by the presence of a stable, covalent carbon-to-phosphorus (C—P) bond, are of widespread occurrence in the environment. Natural and synthetic organophosphonates are of importance, with the latter being utilized extensively in the chemical industry (26). By far the most important use of synthetic organophosphonates, however, is as herbicides, with glyphosate (16), the world's leading agrochemical, worth in excess of $1 billion per year to its manufacturer, Monsanto Company, St. Louis, Mo.
The organophosphonate C—P bond may be cleaved by a range of enzymes, including C—P lyase (27) and various hydrolases (24, 26). Additionally, the C—P bond of phosphonopyruvate can be intramolecularly rearranged to form a phosphate ester, phosphoenolpyruvate, by the action of the enzyme phosphoenolpyruvate phosphomutase (17). However, C—P bond cleavage is not the only route by which organophosphonate biodegradation may proceed with both transaminases (22) and oxidoreductases (4), acting on parts of organophosphonate molecules other than the C—P bond; indeed, microorganisms that degrade organophosphonates without C—P bond cleavage have been described in recent years (15, 23).
Organophosphonate metabolism has traditionally been studied in greatest detail within soil and soil microorganisms, largely due to scientific interest in the environmental fate of the herbicide glyphosate. Few attempts have been made to investigate the biodiversity of microorganisms capable of degrading organophosphonates (26), and studies have concentrated mainly on gram-negative, mesophilic bacteria, although recent work has attempted to redress this imbalance (9, 14, 19). While the isolation, biochemical characterization, and taxonomic description of thermophilic microbial strains have proceeded apace in recent years (18), biodegradation studies with such microorganisms are relatively scarce, and studies with organophosphonates are nonexistent. Here, we report for the first time the ability of a thermophilic bacterium to cleave the C—P bond of a number of organophosphonates and demonstrate a thermotolerant glyphosate oxidoreductase activity in cell extracts of the same.
The thermus isolation medium of Atlas (2) was prepared and solidified with 1.5% purified agar (Oxoid, Basingstoke, United Kingdom). Samples of domestic central heating system water were serially diluted in sterile 0.9% NaCl and plated (100 μl) at 60°C. Following incubation for 12 h, a number of morphologically distinct colonies were picked and screened at 60°C for organophosphonate utilization in the liquid medium of Ternan et al. (25), modified to contain the trace element solution of Atlas (2), with a range of magnesia-treated (28) organophosphonates supplied as the sole P, N, or C source at final concentrations of 1.0, 5.0, and 10.0 mM, respectively. Microbial growth (50-ml cultures in 250-ml Erlenmeyer flasks at 150 rpm in a Stuart Scientific Co. [Staffordshire, United Kingdom] SI 150 orbital incubator) was monitored by the increase in the optical density of the culture at 650 nm, and phosphate release to the culture supernatants was determined by the method of Fiske and SubbaRow (7). Only one isolate, a gram-positive rod designated T20, could grow on a range of organophosphonates as the sole P source (Table 1) and was therefore chosen for further study.
TABLE 1.
Range of organophosphonate substrates utilized by G. caldoxylosilyticus T20 as the sole phosphorus or nitrogen source
| Organophosphonate substrate | Growth (μg of protein ml−1) on substrate asa:
|
|
|---|---|---|
| Sole P source (1 mM) | Sole N source (5 mM) | |
| Positive control | 315 | 300 |
| Negative control | 30 | 20 |
| Methylphosphonate | 30 | − |
| Ethylphosphonate | 40 | − |
| Phenylphosphonate | 45 | − |
| Aminomethylphosphonate | 25 | 20 |
| 2-Aminoethylphosphonate | 45 | 25 |
| 2-Amino-3-phosphono propionate | 75 | 20 |
| Phosphomycin | 20 | − |
| Phosphono formate | 260 | − |
| Phosphono acetate | 50 | − |
| N-[Phosphonomethyl]-glycine (glyphosate) | 310 | 20 |
| Phosphonomethyl-iminodiacetate | 320 | 20 |
| 2-Phosphonopropionate | 60 | − |
| 3-Phosphonopropionate | 40 | 15 |
| 2-Amino-4-phosphono butyrate | 55 | 20 |
| 2-Phosphono butyrate | 110 | − |
| 4-Phosphono butyrate | 40 | − |
Results were scored negative if the protein yield, as measured by the method of Ahmad et al. (1), was less than 20% of that of the positive control containing 1 mM inorganic phosphate. Results are the mean of duplicates, which on no occasion varied by more than 5%.
Isolate T20 was identified by 16S ribosomal DNA (rDNA) sequencing (National Collections of Industrial, Food, and Marine Bacteria, Ltd., Aberdeen, United Kingdom) as having the trivial name “Bacillus caldoxylolyticus.” Recently there has been some controversy over the classification of this isolate, with Ahmad et al. (1) initially assigning it as Saccharococcus caldoxylosilyticus sp. nov. Subsequently, Nazina et al. (18) and Fortina et al. (7a) assigned it to the genus Geobacillus, along with all of the other thermophilic species previously assigned to the genus Bacillus. For the purpose of this study, we have described the isolate as Geobacillus caldoxylosilyticus T20.
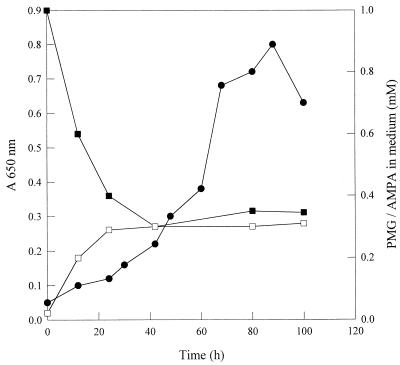
G. caldoxylosilyticus T20 formed spores at temperatures below 50°C, grew most quickly at 60°C, and did not grow at temperatures above 70°C, observations consistent with other members of the Geobacillus genus (18). No phosphate release to the culture medium occurred during growth of G. caldoxylosilyticus T20 on organophosphonates as the sole P source. However, release to the culture supernatant of 0.3 mM aminomethylphosphonate (AMPA), equivalent to some 30% of the substrate phosphorus supplied, was observed during growth on glyphosate (Fig. 1). Release of AMPA, detected as the tosylated derivative (11), occurred concomitantly with culture growth and removal of glyphosate (0.7 mM), suggesting that the microorganism was utilizing some 0.4 mM glyphosate-derived-phosphorus for growth under these conditions. The production of AMPA was confirmed by spiking high-performance liquid chromatography (HPLC) samples with authentic AMPA (Sigma-Aldrich Chemical Co., Poole, United Kingdom) and also by carrying out 1H- and 31P-nuclear magnetic resonance analyses of culture supernatant samples concentrated 20-fold as previously described (24). Spectra were recorded at room temperature in D2O on a Bruker DRX spectrometer (Karlsruhe, Germany) operating at 300.13 MHz for 1H and 121.50 MHz for 31P. No AMPA production was observed in uninoculated control flasks, nor did decomposition of the organophosphonates used occur at 60°C, as measured by HPLC and inorganic phosphate determination.
FIG. 1.
Growth of G. caldoxylosilyticus T20 on glyphosate (1.0 mM) as the sole phosphorus source in defined thermophile medium with an (NH4)2SO4 nitrogen source (2.6 g liter−1) and a glucose-glycerol-succinate carbon source (3 g liter−1 each). •, A650; ▪, glyphosate; □, aminomethylphosphonate. PMG, glyphosate.
Organophosphonate biodegradation by a thermophilic bacterium has not previously been reported. The present study proves that an obligately thermophilic microorganism can cleave C—P bonds and utilize a number of organophosphonates for growth. Notably, the strain did not utilize either 2-aminoethylphosphonate or 2-amino-3-phosphonopropionate, two natural organophosphonates, the biodegradation of which is facilitated by a large majority of environmental isolates (26). Because G. caldoxylosilyticus T20 is a thermophile, it is unsurprising that the range of organophosphonates utilized is different from those of previously studied mesophilic microorganisms. As previously reported for many isolates (26), however, phosphate starvation was required for organophosphonate biodegradation. G. caldoxylosilyticus T20 was unable to utilize AMPA as the sole P source, suggesting that while AMPA produced intercellularly from glyphosate may be metabolized, the strain is incapable of transporting and utilizing exogenously supplied AMPA. This observation may be explained if organophosphonate biodegradation in this microbe is controlled at the level of the transporter (12), rather than by the specificity of C—P bond cleavage enzymes.
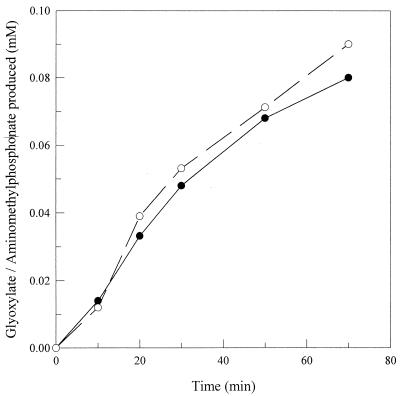
G. caldoxylosilyticus T20 was grown on glyphosate as the sole P source, cell extracts were prepared by sonication, and when assayed for glyphosate oxidoreductase (GOX) by the method of Barry and Kishore (4) at 50°C, the level of release of dinitrophenylhydrazine (DNPH)-reactive material (corresponding to an activity of 0.6 nmol min−1 mg−1) was above that in control experiments lacking glyphosate substrate or cell extract. This DNPH-reactive material was confirmed as glyoxylate by the HPLC method of Qureshi et al. (21) following extraction with ethyl acetate. AMPA production and glyoxylate release were stoichiometric and linear with time up to 60 min (Fig. 2). Unlike the GOX activity described by Barry and Kishore (4), which occurred at 30°C, no activity was detectable in T20 cell extracts at less than 50°C, which is unsurprising, because the source organism does not grow below this temperature. However, the level of activity in isolate T20 is comparable to that reportedly obtained in cell lysates of a number of microorganisms obtained from a glyphosate waste treatment plant described in U.S. patent no. 5776760 (4)
FIG. 2.
Release of equimolar amounts of aminomethylphosphonate (•) and glyoxylate (○) from glyphosate with time by cell extract from G. caldoxylosilyticus T20. The glyphosate oxidoreductase assay contained 2.0 mg of cell extract protein ml−1; samples were removed at various times, and the values reported are the means of duplicate determinations.
In order to assess the similarity of G. caldoxylosilyticus GOX to published sequences, specific oligonucleotide primers targeting an internal region of the sequence of the wild-type GOX gene (4) from isolate LBAA (sequence ID no. 3) were designed with the aid of the Oligo Primer Analysis Software (Oligo version 5; NBI). The designations and sequences of the forward and reverse primers were as follows: 18-mer NGT1 (5′-CCTTGATTGACCCGAACC-3′) and 18-mer NG2 (5′-CGATAAAACGCCGAAACA-3′), which correspond, respectively, to positions 214 to 232 on the coding strand and positions 745 to 763 on the negative strand, generating a 552-bp PCR amplicon. In addition, primers specific for claimed conserved flanking regions upstream and downstream of the wild-type GOX gene from isolate LBAA (sequence ID no. 13 and 14) (4) were used, from which an approximately 1.3-kbp PCR amplicon was expected. DNA was extracted with an Anachem (Bedfordshire, United Kingdom) FastDNA SPIN kit for soil as per the manufacturer's instructions. A 1-μl volume of the extracted DNA was amplified by PCR in a 100-μl reaction carried out on a Biometra T gradient thermocycler for 30 cycles with the following programmed profile: initial denaturation for 1 min at 94°C and 30 cycles of amplification (annealing for 1 min at 60°C, extension for 3 min at 72°C, and denaturation at 94°C). A final extension for 10 min at 72°C was carried out, and PCR products were visualized under UV light following electrophoresis on 1% polyacrylamide gel stained with ethidium bromide.
No PCR products were detected with either set of primers, even when a range of lower annealing temperatures, allowing a degree of mismatch between the primers and the target DNA, were used. DNA extracted from a mixed culture (ATCC 55050) derived from a glyphosate waste treatment stream and deposited with American Type Culture Collection by Monsanto was used as a control and gave PCR-positive bands when probed. This suggests that the putative G. caldoxylosilyticus T20 GOX gene sequence is different from that described in the literature, which is not surprising, given the site of isolation of T20 and the evolutionary unrelatedness of bacterial strains from which a GOX gene has been isolated.
The biodegradation of the herbicide glyphosate via the AMPA pathway by a thermophilic microorganism has not been reported before now. While conversion of glyphosate to AMPA is the accepted mechanism for detoxification of this herbicide in soil (15), no microorganism that conclusively exhibits this capability has been isolated from soil (6). To date, our understanding of this phenomenon is based almost exclusively upon work carried out by Monsanto on microbes within a glyphosate waste treatment plant, which also metabolize the herbicide via this pathway (3, 4, 5, 8, 10, 13). The present study shows for the first time the conclusive production of AMPA from glyphosate both in vivo and in vitro by a microorganism not obtained from an industrial source.
It has previously been shown that Arthrobacter atrocyaneus ATCC 13752 could degrade glyphosate via an AMPA intermediate (20), despite this microorganism being deposited in culture collection prior to the invention of glyphosate. This suggests that the enzyme or enzymes responsible for glyphosate biodegradation via the AMPA pathway have a different, natural substrate rather than having evolved to facilitate glyphosate biodegradation since the introduction of the chemical and its widespread use. As with A. atrocyaneus ATCC 13752, isolate T20 was isolated from a source unlikely to have been exposed to glyphosate. It would appear therefore that the ability to degrade glyphosate to AMPA is present in a range of genetically diverse bacteria. Future studies will examine whether this is the result of one or many different enzymes.
REFERENCES
- 1.Ahmad, S., R. K. Scopes, G. Rees, and B. K. C. Patel. 2000. Saccharococcus caldoxylosilyticus sp. nov., an obligately thermophilic, xylose utilizing, endospore forming bacterium. Int. J. Syst. Evol. Microbiol. 50:517-523. [DOI] [PubMed] [Google Scholar]
- 2.Atlas, R. M. 1996. Thermus agar, p. 1392. In L. C. Parks (ed.), Handbook of microbiological media, 2nd ed. Life Science Microbiology, CRC Press, Boca Raton, Fla.
- 3.Balthazor, T. M., and L. E. Hallas. 1986. Glyphosate-degrading microorganisms from industrial activated sludge. Appl. Environ. Microbiol. 51:432-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barry, G. F., and G. M. Kishore. 1998. Glyphosate tolerant plants. U.S. patent no. 5776760.
- 5.Carson, D. B., M. A. Heitkamp, and L. E. Hallas. 1997. Biodegradation of N-phosphonomethyliminodiacetic acid by microorganisms from industrial activated sludge. Can. J. Microbiol. 43:97-101. [DOI] [PubMed] [Google Scholar]
- 6.Dick, R. E., and J. P. Quinn. 1995. Glyphosate-degrading isolates from environmental samples: occurrence and pathways of degradation. Appl. Microbiol. Biotechnol. 43:545-550. [DOI] [PubMed] [Google Scholar]
- 7.Fiske, C. H., and Y. SubbaRow. 1925. The colourimetric determination of phosphorus. J. Biol. Chem. 66:375-400. [Google Scholar]
- 7a.Fortina, M. G., D. Mora, P. Schumann, C. Parini, P. L. Manachini, and E. Stackebrandt. 2001. Reclassification of Saccharococcus caldoxylosilyticus as Geobacillus caldoxylosilyticus (Ahmed et al. 2000) comb. nov. Int. J. Syst. Evol. Microbiol. 51:2063-2071. [DOI] [PubMed] [Google Scholar]
- 8.Hallas, L. E., M. H. Hahn, and C. Korndorfer. 1988. Characterization of microbial traits associated with glyphosate degradation in industrial activated sludge. J. Ind. Microbiol. 3:377-385. [Google Scholar]
- 9.Hayes, V. E. A., N. G. Ternan, and G. McMullan. 2000. Organophosphonate metabolism by a moderately halophilic bacterial isolate. FEMS Microbiol. Lett. 186:171-175. [DOI] [PubMed] [Google Scholar]
- 10.Jacob, G. S., J. Garbow, L. E. Hallas, G. M. Kishore, and J. Schaefer. 1988. Metabolism of glyphosate in Pseudomonas sp. strain L. Br. Appl. Environ. Microbiol. 54:2953-2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai, S., B. Uno, and M. Tomita. 1991. Determination of glyphosate and its major metabolite aminomethylphosphonic acid by high-performance liquid chromatography after derivatization with p toluenesulfonyl chloride. J. Chromatogr. 540:411-415. [Google Scholar]
- 12.Kertesz, M., A. Elegora, and N. Amrhein. 1991. Evidence for two distinct phosphonate-degrading enzymes (C-P lyases) in Arthrobacter sp. GLP-1. Biodegradation 2:53-59. [DOI] [PubMed] [Google Scholar]
- 13.Kishore, G. H., and G. S. Jacob. 1987. Degradation of glyphosate by Pseudomonas sp. PG2982 via a sarcosine intermediate. J. Biol. Chem. 262:12164-12168. [PubMed] [Google Scholar]
- 14.Krzysko-Lupicka, T., W. Strof, K. Kubs, M. Skorupa, P. Wieczorek, B. Lejczak, and P. Kafarski. 1997. The ability of soil-borne fungi to degrade organophosphonate carbon-to-phosphorus bonds. Appl. Microbiol. Biotechnol. 48:549-552. [DOI] [PubMed] [Google Scholar]
- 15.Malik, J., G. Barry, and G. M. Kishore. 1989. The herbicide glyphosate. Biofactors 2:17-25. [PubMed] [Google Scholar]
- 16.McMullan, G., and J. P. Quinn. 1994. The utilization of aminoalkylphosphonic acids as sole nitrogen sources by an environmental bacterial isolate. Lett. Appl. Microbiol. 17:135-138. [Google Scholar]
- 17.Nakashita, H., A. Shimazu, T. Hidaka, and H. Seto. 1992. Purification and characterization of phosphoenolpyruvate phosphomutase from Pseudomonas gladioli B-1. J. Bacteriol. 174:6857-6861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nazina, T. N., T. P. Tourova, A. B. Poltaraus, E. V. Novikova, A. A. Grigoryan, A. E. Ivanova, A. M. Lysenko, V. V. Petrunyaka, G. A. Osipov, S. S. Belyaev, and M. V. Ivanov. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. kaustophilus, G. thermoglucosidasius and G. thermodentrificans. Int. J. Syst. Evol. Microbiol. 51:433-446. [DOI] [PubMed] [Google Scholar]
- 19.Obojska, A., B. Lejczak, and M. Kubrak. 1999. Degradation of phosphonates by streptomycetes isolates. Appl. Microbiol. Biotechnol. 51:872-876. [DOI] [PubMed] [Google Scholar]
- 20.Pipke, R., and N. Amrhein. 1988. Degradation of the phosphonate herbicide glyphosate by Arthrobacter atrocyaneus ATCC 13752. Appl. Environ. Microbiol. 54:1293-1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qureshi, A. A., C. E. Elson, and L. A. Lebeck. 1982. Application of high-performance liquid-chromatography to the determination of glyoxylate synthesis in chick-embryo liver. J. Chromatogr. 249:333-345. [DOI] [PubMed] [Google Scholar]
- 22.Tebbe, C. C., and H. H. Reber. 1988. Utilization of the herbicide phosphinothricin as a nitrogen source by soil bacteria. Appl. Microbiol. Biotechnol. 29:103-105. [Google Scholar]
- 23.Ternan, N. G., and G. McMullan. 2000. The utilization of 4-aminobutylphosphonate as sole nitrogen source by a strain of Kluyveromyces fragilis. FEMS Microbiol. Lett. 184:237-240. [DOI] [PubMed] [Google Scholar]
- 24.Ternan, N. G., J. T. G. Hamilton, and J. P. Quinn. 2000. Initial in vitro characterisation of phosphonopyruvate hydrolase, a novel phosphate starvation independent, carbon-phosphorus bond cleavage enzyme in Burkholderia cepacia Pal6. Arch. Microbiol. 173:35-41. [DOI] [PubMed] [Google Scholar]
- 25.Ternan, N. G., J. W. McGrath, and J. P. Quinn. 1998. Phosphoenolpyruvate phosphomutase activity in an l-phosphonoalanine-mineralizing strain of Burkholderia cepacia. Appl. Environ. Microbiol. 64:2291-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ternan, N. G., J. W. McGrath, G. McMullan, and J. P. Quinn. 1988. Organophosphonates: occurrence, synthesis and biodegradation by microorganisms. World J. Microbiol. Biotechnol. 14:635-647. [Google Scholar]
- 27.Wackett, L. P., S. L. Shames, C. P. Venditti, and C. T. Walsh. 1987. Bacterial carbon-phosphorus lyase: products, rates, and regulation of phosphonic and phosphinic acid metabolism. J. Bacteriol. 169:710-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weimberg, R., and W. L. Orton. 1963. Repressible acid phosphomonoesterase and constitutive pyrophosphatase of Saccharomyces mellis. J. Bacteriol. 86:805-813. [DOI] [PMC free article] [PubMed] [Google Scholar]