Abstract
The history of the rabies virus dates back four millennia, with the virus being considered by many to be the first known transmitted between animals and humans. In Brazil, rabies virus variants associated with terrestrial wild animals, marmosets, and different bat species have been identified. In this study, bat samples from different regions of São Paulo State, in Southeast Brazil, were analyzed to identify their genetic variability and patterns. A total of 51 samples were collected over ten years (1999–2009) and submitted to the immunofluorescent technique using monoclonal antibodies for antigenic profile detection (the diagnostic routine used in Latin American countries) and genetic evolution analysis through maximum likelihood approaches. Three antigenic profiles were detected: one related to the rabies virus maintained by hematophagous bat populations (AgV3), part of the monoclonal antibody panel used, and two other profiles not included in the panel (called NC1 and NC2). These antigenic profiles were genetically distributed in five groups. Group I was related to hematophagous bats (AgV3), Groups II and III were related to insectivorous bats (NC1) and Groups IV and V were also related to insectivorous bats (NC2). The results presented herein show that genetic lineages previously restricted to the northwest region of São Paulo State are now found in other state regions, highlighting the need for a comprehensive genetic study of bat rabies covering geographic and temporal space, through expanded genomic analysis using a standard genomic fragment.
Keywords: rabies virus, non-hematophagous bats, viral diversity, antigenic and genetic characterization
1. Introduction
Rabies is one of the most important viral infectious diseases and, with a history dating back four millennia, is considered by many to be the first known disease transmitted between animals and humans [1]. Rabies was initially described in humans and carnivores, but studies of bats and rabies in Brazil and Trinidad in the 1920s and 1930s showed the existence of a rabies virus aerial cycle. The existence of the aerial cycle explained epidemics that occur without the presence of carnivorous animals and how the virus continues to circulate in places where rabies in domestic animals has been controlled, showing the interrelationship of this aerial cycle with the terrestrial cycle [1,2,3].
The rabies virus (RABV) belongs to the Rhabdoviridae family and Lyssavirus genus, which contains 18 viral species [4], most of them associated with bats from the Old World. The Lyssavirus rabies species is the only one that circulates among numerous mammals, including bats, carnivores, and nonhuman primates, such as marmosets in Brazil [5,6,7]. Cross-species transmission has been observed among non-canid carnivores, bats, and other mammal species, leading to the emergence of new lineages also related to American bats [5].
Antigenic characterization studies have been conducted in several Latin American countries using a panel of monoclonal antibodies (MAbs) produced by the Centers for Disease Control and Prevention (CDC), Atlanta, USA. The use of these MAbs, from the 1980s onwards, established a new era in the knowledge of RABV reservoirs and transmissibility, resulting in immediate advances in epidemiological surveillance in those countries before laboratories implemented sequencing capability. Over the years, this tool has identified several other antigenic profiles not included in the original panel [6] established by Diaz et al. [8]. These additional profiles were detected in many countries, including Brazil [9,10,11,12,13,14,15]. In a certain way, this was already expected, considering that among the samples analyzed for establishing the panel profiles, there were no varieties of isolates from different South American bat species. This identification technique continues to be used routinely in most Latin American countries.
The genetic analysis of the N gene, with a chosen genome fragment (between position 1157 and 1476 of PV-NC_001542), not only allowed the correlation of host species with their geographic distributions but also confirmed the differences observed in the reactivity pattern in the antigenic tests among samples associated with different species of bats in different countries [9,16,17,18].
In Brazil, the first antigenic [11] and genetic [19] studies showed that, as in other Latin American countries, the two predominant antigenic variants/viral lineages were associated with viruses maintained by dogs (called AgV1 and AgV2) and the Desmodus rotundus variant (AgV3). Later, antigenic variants/viral lineages associated with terrestrial wild animals, such as foxes and wild dogs [20,21], marmosets [6], and different species of insectivorous and frugivorous bats, were identified [13,22,23,24,25,26].
Both the genetic and antigenic characterizations of bat RABV isolated in the State of São Paulo in southeastern Brazil have shown the existence of variants/lineages related to D. rotundus (AgV-3), Tadarida brasiliensis (AgV4), and Lasiurus sp. (AgV6), also circulating in other species of insectivorous and frugivorous bats, along with other antigenic profiles that were not included in the CDC monoclonal panel [6,8,10,21,22].
In this study, bat samples from different regions of São Paulo State were analyzed using evolutionary approaches to identify genetic variability of the RABV in São Paulo. These isolates are compared with isolates described in previous publications, characterized antigenically and genetically, for which sequences have been deposited in Genbank (Table A1, Appendix A).
2. Materials and Methods
2.1. Samples
This study included RABV isolates from 32 municipalities in different administrative regions of São Paulo State in Southeast Brazil (Figure 1), between 1999 and 2010.
Figure 1.
Geographic location of sample collection. Colors are related with antigenic variant/profile: red for Group I (AgV3) related to D. rotundus, orange for Group II and III (related to antigenic Non-compatible 1-NC1 profile), and dark blue for Groups IV and V (NC2). Cities where more than one genetic group and/or antigenic variant was detected are in pale blue with a graphic following determined pattern of colors. GenBank sequences used in phylogenetic tree reconstruction can be observed as red dots for Group I, orange stars for samples clustered in Groups II and III, and dark blue dots for Groups IV and V. The map was modified for this study using Inkscape software version 1.3.2 (available at www.inkscape.org accessed on 4 July 2025). The original map is available at https://pt.m.wikipedia.org/wiki/Ficheiro:SaoPaulo_MesoMicroMunicip.svg (accessed on 19 February 2025).
All the samples had been previously diagnosed as positive for rabies by means of the fluorescent antibody test (FAT) [27] and mouse inoculation test (MIT) [28], considered the gold standard at the time of receiving samples for diagnosis between 1995 and 2010. In all, 48 bat samples were studied: 13 frugivorous (11 Artibeus lituratus, 01 Artibeus planirostris, 01 Artibeus fimbriatus) and 35 insectivorous (04 Myotis nigricans, 10 Neoeptesicus furinalis, 03 Neoeptesicus diminutus, 01 Neoeptesicus sp., 05 Molossus fluminensis, 03 Molossus molossus, 01 Cynomops abrasus, 02 Nyctinomops laticaudatus, 01 Nyctinomops macrotis, 01 Lasiurus blossevillii, 01 Lasiurus ega, 01 Eumops glaucinus and 02 non-hematophagous (NH) bats not identified). Samples from one cat, one bovine, and one horse were also included, giving 51 samples in total (Table 1).
Table 1.
Sequences obtained in the study, including environmental and geographical information.
| GenBank Access | ID Sample/ Year |
Species | Place of Origin | Antigenic Variant/Profile |
Genetic Lineage |
|---|---|---|---|---|---|
| HQ666824 | IB 346/99 | Artibeus lituratus | São José do Rio Preto | V-3 | D. rotundus |
| HQ666825 | IB 777/00 | Cynomops abrasus | Ipiguá | NC2 | Insect. Bats |
| HQ666826 | IB 249/01 | Nyctinomops macrotis | São José do Rio Preto | NC1 | Insect. bats |
| HQ666827 | IB 250/01 | Nyctinomops laticaudatus | São José do Rio Preto | NC1 | Insect. Bats |
| HQ666828 | IB 636/01 | Neoeptesicus furinalis * | Olímpia | NC1 | Insect. Bats |
| HQ666829 | IB 808/01 | Artibeus lituratus | São José do Rio Preto | V-3 | D. rotundus |
| HQ666830 | IB 1019/01 | Neoeptesicus sp. * | Cardoso | NC1 | Insect. Bats |
| HQ666831 | IB 1070/01 | Neoeptesicus furinalis * | São José do Rio Preto | NC1 | Insect. Bats |
| HQ666832 | IB 62/02 | Neoeptesicus furinalis * | Catanduva | NC1 | Insect. Bats |
| HQ666833 | IB 109/02 | Molossus molossus | Ilha Solteira | NC2 | Insect. Bats |
| HQ666834 | IB 835/02 | Myotis nigricans | Cajobi | NC1 | Insect. Bats |
| HQ666835 | IB 992/02 | Artibeus lituratus | Dracena | V-3 | D. rotundus |
| HQ666836 | IB 1021/02 | Molossus fluminensis * | Presidente Venceslau | V-3 | D. rotundus |
| HQ666837 | IB 1141/02 | Neoeptesicus furinalis * | Santo Anastácio | NC1 | Insect. Bats |
| HQ666838 | IB 1256/02 | NH bat (not identified) | Martinópolis | V-3 | D. rotundus |
| HQ666839 | IB 1371B/02 | Artibeus lituratus | Presidente Venceslau | V-3 | D. rotundus |
| HQ666840 | IB 1535/02 | Artibeus lituratus | Taciba | V-3 | D. rotundus |
| HQ666841 | IB 1539/02 | Molossus molossus | Presidente Prudente | V-3 | D. rotundus |
| HQ666842 | IB 1782/02 | Lasiurus ega | Presidente Prudente | NC1 | D. rotundus |
| HQ666843 | IB 349/03 | Artibeus planirostris | Santa Fé do Sul | V-3 | D. rotundus |
| HQ666844 | IB 350/03 | NH bat (not identified) | Catanduva | V-3 | D. rotundus |
| HQ666845 | IB 791/03 | Artibeus lituratus | Presidente Prudente | V-3 | D. rotundus |
| - | IB 826/03 | Artibeus fimbriatus | São José do Rio Preto | V-3 | ND |
| HQ666846 | IB 168/04 | Myotis nigricans | Campinas | NC1 | Insect. Bats |
| HQ666847 | IB 184/04 | Nyctinomops laticaudatus | São José do Rio Preto | NC1 | Insect. Bats |
| HQ666848 | IB 550/04 | Artibeus lituratus | Caçapava | V-3 | D.rotundus |
| HQ666849 | LRU 329/05 | Eumops glaucinus | Araçatuba | V-3 | D.rotundus |
| - | LRU 397/05 | Artibeus lituratus | Araçatuba | V-3 | ND |
| HQ666850 | LRU 43/09 | Neoeptesicus diminutus * | Pereira Barreto | NC1 | Insect. Bats |
| HQ666851 | LRU 84/09 | Neoeptesicus diminutus * | Araçatuba | NC1 | Insect. Bats |
| HQ666852 | LRU 149/09 | Myotis nigricans | Coroados | NC1 | Insect. Bats |
| HQ666853 | LRU 181/09 | Artibeus lituratus | Penápolis | V-3 | D. rotundus |
| HQ666856 | LRU 325/09 | Artibeus lituratus | Birigui | V-3 | D. rotundus |
| HQ666857 | LRU 374/09 | Artibeus lituratus | Guararapes | V-3 | D. rotundus |
| HQ666858 | LRU 389/09 | Molossus fluminensis * | Guararapes | V-3 | D. rotundus |
| HQ666859 | LRU 433/09 | Myotis nigricans | Penápolis | NC1 | Insect. Bats |
| HQ666860 | LRU 589/09 | Neoeptesicus furinalis * | Penápolis | NC1 | Insect. Bats |
| HQ666854 | LRPP 199/09 | Neoeptesicus furinalis * | Dracena | NC1 | Insect. Bats |
| HQ666855 | LRPP 224/09 | Neoeptesicus furinalis * | Parapuã | V-3 | D. rotundus |
| HQ666861 | LRPP 672/09 | Bovine/Cattle | Narandiba | V-3 | D. rotundus |
| HQ666862 | LRU 17/10 | Molossus fluminensis * | Penápolis | NC1 | Insect. Bats |
| KU299782 | LRPP 28/10 | Horse | Taciba | V-3 | D. rotundus |
| HQ666864 | LRPP 43/10 | Neoeptesicus diminutus * | Osvaldo Cruz | V-3 | D. rotundus |
| HQ666865 | LRU 60/10 | Neoeptesicus furinalis * | Birigui | NC1 | Insect. Bats |
| HQ666866 | LRU 76/10 | Neoeptesicus furinalis * | Penápolis | NC1 | Insect. Bats |
| HQ666867 | LRU 169/10 | Molossus molossus | Araçatuba | NC2 | Insect. Bats |
| HQ666868 | LRU 171/10 | Cat | Araçatuba | V-3 | D. rotundus |
| HQ666869 | LRU 177/10 | Molossus fluminensis * | Birigui | NC2 | Insect. Bats |
| HQ666870 | LRPP 198/10 | Lasiurus blossevillii | Teodoro Sampaio | NC2 | Insect. Bats |
| HQ666871 | LRU 299/10 | Neoeptesicus furinalis * | Penápolis | NC1 | Insect. Bats |
| HQ666872 | LRU 300/10 | Molossus fluminensis * | Penápolis | V-3 | D. rotundus |
ND = not done; NC = not compatible; NH = non-hematophagous; V-3 = variant 3. IB—Rabies Laboratory of “Instituto Biológico de São Paulo”; LRU—Rabies Laboratory of UNESP (São Paulo State University), Araçatuba; LRPP—Rabies Laboratory of APTA (São Paulo Agribusiness Technology Agency) of Presidente Prudente; * new taxonomy bat species classification according to [29,30,31].
2.2. Antigenic Characterization
Antigenic characterization was performed using the CDC (Atlanta, GA, USA) monoclonal antibodies (MAbs) panel according to the protocol determined by Favoretto et al. [11]. These eight MAbs against the RABV nucleoprotein, developed by CDC, can identify different RABV variants through different reactivity patterns.
2.3. Sequencing and Genetic Characterization
For genetic characterization, the initial steps (i.e., extracting RNA and obtaining cDNA) were performed using methods described in previous studies [13]. Molecular reactions were performed using primers described previously by Smith et al. [32] and Campos et al. [33] to amplify 320 base pairs from the coding and non-coding region of the nucleoprotein (between positions 1157 and 1476 of PV-NC_001542). Double-strand PCR-amplified products were purified using the ExoSAP-IT system (GE Healthcare Bio-Sciences Ltd.—USB Corporation, Cleveland, OH, USA) according to the manufacturer’s instructions, and Sanger sequencing was performed as previously described by Campos et al. [33]. The excess dideoxynucleotide terminators were removed with the Applied Biosystems Big Dye XTerminatorTM Purification Kit (Applied Biosystems, Foster City, CA, USA), following the manufacturer’s recommendations. Purified samples were subjected to electrophoresis in POP6 polymer using an ABI-PRISM model 3100 automatic sequencer (Applied Biosystems, Foster City, CA, USA). The samples were tracked automatically using the Automatic DNA Analyzer software package of the ABI-PRISM model 3100.
2.4. Phylogenetic Analysis
The obtained nucleotide sequences were pre-analyzed using the BLASTn program (https://blast.ncbi.nlm.nih.gov/Blast.cgi, accessed on 19 February 2025) to confirm amplification of the specific product and then aligned with available GenBank sequences (Appendix A) using Geneious Prime software version 2019.2.3. The chosen GenBank sequences were selected based on full information like the host, place, and year of collection. Pairwise distances were calculated by MEGA 11 version 11.0.13 (available at https://www.megasoftware.net/ accessed on 19 February 2025) and phylogenetic trees were reconstructed using IQTREE software version 2.4.0 (available at http://www.iqtree.org/ accessed on 18 February 2025). The best model fit determined by IQTree was TIM + F + I + G4. To analyze the temporal virus variability, we used TempEst v1.5.3 (available at http://tree.bio.ed.ac.uk/software/tempest/ accessed on 19 February 2025) in the phylogenetic tree and root-to-tip method with best-fitting root in correlation function. The time-scaled phylogenetic tree was analyzed via Augur version 21.1.0 and auspice version 2.62.0 implemented on Nextstrain [34,35].
2.5. Phylogenetic and Antigenic Site Amino Acid Visualization
Phylogenetic visualization of RABV sequences was conducted using R version 4.4.1 (14 June 2024, ucrt). A phylogenetic tree in nexus format was imported using the ape package and further processed and visualized with the ggtree, treeio, and igraph packages. The final annotated phylogenetic tree was visualized with tip points colored with regard to geographic location and a scale bar indicating time (years) and genetic distance (substitutions/site). The final figure editing and layout adjustments were performed using the Inkscape program version 1.3.2. The alignment used for phylogenetic tree reconstruction was translated to amino acid and used to prepare one Figure with the region of antigenic site I present in the nucleoprotein.
3. Results
Among the fifty-one samples submitted to antigenic characterization, twenty-five (49%) were characterized as AgV-3 (RABV maintained by D. rotundus hematophagous bat populations) and twenty-six were characterized as RABV maintained by non-hematophagous bat populations (NC1 and NC2); out of these, twenty-one (41.2%) presented the antigenic profile NC1 and five (9.8%) presented the antigenic profile NC2.
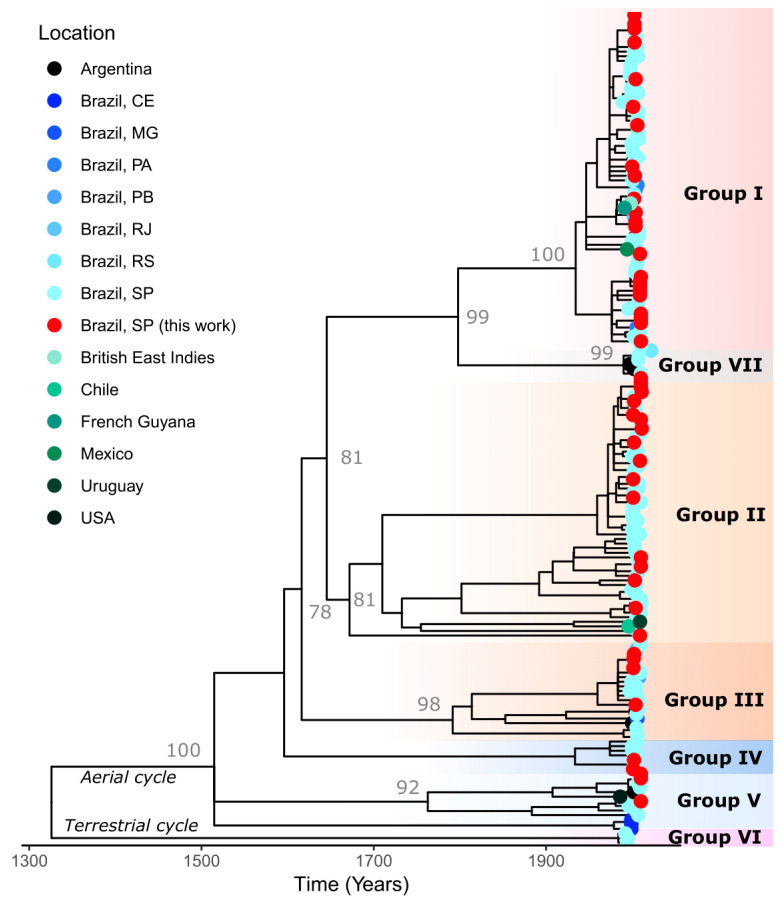
The samples from this study were segregated into five different phylogenetic groups, highlighted in colors according to their genetic and antigenic patterns (Figure 2). The group called Group I showed samples with a genetic lineage associated with the virus maintained by D. rotundus hematophagous bats and antigenic variant AgV3. The virus groups isolated from insectivorous bats presented four independent phylogenetic clades, called Group II, Group III, Group IV, and Group V, with the antigenic profile NC1 in Groups II and III, and antigenic profile NC2 in Groups IV and V. Group II presented the greatest diversity of its host species, consisting of Eptesicus spp. (currently called Neoeptesicus), Eumops spp., Myotis spp., Nyctinomops spp., and Lasiurus spp. Groups VI and VII, with isolates external to the present study, presented isolates from marmosets (Group VI) and bat isolates related to Tadarida brasiliensis (Group VII).
Figure 2.
Time-scaled phylogenetic tree reconstructed using 320 nucleotides from nucleoprotein terminal gene using IQ-TREE software version 2.4.0, visualized and edited using FigTree software version 1.4.4 and plotted in RStudio version 2024.12.1+563 using ggtree version 3.14.0, treeio version 1.30.0, and igraph version 2.1.4 packages. The samples from this study can be observed in the tree in red dots; other samples from Brazil, available in GenBank, are marked in a blue pallet of colors while samples from other countries are marked in a green pallet of colors. The groups determined in this study are delineated vertically by red (Group I), orange (Groups II and III), and dark blue (Groups IV and V) shading. The groups without segregated samples from this study are shown with pink (Group VI) and gray (Group VII) shadings.
The lowest percentage of identity was observed in comparison with the clade related to marmosets (Group VI in the phylogenetic tree). Estimates of evolutionary divergence over sequence pairs between groups, obtained using the maximum likelihood method in MEGA 11, were used to calculate the distances between groups (Table A2, Appendix A), showing a range from 8.7% between Groups I and VII to 17.9% between Groups II and VI. Among the groups detected in this study, the highest within-group distances (Table A3, Appendix A) were observed in Groups II (6.7%), III (4.7%), and V (5.7%) while minor within-group distances were detected in Groups IV (1.8%) and I (2.5%).
In Figure 1, we can also observe the geographic distribution of the samples from this study and from Genbank, used for the reconstruction of the phylogenetic tree, according to the resulting groups (genetic characterization) and antigenic variants/profiles (AgV-3, NC1, and NC2).
During the analysis of the antigenic site present in the nucleoprotein, we identified genetic signatures for some groups in the phylogenetic tree. Groups I, IV, V, VI, and VII and the root group showed recognized patterns (AET, AEV, TEV, TEA, TEM, and TDV, respectively), indicating the stability of these genetic groups. On the other hand, we could not identify any pattern for Groups II (TEA, TDE, IDT, TEV, and TDV) and III (TEA, TEL, and TEV). In these groups, we found higher variability that was confirmed by tree topology and can be seen in Figure 3. The full map of the antigenic site was produced and can be accessed in the Supplementary Materials.
Figure 3.
A partial amino-acid alignment showing the presence of antigenic site I in the nucleoprotein. The color match those in the phylogenetic tree presented in Figure 2. For this figure, the sequences used in the phylogenetic tree reconstruction were employed, maintaining only the variability in the antigenic site region, with a preference for sequences from this study. The genetic signature AET is shown in red (and also highlighted with red shading) for phylogenetic Group I related to hematophagous bat species D. rotundus; AEV is shown in dark blue (and also highlighted with dark blue shading) for phylogenetic Group IV related to non-hematophagous bats; TEV is shown in a gradient of dark blue (and also highlighted in a gradient dark blue shading) for phylogenetic Group V; TEA is shown in magenta/pink (and also highlighted with pink shading) for phylogenetic Group VI related to marmosets; TEM in black (and also highlighted with gray shading) for phylogenetic Group VII related to the bat species T. brasiliensis; TDV is shown in green for the root group related to the terrestrial cycle of transmission of RABV. For phylogenetic groups II and III (highlighted with gradient orange shading), it was not possible to find one genetic signature in the antigenic site I; in fact, in these groups, the variability was diverse and is noted in different colors: TEA in orange for the major antigenic site found (which was the same genetic signature found in Group VI related to marmosets), purple for variations (TEL, TDE, IDT), dark blue for the same signature present in Group V (TEV), and green for the same signature present in the terrestrial cycle of transmission (TDV) in one sequence available at GenBank (AF396064). The amino acid letters and class are outlined above.
The Pearson correlation coefficient calculated in the analysis of temporal virus variability in this dataset was 0.18 (p = 0.015), as shown in Figure 4. Although the correlation coefficient is slightly positive, the TempEst analysis showed the stable evolution rate to the dataset during the sampling period considered in the analysis.
Figure 4.
Correlation between genetic divergence and sampling time was obtained by a root-to-tip analysis using the RABV sequences and plotted using R command line. The dots are related with clades from phylogenetic tree, the red dots are related with Group I, the orange with Group II, the brown with Group III, the blue with Group IV, the clear blue with Group V, the pink with Group VI, the clear gray with the Group VII and the dark gray dots are related with the terrestrial cycle of transmission. The dashed line (colored dark gray) represents the regression line, and the light gray area around the dashed line is the confidence interval.
4. Discussion
All 13 fruit bats of the genus Artibeus presented antigenic variant 3 (AgV-3), as did bats from the genera Molossus (04), Eumops (1), and Neoeptesicus (2) and non-identified bats (2). Similar results were observed in rabies-positive bat samples from other regions of São Paulo State [11,15,19,24,36]. Previous studies had demonstrated that the frequency of RABV in Artibeus was higher than that in Desmodus bats in the study area [36,37,38,39], which could explain how AgV-3 is present in species that do not co-inhabit with D. rotundus species. This is corroborated by the finding that the genetic lineage of D. rotundus is not exclusive to the species since this lineage has been detected in non-hematophagous bats such as the fruit bat Artibeus lituratus [40] and insectivorous bats in this study, in addition to other previous studies [13].
The samples antigenically described as NC1 in phylogenetic group II were segregated with samples from the same geographic region and with one sample (EU981922) from Uruguay, with a geographical distance of more than 1,200 miles. The samples in the phylogenetic group III were segregated with samples from the same geographical region and with one sample (AB297647) from Rio de Janeiro State, more than 300 m away, and another sample (AB618034) from Paraiba State, more than 1600 m away. This antigenic profile was previously described in São Paulo State [11,12,13,14,15,41]. These groups were previously related particularly with host species described by Oliveira et al. [42]; nonetheless, in the present study, we observed different species in the same clade/phylogenetic group. Bats play an important role in virus transmission and spread in the Americas [42,43,44], and it was clearly demonstrated in this study that insectivorous bats present a heterogeneous genetic distribution independent of host species.
The NC2 antigenic profile detected in five samples from the Molossus and Lasiurus genera was previously described in the same geographical region [7] and was observed in three monophyletic clades with high bootstrap value support (81 to 98%). In a previous study [13], two samples (GU646777 and HM854031) were segregated independently as subgroups; in the present study, these samples were segregated as part of phylogenetic groups IV and V, confirming the importance of including more sequences and information from other regions of the state and the country, as well as from other bat host species. This was also visualized on the map (Figure 1), where the geographical distribution of the variants was described. With the inclusion of new samples, a more homogeneous distribution of antigenic profiles and genetic lineages across Sao Paulo State can be observed since, previously, these profiles and lineages were restricted to the northwest region of the state [13]. Nonetheless, important data from previous publications regarding the central region of this state and from other states of Brazil could not be compared with the isolates from this study, considering that the authors sequenced a different genome region or only a coding region [15,40,42,45,46,47]. This reinforces the idea that the same region of the genome should be analyzed and standardized by researchers in future studies.
Currently, the term “antigenic variants” and the CDC MAbs panel still are used mainly in Latin America and only in a few accredited laboratories. The results obtained in the present sample’s dataset corroborated previous results [13,15] that showed that this panel, despite its valuable importance in the past, does not have sufficient resolution as high as that obtained using genetic tools to characterize RABV variants from non-hematophagous bats, leading to divergences. An approach of comparing samples from different geographical regions using antigenic and genetic characterization is no longer ideal, as previously highlighted in other studies [15]. In any case, the term ‘antigenic variant’ will not become extinct immediately since it is still the language used in Ministry of Health reports in Brazil, for example. As found in this study, the antigenic characterization was realized during sample processing almost twenty years ago, providing useful information that could be used for future studies and to better understand rabies epidemiology. Therefore, future RABV studies must be focused on genetic analyses to provide a deeper and more comprehensive understanding of the virus, explaining its epidemiology, its dynamics, and possible interventions. This could lead to significant advances in rabies surveillance, prevention, and control at both population and individual levels.
Antigenic site I was presented here using nucleoprotein amino acid alignments, and the authors observed a genetic signature that had previously been described [6]. However, for genetic groups II and III, these signatures could not be observed; in fact, they are genetically and antigenically diverse, independent of the comparison between these two approaches.
In the first RABV genetic studies in Latin America, it was established that the 320 nucleotides in the nucleoprotein carboxi-terminus region, including the nucleo- and phosphoprotein intergenic regions (non-coding) between genome positions 1157 and 1476 (based in PV genome NC_001542), could be the standard for phylogenetic studies because this region presents the large nucleotide variability, 1.9 times greater than in the coding region [17]. For phylogenetic analyses, according to Smith et al. [46], groups that present a distance higher than 5% from other groups can be considered a distinct genetic lineage. Thus, for this study, the groups in the phylogenetic tree were determined by following this consideration.
In the phylogenetic tree (Figure 3 and Figure S1), the high bootstrap values (100% for aerial cycle of transmission; 78% for genetic lineages segregated into Groups I, II, III and VII; and 81% for Groups I, II and VII) support the presence of basal genotypes of the virus. For example, sample HQ666860 from an insectivorous bat, Neoeptesicus furinalis, collected in 2009 presented a long branch in Group II (bootstrap 81%), indicating a high number of nucleotide substitutions. Group I, Group III, Group IV, and Group V showed bootstrap values of 100%, 98%, 78% and 92%, respectively; this tree topology could also explain how and why samples in the antigenic analysis from genetic groups II to V presented a different antigenic profile.
The positive but low correlation coefficient (0.18) associated with the phylogenetic analysis suggests that during the short period of sequence sampling, between the years of 1986 and 2022, there was an accumulation of diversity, but the occurrence of the common ancestor to all sequences was distant in the past. This had been previously shown by other researchers [42], and this result means a stable RABV evolution rate in the analyzed period. This result reflects the profile of zoonotic viruses such as rabies, considering only the aerial cycle, and agrees with the TMRCA (time to the most recent common ancestor) of approximately 170 years determined by de Souza et al. [40] when analyzing the D. rotundus/A. lituratus genetic lineage (both antigenically AgV3).
5. Conclusions
Despite some limitations, such as analyzing only a 320-nucleotide fragment; the limited number of sequences available in GenBank for this same fragment; the absence of relevant information such as on the date, species, and collection location of these available sequences in the GenBank; and the retrospective nature of this study performed over a decade ago, these data provide valuable insights into RABV among bats. The key findings of this study are as follows: (i) antigenic profiles and genetic lineages previously restricted to the northwest region of the state of São Paulo are now found in other state regions, (ii) future rabies studies must be focused only on genetic analysis, and (iii) there is a need for a comprehensive genetic study of bat rabies in São Paulo State and greater Brazil with diverse sample locations and expanded genomic analyses using a standard genomic fragment or full genome when possible. Moreover, focusing only on host species could lead to misleading conclusions about RABV evolution and dispersal concerning time and geography.
Acknowledgments
The authors thank Adriana Ruckert da Rosa for bat species revision.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/v17081063/s1, Figure S1: Conventional phylogenetic tree, Figure S2: Partial amino acid antigenic site I for all sequences used in phylogenetic tree reconstruction.
Appendix A
Table A1.
| Sequence ID | Group | Year | Host | Site Origin | Country | Genbank Access | Reference |
|---|---|---|---|---|---|---|---|
| AB201802_BR_AL4 | I | 2002 | Artibeus lituratus | Dracena, SP | Brazil | AB201802 | [24] |
| AB201803_BR_DR1 | I | 2000 | Desmodus rotundus | Lindóia, SP | Brazil | AB201803 | [24] |
| AB201805_BR_DR3 | I | 2001 | Desmodus rotundus | São José do Barreiro, SP | Brazil | AB201805 | [24] |
| AB201806_BR_NL1 | III | 1998 | Nyctinomops laticaudatus | São José do Rio Preto, SP | Brazil | AB201806 | [24] |
| AB201807_BR_NL2 | II | 1999 | Nyctinomops laticaudatus | São José do Rio Preto, SP | Brazil | AB201807 | [24] |
| AB201808_BR_NL3 | III | 2001 | Nyctinomops laticaudatus | Nova Granada, SP | Brazil | AB201808 | [24] |
| AB201812_BR_EF2 | II | 2001 | Eptesicus furinalis | Olimpia, SP | Brazil | AB201812 | [24] |
| AB201813_BR_EF3 | II | 2001 | Eptesicus furinalis | São José do Rio Preto, SP | Brazil | AB201813 | [24] |
| AB201814_BR_EF4 | II | 2002 | Eptesicus furinalis | Catanduva, SP | Brazil | AB201814 | [24] |
| AB201815_BR_MM1 | IV | 1999 | Molossus molossus | Jales, SP | Brazil | AB201815 | [24] |
| AB201816_BR_MM2 | IV | 2002 | Molossus molossus | Ilha Solteira, SP | Brazil | AB201816 | [24] |
| AB201817_BR_MR1 | I | 2002 | Molossus rufus | Presidente Venceslau, SP | Brazil | AB201817 | [24] |
| AB201818_BR_MA1 | IV | 2000 | Molossus abrasus | Itapira, SP | Brazil | AB201818 | [24] |
| AB297630_BR_AL6 | I | 2001 | Artibeus lituratus | Rio de Janeiro, RJ | Brazil | AB297630 | [48] |
| AB297631_BR_AL7 | I | 2004 | Artibeus lituratus | Vargem Grande Paulista, SP | Brazil | AB297631 | [48] |
| AB297647_BR_NL4 | III | 2004 | Nyctinomops laticaudatus | Rio de Janeiro, RJ | Brazil | AB297647 | [48] |
| AB618034_strain_MPVI | III | 2007 | Molossus molossus | Santo Antonio, Paraíba | Brazil | AB618034 | Unpublished |
| AF394886_2085 | V | 1986 | Lasiurus borealis | Walker County, Texas | USA | AF394886 | [49] |
| AF396064_cym3941_1995 | II | 1995 | Myotis chiloensis | - | Chile | AF396064 | [50] |
| AY233427_Batbbt123 | VII | 2001 | Tadarida brasiliensis | Buenos Aires | Argentine | AY233427 | [44] |
| AY233448_Stchmbt80 | III | 2000 | Histiotus montanus | Rio Turbio, Santa Cruz | Argentine | AY233448 | [44] |
| AY233451_Batbbt125 | V | 2001 | Tadarida brasiliensis | Buenos Aires | Argentine | AY233451 | [44] |
| AY654585_Brhm4097 | VI | 1998 | Human | Ceará | Brazil | AY654585 | [6] |
| AY654586_Brsg4108 | VI | 1998 | Callithrix jacchus jacchus | Ceará | Brazil | AY654586 | [6] |
| AY654587_Brhm4138 | VI | 1998 | Human | Ceará | Brazil | AY654587 | [6] |
| AY877435_V920 | I | 1993 | Bovine | Chiapas | Mexico | AY877435 | [51] |
| DQ631835_bref8150_05 | III | 2005 | Eptesicus furinalis | Jundiaí, SP | Brazil | DQ631835 | [41] |
| EF363743_IP306_Portel_PA_2004 | I | 2004 | Human | Portel, PA | Brazil | EF363743 | [52] |
| EF363751_IP5214Viseu_PA_2004 | I | 2004 | Human | Viseu, PA | Brazil | EF363751 | [52] |
| EF363757_IP7541Viseu_PA_2005 | I | 2005 | Human | Viseu, PA | Brazil | EF363757 | [52] |
| EU293113_9001FRA | I | 1990 | Fox | - | French Guyana | EU293113 | [53] |
| EU293116_9704ARG | VII | 1997 | bat | - | Argentine | EU293116 | [53] |
| EU873001_brctSP4551_96 | III | 1996 | cat | São Paulo State | Brazil | EU873001 | Unpublished, Favoretto |
| EU981922_IP6773U_2008 | II | 2008 | Myotis sp. | - | Uruguay | EU981922 | [43] |
| GU552788_IP2989_2007 | VII | 2007 | Nyctnomops laticaudatus | Joanópolis, SP | Brazil | GU552788 | [25] |
| GU552789_IP1779_2006 | IV | 2006 | Molossus rufus | Ribeirão Preto, SP | Brazil | GU552789 | [25] |
| GU552790_IP1992_2005 | III | 2005 | Histiotus velatus | Vargem Grande Paulista, SP | Brazil | GU552790 | [25] |
| GU552791_IP6883_2006 | III | 2006 | Histiotus velatus | Campo Limpo Paulista, SP | Brazil | GU552791 | [25] |
| GU552792_IP3321_2005 | III | 2005 | Histiotus sp. | Belo Horizonte, MG | Brazil | GU552792 | [25] |
| GU552795_IP10529_2005 | III | 2005 | Nyctnomops laticaudatus | Ribeirão Preto, SP | Brazil | GU552795 | [25] |
| GU552796_IP4359_2007 | III | 2007 | Molossus molossus | Campinas, SP | Brazil | GU552796 | [25] |
| GU552798_IP8089_2005 | III | 2005 | Nyctnomops laticaudatus | São Sebastião, SP | Brazil | GU552798 | [25] |
| GU552807_IP8061_2006 | II | 2006 | Eptesicus furinalis | Campinas, SP | Brazil | GU552807 | [25] |
| GU552810_IP3056_2007 | II | 2007 | Eptesicus furinalis | Barretos, SP | Brazil | GU552810 | [25] |
| GU552815_IP8665_2005 | II | 2005 | Myotis nigricans | Ribeirão Preto, SP | Brazil | GU552815 | [25] |
| GU552820_IP4157_2005 | II | 2005 | Myotis nigricans | Águas de Lindóia, SP | Brazil | GU552820 | [25] |
| GU552821_IP4896_2005 | II | 2005 | Myotis nigricans | Caçapava, SP | Brazil | GU552821 | [25] |
| GU552824_IP2654_2006 | V | 2006 | Lasiurus cinereus | Garça, SP | Brazil | GU552824 | [25] |
| GU592648_brdrusp100_07 | I | 2007 | Desmodus rotundus | São José do Barreiro, SP | Brazil | GU592648 | [33] |
| GU646775_brmn131_03 | II | 2003 | Myotis nigricans | Araçatuba, SP | Brazil | GU646775 | [13] |
| GU646776_brmn45_03 | II | 2003 | Myotis nigricans | Araçatuba, SP | Brazil | GU646776 | [13] |
| GU646777_brmm95_03 | V | 2003 | Molossus molossus | Nova Independência, SP | Brazil | GU646777 | [13] |
| GU646778_brmn38_03 | II | 2003 | Myotis nigricans | Penápolis, SP | Brazil | GU646778 | [13] |
| GU646779_bral268_98 | I | 1998 | Artibeus lituratus | Mirandópolis, SP | Brazil | GU646779 | [13] |
| GU646780_bral452_99 | I | 1999 | Artibeus lituratus | Ilha Solteira, SP | Brazil | GU646780 | [13] |
| GU646781_bref431_04 | II | 2004 | Eptesicus furinalis | Bilac, SP | Brazil | GU646781 | [13] |
| GU646782_brlb46_04 | II | 2004 | Lasiurus blossevillii | Valparaíso, SP | Brazil | GU646782 | [13] |
| GU646783_bral311_03 | I | 2003 | Artibeus lituratus | Penápolis, SP | Brazil | GU646783 | [13] |
| GU646784_bral304_03 | I | 2003 | Artibeus lituratus | Penápolis, SP | Brazil | GU646784 | [13] |
| GU646785_brmn150_03 | II | 2003 | Myotis nigricans | Araçatuba, SP | Brazil | GU646785 | [13] |
| GU646786_brmn234_02 | II | 2002 | Myotis nigricans | Bilac, SP | Brazil | GU646786 | [13] |
| GU646787_bral625_01 | I | 2001 | Artibeus lituratus | Birigui, SP | Brazil | GU646787 | [13] |
| GU646788_brmn610_01 | II | 2001 | Myotis nigricans | Sud Menucci, SP | Brazil | GU646788 | [13] |
| GU646789_bral499_01 | I | 2001 | Artibeus lituratus | Guararapes, SP | Brazil | GU646789 | [13] |
| GU646790_bref126_01 | II | 2001 | Eptesicus furinalis | Penápolis, SP | Brazil | GU646790 | [13] |
| GU646791_bral84_01 | I | 2001 | Artibeus lituratus | Guararapes, SP | Brazil | GU646791 | [13] |
| GU646792_bref40_01 | II | 2001 | Epitesicus furinalis | Bilac, SP | Brazil | GU646792 | [13] |
| GU646793_breg01_01 | II | 2001 | Eumops glaucinus | Bilac, SP | Brazil | GU646793 | [13] |
| GU646794_brmn839_00 | II | 2000 | Myotis nigricans | Bilac, SP | Brazil | GU646794 | [13] |
| GU646795_bral566_00 | I | 2000 | Artibeus lituratus | Penápolis, SP | Brazil | GU646795 | [13] |
| GU646796_bref213_00 | II | 2000 | Eptesicus furinalis | Araçatuba, SP | Brazil | GU646796 | [13] |
| GU646818_brbv119_03 | I | 2003 | Bovine/Cattle | José Bonifácio, SP | Brazil | GU646818 | [13] |
| GU646828_brbv356_97 | I | 1997 | Bovine/Cattle | Guararapes, SP | Brazil | GU646828 | [13] |
| GU646833_brbv32_94 | T | 1994 | Bovine/Cattle | Araçatuba, SP | Brazil | GU646833 | [13] |
| GU646835_brdg70_93 | T | 1993 | Dog | Araçatuba, SP | Brazil | GU646835 | [13] |
| GU646841_brhr308_00 | I | 2000 | Horse | Barbosa, SP | Brazil | GU646841 | [13] |
| GU646842_bral5341 | I | 1999 | Artibeus lituratus | Birigui, SP | Brazil | GU646842 | [13] |
| GU646843_brmm4105 | I | 1998 | Molossus molossus | Mirandópolis, SP | Brazil | GU646843 | [13] |
| GU646844_brle4132 | I | 1988 | Lasiurus ega | Glicério, SP | Brazil | GU646844 | [13] |
| GU646845_brmr4114 | I | 1998 | Molossus molossus | Penápolis, SP | Brazil | GU646845 | [13] |
| GU646846_brmr4095 | I | 1998 | Molossus rufus | Araçatuba, SP | Brazil | GU646846 | [13] |
| GU646849_brdg5356 | I | 2000 | Dog | Ilha Solteira, SP | Brazil | GU646849 | [13] |
| GU646855_brct60_92 | T | 1992 | Cat | Andradina, SP | Brazil | GU646855 | [13] |
| GU646856_brmr298_07 | I | 2007 | Molossus rufus | Ilha Solteira, SP | Brazil | GU646856 | [13] |
| GU646857_brmn182_07 | II | 2007 | Myotis nigricans | Bilac, SP | Brazil | GU646857 | [13] |
| GU646858_brmr350_06 | I | 2006 | Molossus rufus | Andradina, SP | Brazil | GU646858 | [13] |
| GU646859_bral309_06 | I | 2006 | Artibeus lituratus | Ilha Solteira, SP | Brazil | GU646859 | [13] |
| GU646860_bral195_06 | I | 2006 | Artibeus lituratus | Araçatuba, SP | Brazil | GU646860 | [13] |
| GU646861_bref341_02 | II | 2002 | Epitesicus furinalis | Birigui, SP | Brazil | GU646861 | [13] |
| HM173087_brmn100_05 | II | 2005 | Myotis nigricans | Guararapes, SP | Brazil | HM173087 | [13] |
| HM173088_bral239_05 | I | 2005 | Artibeus lituratus | Birigui, SP | Brazil | HM173088 | [13] |
| HM854029_brdg354_95 | I | 1995 | Dog | Birigui, SP | Brazil | HM854029 | [13] |
| HM854030_brct100_04 | I | 2004 | Cat | Guararapes, SP | Brazil | HM854030 | [13] |
| HM854031_brmr178_05 | IV | 2005 | Molossus rufus | Andradina, SP | Brazil | HM854031 | [13] |
| HM854032_brmn391_05 | II | 2005 | Myotis nigricans | Ilha Solteira, SP | Brazil | HM854032 | [13] |
| HM854033_bral540_05 | I | 2005 | Artibeus lituratus | Araçatuba, SP | Brazil | HM854033 | [13] |
| HM014315_brmm1994_08 | V | 2008 | Molossus molossus | São Paulo, SP | Brazil | HM014315 | [14] |
| HM014316_brmng2449_05 | III | 2005 | Molossops neglectus | São Paulo, SP | Brazil | HM014316 | [14] |
| HM014317_brmr6464_05 | II | 2005 | Myotis riparius | São Paulo, SP | Brazil | HM014317 | [14] |
| HQ666824_bral346_99 | I | 1999 | Artibeus lituratus | São José do Rio Preto, SP | Brazil | HQ666824 | This work |
| HQ666825_brma777_00 | IV | 2000 | Cynomops abrasus | Ipiguá, SP | Brazil | HQ666825 | This work |
| HQ666826_brnl249_01 | III | 2001 | Nyctinomops macrotis | São José do Rio Preto, SP | Brazil | HQ666826 | This work |
| HQ666827_brnl250_01 | III | 2001 | Nyctinomops laticaudatus | São José do Rio Preto, SP | Brazil | HQ666827 | This work |
| HQ666828_bref636_01 | II | 2001 | Neoepitesicus furinalis | Olímpia, SP | Brazil | HQ666828 | This work |
| HQ666829_bral808_01 | I | 2001 | Artibeus lituratus | São José do Rio Preto, SP | Brazil | HQ666829 | This work |
| HQ666830_bresp1019_01 | II | 2001 | Neoepitesicus sp. | Cardoso, SP | Brazil | HQ666830 | This work |
| HQ666831_bref1070_01 | II | 2001 | Neoepitesicus furinalis | São José do Rio Preto, SP | Brazil | HQ666831 | This work |
| HQ666832_bref62_02 | II | 2002 | Neoepitesicus furinalis | Catanduva, SP | Brazil | HQ666832 | This work |
| HQ666833_brmm109_02 | IV | 2002 | Molossus molossus | Ilha Solteira, SP | Brazil | HQ666833 | This work |
| HQ666834_brmn835_02 | II | 2002 | Myotis nigricans | Cajobi, SP | Brazil | HQ666834 | This work |
| HQ666835_bral992_02 | I | 2002 | Artibeus lituratus | Dracena, SP | Brazil | HQ666835 | This work |
| HQ666836_brmr1021_02 | I | 2002 | Molossus fluminensis | Presidente Venceslau, SP | Brazil | HQ666836 | This work |
| HQ666837_bref1141_02 | II | 2002 | Neoepitesicus furinalis | Santo Anastácio, SP | Brazil | HQ666837 | This work |
| HQ666838_brbat1256_02 | I | 2002 | Non- hematophagous bat | Martinópolis, SP | Brazil | HQ666838 | This work |
| HQ666839_bral1371_02 | I | 2002 | Artibeus lituratus | Presidente Venceslau, SP | Brazil | HQ666839 | This work |
| HQ666840_bral1535_02 | I | 2002 | Artibeus lituratus | Taciba, SP | Brazil | HQ666840 | This work |
| HQ666841_brmm1539_02 | I | 2002 | Molossus molossus | Presidente Prudente, SP | Brazil | HQ666841 | This work |
| HQ666842_brle1782_02 | III | 2002 | Lasiurus ega | Presidente Prudente, SP | Brazil | HQ666842 | This work |
| HQ666843_braj349_03 | I | 2003 | Artibeus planirostris | Santa Fé do Sul, SP | Brazil | HQ666843 | This work |
| HQ666844_brbat350_03 | I | 2003 | Non-hematophagous bat | Catanduva, SP | Brazil | HQ666844 | This work |
| HQ666845_bral791_03 | I | 2003 | Artibeus lituratus | Presidente Prudente, SP | Brazil | HQ666845 | This work |
| HQ666846_brmn168_04 | II | 2004 | Myotis nigricans | Campinas, SP | Brazil | HQ666846 | This work |
| HQ666847_brnl184_04 | III | 2004 | Nyctinomops laticaudatus | São José do Rio Preto, SP | Brazil | HQ666847 | This work |
| HQ666848_bral550_04 | I | 2004 | Artibeus lituratus | Caçapava, SP | Brazil | HQ666848 | This work |
| HQ666849_breg329_05 | I | 2005 | Eumops glaucinus | Araçatuba, SP | Brazil | HQ666849 | This work |
| HQ666850_bred43_09 | II | 2009 | Neoepitesicus diminutus | Pereira Barreto, SP | Brazil | HQ666850 | This work |
| HQ666851_bred84_09 | II | 2009 | Neoepitesicus diminutus | Araçatuba, SP | Brazil | HQ666851 | This work |
| HQ666852_brmn149_09 | II | 2009 | Myotis nigricans | Coroados, SP | Brazil | HQ666852 | This work |
| HQ666853_bral181_09 | I | 2009 | Artibeus lituratus | Penápolis, SP | Brazil | HQ666853 | This work |
| HQ666854_bref199_09 | II | 2009 | Neoeptesicus furinalis | Dracena, SP | Brazil | HQ666854 | This work |
| HQ666855_bref224_09 | I | 2009 | Neoeptesicus furinalis | Parapuã, SP | Brazil | HQ666855 | This work |
| HQ666856_bral325_09 | I | 2009 | Artibeus lituratus | Birigui, SP | Brazil | HQ666856 | This work |
| HQ666857_bral374_09 | I | 2009 | Artibeus lituratus | Guararapes, SP | Brazil | HQ666857 | This work |
| HQ666858_brmr389_09 | I | 2009 | Molossus fluminensis | Guararapes, SP | Brazil | HQ666858 | This work |
| HQ666859_brmn433_09 | II | 2009 | Myotis nigricans | Penápolis, SP | Brazil | HQ666859 | This work |
| HQ666860_bref589_09 | II | 2009 | Neoeptesicus furinalis | Penápolis, SP | Brazil | HQ666860 | This work |
| HQ666861_brbv672_09 | I | 2009 | Bovine | Narandiba, SP | Brazil | HQ666861 | This work |
| HQ666862_brmr17_10 | II | 2010 | Molossus fluminensis | Penápolis, SP | Brazil | HQ666862 | This work |
| HQ666863_breq28_10 | I | 2010 | Equine | Taciba, SP | Brazil | HQ666863 | This work |
| HQ666864_bred43_10 | I | 2010 | Neoeptesicus diminutus | Osvaldo Cruz, SP | Brazil | HQ666864 | This work |
| HQ666865_bref60_10 | II | 2010 | Neoeptesicus furinalis | Birigui, SP | Brazil | HQ666865 | This work |
| HQ666866_bref76_10 | II | 2010 | Neoeptesicus furinalis | Penápolis, SP | Brazil | HQ666866 | This work |
| HQ666867_brmm169_10 | V | 2010 | Molossus molossus | Araçatuba, SP | Brazil | HQ666867 | This work |
| HQ666868_brct171_10 | I | 2010 | Feline | Araçatuba, SP | Brazil | HQ666868 | This work |
| HQ666869_brmr177_10 | V | 2010 | Molossus fluminensis | Birigui, SP | Brazil | HQ666869 | This work |
| HQ666870_brlbl198_10 | V | 2010 | Lasiurus blossevillii | Teodoro Sampaio, SP | Brazil | HQ666870 | This work |
| HQ666871_bref299_10 | II | 2010 | Neoeptesicus furinalis | Penápolis, SP | Brazil | HQ666871 | This work |
| HQ666872_brmr300_10 | I | 2010 | Molossus fluminensis | Penápolis, SP | Brazil | HQ666872 | This work |
| JF916647_BRLB4096_1995 | V | 1995 | Lasiurus blossevillii | Unknown | Brazil | BRLB4096 | Unpublished, Favoretto |
| JF916650_brmyaSP4115_1998 | II | 1998 | Myotis albecens | Unknown | Brazil | JF916650 | Unpublished, Favoretto |
| JF916652_bralusp041_05 | I | 2005 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916652 | [26] |
| JF916655_brlbusp040_07 | V | 2007 | Lasiurus blossevillii | Presidente Prudente, SP | Brazil | JF916655 | [26] |
| JF916656_bralusp042_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916656 | [26] |
| JF916657_brmmusp043_07 | I | 2007 | Molossus molossus | Presidente Prudente, SP | Brazil | JF916657 | [26] |
| JF916659_bralusp047_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916659 | [26] |
| JF916661_bralusp049_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916661 | [26] |
| JF916662_bralusp050_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916662 | [26] |
| JF916663_bralusp052_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916663 | [26] |
| JF916664_bralusp054_07 | VII | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916664 | [26] |
| JF916667_brmnusp058_07 | II | 2007 | Myotis nigricans | Presidente Prudente, SP | Brazil | JF916667 | [26] |
| JF916668_brmnusp061_07 | II | 2007 | Myotis nigricans | Presidente Prudente, SP | Brazil | JF916668 | [26] |
| JF916669_bralusp062_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916669 | [26] |
| JF916671_brefusp064_07 | II | 2007 | Eptesicus furinalis | Presidente Prudente, SP | Brazil | JF916671 | [26] |
| JF916672_bralusp069_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916672 | [26] |
| JF916673_bralusp071_07 | I | 2007 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916673 | [26] |
| JF916674_bralusp001_08 | I | 2008 | Artibeus lituratus | Presidente Prudente, SP | Brazil | JF916674 | [26] |
| JF916678_brefusp008_09 | II | 2009 | Eptesicus furinalis | Presidente Prudente, SP | Brazil | JF916678 | [26] |
| KM594026_IP_512_09 | II | 2009 | Eptesicus furinalis | Ribeirão Preto, SP | Brazil | KM594026 | [42] |
| KM594027_IP_230_10 | II | 2010 | Eptesicus furinalis | Valinhos, SP | Brazil | KM594027 | [42] |
| KM594028_IP_346_10 | II | 2010 | Eptesicus furinalis | Tambaú, SP | Brazil | KM594028 | [42] |
| KM594029_IP_3208_06 | III | 2006 | Eptesicus furinalis | Vinhedo, SP | Brazil | KM594029 | [42] |
| KM594030_IP_1400_10 | II | 2010 | Myotis nigricans | Campinas, SP | Brazil | KM594030 | [42] |
| KM594031_IP_163_10 | II | 2010 | Myotis nigricans | Caieiras, SP | Brazil | KM594031 | [42] |
| KM594032_IP_497_10 | II | 2010 | Myotis nigricans | Campinas, SP | Brazil | KM594032 | [42] |
| KM594034_IP_350_10 | III | 2010 | Nyctinomops laticaudatus | Conchal, SP | Brazil | KM594034 | [42] |
| KM594035_IP_412_10 | III | 2010 | Nyctinomops laticaudatus | Barretos, SP | Brazil | KM594035 | [42] |
| KM594036_IP_542_10 | III | 2010 | Nyctinomops laticaudatus | Ribeirão Preto, SP | Brazil | KM594036 | [42] |
| KM594037_IP_3176_09 | VII | 2009 | Tadarida brasiliensis | Santo André, SP | Brazil | KM594037 | [42] |
| KM594038_IP_1586_10 | VII | 2010 | Tadarida brasiliensis | São Bernardo do Campo, SP | Brazil | KM594038 | [42] |
| MG458314_RV1789 | I | 1997 | Cow | Unknown | British Est Indies | MG458314 | [54] |
| PQ671596_RS61 | VII | 2022 | Tadarida brasiliensis | Rio Grande do Sul | Brazil | PQ671596 | unpublished |
Legend: samples in red are from this study; samples in blue are from our group’s previous study [13].
Table A2.
Estimates of evolutionary divergence over sequence pairs between groups.
| Gp_1 | Gp_3 | Gp_2 | Gp_4 | Gp_5 | Gp_7 | Gp_6 | |
|---|---|---|---|---|---|---|---|
| Gp_1 | |||||||
| Gp_3 | 0.140 | ||||||
| Gp_2 | 0.128 | 0.126 | |||||
| Gp_4 | 0.139 | 0.103 | 0.138 | ||||
| Gp_5 | 0.152 | 0.134 | 0.160 | 0.132 | |||
| Gp_7 | 0.087 | 0.117 | 0.118 | 0.111 | 0.136 | ||
| Gp_6 | 0.181 | 0.148 | 0.179 | 0.152 | 0.158 | 0.167 | |
| outgroup | 0.178 | 0.180 | 0.188 | 0.162 | 0.172 | 0.182 | 0.213 |
Gp 1 means group I Related to hematophagous bats-AgV3, Gp 2 means group II Related to insectivorous bats-NC1, Gp 3 means group III Related to insectivorous bats-NC1, Gp 4 means group IV Related to insectivorous bats-NC2, Gp 5 means group V Related to insectivorous bats-NC2, Gp 6 means group VI Related to marmosets, Gp 7 means group VII Related to Tadarida brasiliensis-AgV4 and outgroup represents the Terrestrial cycle of transmission-root of phylogenetic tree.
Table A3.
Estimates of average evolutionary divergence over sequence pairs within groups.
| Gp 1 | 0.0253 |
| Gp 3 | 0.0467 |
| Gp 2 | 0.0674 |
| Gp 4 | 0.0185 |
| Gp 5 | 0.0566 |
| Gp 7 | 0.0029 |
| Gp 6 | 0.0051 |
| outgroup | 0.0051 |
Gp 1 means group I Related to hematophagous bats-AgV3, Gp 2 means group II Related to insectivorous bats-NC1, Gp 3 means group III Related to insectivorous bats-NC1, Gp 4 means group IV Related to insectivorous bats-NC2, Gp 5 means group V Related to insectivorous bats-NC2, Gp 6 means group VI Related to marmosets, Gp 7 means group VII Related to Tadarida brasiliensis-AgV4 and outgroup represents the Terrestrial cycle of transmission-root of phylogenetic tree.
Author Contributions
Conceptualization: L.H.Q., S.R.F. and A.C.A.C.; methodology, L.H.Q., A.C.A.C., M.C.L., E.M.S.C., A.A., C.d.C., W.A.P., E.C.S., M.S.L., S.V.I., D.B.A. and S.R.F.; software, A.C.A.C., M.P.C. and L.G.B.G.; validation, A.C.A.C., M.C.L., E.C.S., M.S.L., S.V.I. and D.B.A.; formal analysis, A.C.A.C., L.H.Q., M.P.C. and L.G.B.G.; investigation, A.C.A.C. and L.H.Q.; resources, S.R.F., E.L.D. and L.G.B.G.; data curation, A.C.A.C. and L.H.Q.; writing—original draft preparation, A.C.A.C. and L.H.Q.; writing—review and editing, A.C.A.C., L.H.Q., S.R.F., D.B.A., M.P.C. and L.G.B.G.; visualization, A.C.A.C., L.H.Q. and S.R.F.; supervision, L.H.Q., S.R.F. and E.L.D.; project administration, L.H.Q.; funding acquisition, L.H.Q., S.R.F. and E.L.D. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
The study was conducted in accordance with the ethics principles of the Brazilian College of Animal Experimentation (COBEA) and approved by the Animal Experimentation Ethics Committee of the School of Dentistry and Veterinary Medicine of Araçatuba, UNESP (Process No. 00858-2012 and 00902-2016).
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author(s).
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP)—grant number 2008/00976-0 and 2007/01843-0 and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)—grant number 578281/2008-2. Data analysis and publication were supported by LGBG FAPESP 2022-13054-0. Scholarships: A.C.A.C.—CNPq 102474/2022-2 and FAPESP 2024-08821-8; M.C.L.—FAPESP (04/12793-6); E.C.S.—FAPESP (2008/08423-0); M.S.L.—CNPq technical assistance (grant number 578281/2008-2), S.V.I.—FAPESP (2008/00976-0). M.P.C. was supported by the Fund to Support Teaching, Research and Extension (FAEPEX/UNICAMP) (grant #2502/24). L.G.B.G. is supported by Young Research Project FAPESP 2022/13054-0. A.C.A.C. is currently supported by Young Research Project FAPESP 2024/10801-5.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.de Almeida M.F., Queiroz L.H. História Da Raiva No Brasil. 1st ed. Editora UNESP Digital; São Paulo City, Brazil: 2023. [Google Scholar]
- 2.Favoretto S.R., Araújo D.B., Sacramento D.V., Martorelli L.F.A. Raiva. In: Trabulsi A., Altherthum F., editors. Microbiologia—Trabulsi-Alterthum. Atheneu; São Paulo City, Brazil: 2024. pp. 945–956. [Google Scholar]
- 3.Giménez A.L., Zabalza M.J., Novaro L.P., Centurion G.A., Barrios-Benito M.Y., Moncá I., Chaar Letourneau F., Casanovas R., Russo S.E. Diversity of Rabies Virus Variants in Insectivorous Bats (Chiroptera: Vespertilionidae and Molossidae): An Epidemiological Study in Central Argentine Patagonia. Viruses. 2025;17:788. doi: 10.3390/v17060788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.ICTV ICTV-International Committee on Taxonomy of Viruses. [(accessed on 3 June 2025)]. Available online: https://ictv.global/taxonomy/taxondetails?taxnode_id=202401733&taxon_name=Lyssavirus rabies.
- 5.Rupprecht C.E., Mshelbwala P.P., Reeves R.G., Kuzmin I.V. Rabies in a Postpandemic World: Resilient Reservoirs, Redoubtable Riposte, Recurrent Roadblocks, and Resolute Recidivism. Anim. Dis. 2023;3:15. doi: 10.1186/s44149-023-00078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Favoretto S.R., De Mattos C.C., Morais N.B., Araújo F.A.A., Mattos C.C. De Rabies in Marmosets (Callithrix Jacchus) Ceará, Brazil. Emerg. Infect. Dis. 2001;7:1062–1065. doi: 10.3201/eid0706.010630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Favoretto S.R., De Mattos C.C., De Mattos C.A., Campos A.C.A., Sacramento D.R.V., Durigon E.L. The Emergence of Wildlife Species as a Source of Human Rabies Infection in Brazil. Epidemiol. Infect. 2013;141:1552–1561. doi: 10.1017/S0950268813000198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaz A.-M., Papo S., Rodriguez A., Smith J.S. Antigenic Analysis of Rabies-virus Isolates from Latin America and the Caribbean. J. Vet. Med. Ser. B. 1994;41:153–160. doi: 10.1111/j.1439-0450.1994.tb00219.x. [DOI] [PubMed] [Google Scholar]
- 9.Velasco-Villa A., Gómez-Sierra M., Hernández-Rodríguez G., Juárez-Islas V., Meléndez-Félix A., Vargas-Pino F., Velázquez-Monroy O., Flisser A. Antigenic Diversity and Distribution of Rabies Virus in Mexico. J. Clin. Microbiol. 2002;40:951–958. doi: 10.1128/JCM.40.3.951-958.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Páez A., Nũñez C., García C., Bóshell J. Molecular Epidemiology of Rabies Epizootics in Colombia: Evidence for Human and Dog Rabies Associated with Bats. J. Gen. Virol. 2003;84:795–802. doi: 10.1099/vir.0.18899-0. [DOI] [PubMed] [Google Scholar]
- 11.Favoretto S.R., Carrieri M.L., Cunha E.M.S., Aguiar E.A.C., Silva L.H.Q., Sodre M.M., Souza M.C.A.M., Kotait I. Antigenic Typing of Brazilian Rabies Virus Samples Isolated from Animals and Humans, 1989–2000. Rev. Inst. Med. Trop. Sao Paulo. 2002;44:91–95. doi: 10.1590/S0036-46652002000200007. [DOI] [PubMed] [Google Scholar]
- 12.Castilho J.G., Canello F.M., Scheffer K.C., Achkar S.M., Carrieri M.L., Kotait I. Antigenic and Genetic Characterization of the First Rabies Virus Isolated from the Bat Eumops Perotis in Brazil. Rev. Inst. Med. Trop. Sao Paulo. 2008;50:95–99. doi: 10.1590/S0036-46652008000200006. [DOI] [PubMed] [Google Scholar]
- 13.Queiroz L.H., Favoretto S.R., Cunha E.M.S., Campos A.C.A., Lopes M.C., de Carvalho C., Iamamoto K., Araújo D.B., Venditti L.L.R., Ribeiro E.S., et al. Rabies in Southeast Brazil: A Change in the Epidemiological Pattern. Arch. Virol. 2012;157:93–105. doi: 10.1007/s00705-011-1146-1. [DOI] [PubMed] [Google Scholar]
- 14.da Rosa A.R., de Arruda Geraldes Kataoka A.P., Favoretto S.R., Sodré M.M., Netto J.T., de Almeida Campos A.C., Durigon E.L., Martorelli L.F.A. First Report of Rabies Infection in Bats, Molossus Molossus, Molossops Neglectus and Myotis Riparius in the City of São Paulo, State of São Paulo, Southeastern Brazil. Rev. Soc. Bras. Med. Trop. 2011;44:146–149. doi: 10.1590/S0037-86822011005000018. [DOI] [PubMed] [Google Scholar]
- 15.Menozzi B.D., de Novaes Oliveira R., Paiz L.M., Richini-Pereira V.B., Langoni H. Antigenic and Genotypic Characterization of Rabies Virus Isolated from Bats (Mammalia: Chiroptera) from Municipalities in São Paulo State, Southeastern Brazil. Arch. Virol. 2017;162:1201–1209. doi: 10.1007/s00705-017-3220-9. [DOI] [PubMed] [Google Scholar]
- 16.Badrane H., Tordo N. Host Switching in Lyssavirus History from the Chiroptera to the Carnivora Orders. J. Virol. 2001;75:8096–8104. doi: 10.1128/JVI.75.17.8096-8104.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kissi B., Tordo N., Bourhy H. Genetic Polymorphism in the Rabies Virus Nucleoprotein Gene. Virology. 1995;209:526–537. doi: 10.1006/viro.1995.1285. [DOI] [PubMed] [Google Scholar]
- 18.Nadin-Davis S.A. Polymerase Chain Reaction Protocols for Rabies Virus Discrimination. J. Virol. Methods. 1998;75:1–8. doi: 10.1016/S0166-0934(98)00106-2. [DOI] [PubMed] [Google Scholar]
- 19.Ito M., Arai Y.T., Itou T., Sakai T., Ito F.H., Takasaki T., Kurane I. Genetic Characterization and Geographic Distribution of Rabies Virus Isolates in Brazil: Identification of Two Reservoirs, Dogs and Vampire Bats. Virology. 2001;284:214–222. doi: 10.1006/viro.2000.0916. [DOI] [PubMed] [Google Scholar]
- 20.Favoretto S.R., de Mattos C.C., De Morais N.B., Carrieri M.L., Rolim B.N., Silva L.M., Rupprecht C.E., Durigon E.L., de Mattos C.A. Maintained by Dogs in Humans and Terrestrial Wildlife. Ceará State, Brazil. Emerg. Infect. Dis. 2006;12:1978–1981. doi: 10.3201/eid1212.060429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carnieli P., Fahl W.d.O., Castilho J.G., Oliveira R.d.N., Macedo C.I., Durymanova E., Jorge R.S.P., Morato R.G., Spíndola R.O., Machado L.M., et al. Characterization of Rabies Virus Isolated from Canids and Identification of the Main Wild Canid Host in Northeastern Brazil. Virus Res. 2008;131:33–46. doi: 10.1016/j.virusres.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 22.Favoretto S.R., de Mattos C.C., Mattos C.A., Carrieri M.L., Durigon E.L. Antigenic and Genetic Characterization of Rabies Virus Samples Isolated from Human in Brazil––Preliminary Results; Proceedings of the XIII International Meeting on Research Advances and Rabies Control in the Américas–RITA; Oaxaca Municipality, Mexico. 3–8 November 2002; pp. 25–26. [Google Scholar]
- 23.Martorelli L.F.A. Diagnóstico Laboratorial e Diversidade Genética Do Vírus Rábico Isolado No Estado de São Paulo, 1989–2000. Universidade de São Paulo-USP; São Paulo, Brazil: 2004. [Google Scholar]
- 24.Kobayashi Y., Sato G., Shoji Y., Sato T., Itou T., Cunha E.M.S., Samara S.I., Carvalho A.A.B., Nociti D.P., Ito F.H., et al. Molecular Epidemiological Analysis of Bat Rabies Viruses in Brazil. J. Vet. Med. Sci. 2005;67:647–652. doi: 10.1292/jvms.67.647. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira R.d.N., de Souza S.P., Lobo R.S.V., Castilho J.G., Macedo C.I., Carnieli P., Fahl W.O., Achkar S.M., Scheffer K.C., Kotait I., et al. Rabies Virus in Insectivorous Bats: Implications of the Diversity of the Nucleoprotein and Glycoprotein Genes for Molecular Epidemiology. Virology. 2010;405:352–360. doi: 10.1016/j.virol.2010.05.030. [DOI] [PubMed] [Google Scholar]
- 26.Albas A., Campos A.C.D.A., Araujo D.B., Rodrigues C.S., Sodré M.M., Durigon E.L., Favoretto S.R. Molecular Characterization of Rabies Virus Isolated from Non-Haematophagous Bats in Brazil. Rev. Soc. Bras. Med. Trop. 2011;44:678–683. doi: 10.1590/S0037-86822011000600006. [DOI] [PubMed] [Google Scholar]
- 27.Dean D.J., Abelseth M.K., Atanasiu P. The Fluorescent Antibody Test. In: Meslin F., Kaplan M.M., Koprowski H., editors. Laboratory Techniques in Rabies. WHO—World Health Organization—Licence; Geneva, Switzerland: 1996. p. 47. CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 28.Koprowski H. The Mouse Inoculation Test. In: Meslin F., Kaplan M.M., Koprowski H., editors. Laboratory Techniques in Rabies. WHO—World Health Organization—Licence; Geneva, Switzerland: 1996. p. 476. CC BY-NC-SA 3.0 IGO. [Google Scholar]
- 29.Cláudio V.C., Novaes R.L.M., Gardner A.L., Nogueira M.R., Wilson D.E., Maldonado J.E., Oliveira J.A., Moratelli R. Taxonomic Re-Evaluation of New World Eptesicus and Histiotus (Chiroptera: Vespertilionidae), with the Description of a New Genus. Zoologia. 2023;40:e22029. doi: 10.1590/s1984-4689.v40.e22029. [DOI] [Google Scholar]
- 30.Yi X., Latch E.K. Systematics of the New World Bats Eptesicus and Histiotus Suggest Trans-Marine Dispersal Followed by Neotropical Cryptic Diversification. Mol. Phylogenet. Evol. 2022;175:107582. doi: 10.1016/j.ympev.2022.107582. [DOI] [PubMed] [Google Scholar]
- 31.Chambi Velasquez M.A., Pavé R., Argoitia M.A., Schierloh P., Piccirilli M.G., Colombo V.C., Beltrán F.J., Cisterna D.M., Caraballo D.A. Revisiting Molossus (Mammalia: Chiroptera: Molossidae) Diversity: Exploring Southern Limits and Revealing a Novel Species in Argentina. Vertebr. Zool. 2024;74:397–416. doi: 10.3897/vz.74.e122822. [DOI] [Google Scholar]
- 32.Smith J.S., Orciari L.A., Yager P. Molecular Epidemiology of Rabies in the US. Semin Virol. 1995;6:387–400. doi: 10.1016/S1044-5773(05)80016-2. [DOI] [Google Scholar]
- 33.Campos A.C.d.A., Melo F.L., Romano C.M., Araujo D.B., Cunha E.M.S., Sacramento D.R.V., de Andrade Zanotto P.M., Durigon E.L., Favoretto S.R. One-Step Protocol for Amplification of near Full-Length CDNA of the Rabies Virus Genome. J. Virol. Methods. 2011;174:1–6. doi: 10.1016/j.jviromet.2011.03.030. [DOI] [PubMed] [Google Scholar]
- 34.Hadfield J., Megill C., Bell S.M., Huddleston J., Potter B., Callender C., Sagulenko P., Bedford T., Neher R.A. Nextstrain: Real-Time Tracking of Pathogen Evolution. Bioinformatics. 2018;34:4121–4123. doi: 10.1093/bioinformatics/bty407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagulenko P., Puller V., Neher R.A. TreeTime: Maximum-Likelihood Phylodynamic Analysis. Virus Evol. 2018;4:vex042. doi: 10.1093/ve/vex042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albas A., de Souza E.A.N., Lourenço R.A., Favoretto S.R., Sodré M.M. Antigen Profile of Rabies Virus Isolated from Different Species of Non-Hematophagous Bats in the Region of Presidente Prudente, State of São Paulo. Rev. Soc. Bras. Med. Trop. 2009;42:15–17. doi: 10.1590/S0037-86822009000100004. [DOI] [PubMed] [Google Scholar]
- 37.Cunha E.M.S., da Silva L.H.Q., Lara M.d.C.C.S.H., Nassar A.F.C., Albas A., Sodré M.M., Pedro W.A. Bat Rabies in the North-Northwestern Regions of the State of São Paulo, Brazil: 1997–2002. Rev. Saude Publica. 2006;40:1082–1086. doi: 10.1590/S0034-89102006000700017. [DOI] [PubMed] [Google Scholar]
- 38.Queiroz L.H., de Carvalho C., Buso D.S., Ferrari C.I.d.L., Pedro W.A. Epidemiological Profile of Rabies in the Northwestern Region of São Paulo State, from 1993 to 2007. Rev. Soc. Bras. Med. Trop. 2009;42:9–14. doi: 10.1590/S0037-86822009000100003. [DOI] [PubMed] [Google Scholar]
- 39.Garcia A.B., de Carvalho C., Casagrande D., Picinato M.A.d.C., Pedro W.A., Marinho M., Queiroz L.H. Rabies in Bats (Chiroptera, Mammalia) in Brazil: Prevalence and Potential Risk Factors Based on Twenty Years of Research in the Northwestern Region of São Paulo, Brazil. Vet. Sci. 2023;10:34. doi: 10.3390/vetsci10010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Souza D.N., Oliveira R.N., Asprino P.F., Bettoni F., Macedo C.I., Achkar S.M., Fahl W.O., Brandão P.E., Castilho J.G. Evolution and Divergence of the Genetic Lineage Desmodus rotundus/Artibeus lituratus of Rabies Virus in São Paulo State. Arch. Virol. 2023;168:266. doi: 10.1007/s00705-023-05864-w. [DOI] [PubMed] [Google Scholar]
- 41.de Almeida M.F., Favoretto S.R., Martorelli L.F.A., Trezza-Netto J., Campos A.C.d.A., Ozahata C.H., Sodré M.M., Kataoka A.P.A.G., Veiga Sacramento D.R., Durigon E.L. CHARACTERIZATION OF RABIES VIRUS ISOLATED FROM A COLONY OF Eptesicus Furinalis BATS IN BRAZIL. Rev. Inst. Med. Trop. Sao Paulo. 2011;53:31–37. doi: 10.1590/S0036-46652011000100006. [DOI] [PubMed] [Google Scholar]
- 42.Oliveira R.N., Freire C.C., Iamarino A., Zanotto P.M., Pessoa R., Sanabani S.S., de Souza S.P., Castilho J.G., Batista H.B.C.R., Carnieli P., et al. Rabies Virus Diversification in Aerial and Terrestrial Mammals. Genet. Mol. Biol. 2020;43:e20190370. doi: 10.1590/1678-4685-gmb-2019-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guarino H., Castilho J.G., Souto J., Oliveira R.d.N., Carrieri M.L., Kotait I. Antigenic and Genetic Characterization of Rabies Virus Isolates from Uruguay. Virus Res. 2013;173:415–420. doi: 10.1016/j.virusres.2012.12.013. [DOI] [PubMed] [Google Scholar]
- 44.Cisterna D., Bonaventura R., Caillou S., Pozo O., Andreau M.L., Dalla Fontana L., Echegoyen C., De Mattos C., De Mattos C., Russo S., et al. Antigenic and Molecular Characterization of Rabies Virus in Argentina. Virus Res. 2005;109:139–147. doi: 10.1016/j.virusres.2004.10.013. [DOI] [PubMed] [Google Scholar]
- 45.Castilho J.G., Carnieli P., Oliveira R.N., Fahl W.O., Cavalcante R., Santana A.A., Rosa W.L.G.A., Carrieri M.L., Kotait I. A Comparative Study of Rabies Virus Isolates from Hematophagous Bats in Brazil. J. Wildl. Dis. 2010;46:1335–1339. doi: 10.7589/0090-3558-46.4.1335. [DOI] [PubMed] [Google Scholar]
- 46.Smith J.S., Orciari L.A., Yager P.A., Seidel H.D., Warner C.K. Epidemiologic and Historical Relationships among 87 Rabies Virus Isolates as Determined by Limited Sequence Analysis. J. Infect. Dis. 1992;166:296–307. doi: 10.1093/infdis/166.2.296. [DOI] [PubMed] [Google Scholar]
- 47.de Sousa L.L.F., de Souza T.L., Tibo L.H.S., Moura F.B.P., Junior F.A.S., de Oliveira-Filho E.F., Ludwig-Begall L.F., Cabral-Miranda G., Andreata-Santos R., Janini L.M.R., et al. Rabies Virus Variants from Bats Closely Related to Variants Found in Marmosets (Callithrix Jacchus), a Neglected Source of Human Rabies Infection in Brazil. J. Med. Virol. 2023;95:e29046. doi: 10.1002/jmv.29046. [DOI] [PubMed] [Google Scholar]
- 48.Kobayashi Y., Okuda H., Nakamura K., Sato G., Itou T., Carvalho A.A.B., Silva M.V., Mota C.S., Ito F.H., Sakai T. Genetic Analysis of Phosphoprotein and Matrix Protein of Rabies Viruses Isolated in Brazil. J. Vet. Med. Sci. 2007;69:1145–1154. doi: 10.1292/jvms.69.1145. [DOI] [PubMed] [Google Scholar]
- 49.Rohde R.E., Mayes B.C., Smith J.S., Neill S.U. Bat Rabies, Texas, 1996–2000. Emerg. Infect. Dis. 2004;10:948–952. doi: 10.3201/eid1005.030719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Favi M., de Mattos C.A., Yung V., Chala E., López L.R., de Mattos C.C. First Case of Human Rabies in Chile Caused by an Insectivorous Bat Virus Variant. Emerg. Infect. Dis. 2002;8:79–81. doi: 10.3201/eid0801.010108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nadin-Davis S.A., Loza-Rubio E. The Molecular Epidemiology of Rabies Associated with Chiropteran Hosts in Mexico. Virus Res. 2006;117:215–226. doi: 10.1016/j.virusres.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 52.Castilho J.G., Carnieli P., Durymanova E.A., Fahl W.d.O., Oliveira R.d.N., Macedo C.I., da Rosa E.S.T., Mantilla A., Carrieri M.L., Kotait I. Human Rabies Transmitted by Vampire Bats: Antigenic and Genetic Characterization of Rabies Virus Isolates from the Amazon Region (Brazil and Ecuador) Virus Res. 2010;153:100–105. doi: 10.1016/j.virusres.2010.07.012. [DOI] [PubMed] [Google Scholar]
- 53.Delmas O., Holmes E.C., Talbi C., Larrous F., Dacheux L., Bouchier C., Bourhy H. Genomic Diversity and Evolution of the Lyssaviruses. PLoS ONE. 2008;3:e2057. doi: 10.1371/journal.pone.0002057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fischer S., Freuling C.M., Müller T., Pfaff F., Bodenhofer U., Höper D., Fischer M., Marston D.A., Fooks A.R., Mettenleiter T.C., et al. Defining Objective Clusters for Rabies Virus Sequences Using Affinity Propagation Clustering. PLoS Negl. Trop. Dis. 2018;12:e0006182. doi: 10.1371/journal.pntd.0006182. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the article. Further inquiries can be directed to the corresponding author(s).