Abstract
Complex microbial communities remain poorly characterized despite their ubiquity and importance to human and animal health, agriculture, and industry. Attempts to describe microbial communities by either traditional microbiological methods or molecular methods have been limited in both scale and precision. The availability of genomics technologies offers an unprecedented opportunity to conduct more comprehensive characterizations of microbial communities. Here we describe the application of an established molecular diagnostic method based on the chaperonin-60 sequence, in combination with high-throughput sequencing, to the profiling of a microbial community: the pig intestinal microbial community. Four libraries of cloned cpn60 sequences were generated by two genomic DNA extraction procedures in combination with two PCR protocols. A total of 1,125 cloned cpn60 sequences from the four libraries were sequenced. Among the 1,125 cloned cpn60 sequences, we identified 398 different nucleotide sequences encoding 280 unique peptide sequences. Pairwise comparisons of the 398 unique nucleotide sequences revealed a high degree of sequence diversity within the library. Identification of the likely taxonomic origins of cloned sequences ranged from imprecise, with clones assigned to a taxonomic subclass, to precise, for cloned sequences with 100% DNA sequence identity with a species in our reference database. The compositions of the four libraries were compared and differences related to library construction parameters were observed. Our results indicate that this method is an alternative to 16S rRNA sequence-based studies which can be scaled up for the purpose of performing a potentially comprehensive assessment of a given microbial community or for comparative studies.
The biosphere is dominated by microbial life, and microbes, both in the environment and as commensals of other organisms, exist in complex communities (20, 31). The earliest efforts to examine microbial diversity were direct microscopic observations, such as Antonie van Leeuwenhoek's observations of “very many small Animals” in the human oral cavity (47). Subsequently, microbiologists developed the culture-based techniques which, combined with differentiation of isolates based on numerous physiological and biochemical tests, became the standard method for investigating microbial community composition. A serious limitation of these methods is what has been referred to as the great plate count anomaly (41). That is, only a small fraction of microorganisms present in a population can be cultured in the laboratory, as little as 0.001 to 15%, depending on the community (2).
The development of recombinant DNA methods has led to a proliferation in small-scale studies of complex microbial communities, such as those associated with termite guts (33), rice paddy soil (3), 120-million-year-old amber (17), Antarctic lake ice (16), and leaves of a seagrass in the northern Gulf of Eilat (50). Molecular methods for microbial community analysis (reviewed in references 36, 45, and 48) include denaturing gradient gel electrophoresis, temperature gradient gel electrophoresis, and restriction fragment length polymorphism analysis. While these methods provide rapid comparative analyses of populations and generate population “fingerprints,” they do not identify individual organisms within populations.
Methods that do identify individual members of microbial communities are based on PCR and direct sequencing or cloning and sequencing of specific targets within microbial genomes. By far the most frequently used target is the 16S rRNA gene (30, 32). Our molecular phylogenetic view of the microbial world is dominated by 16S rRNA sequence relationships, and the wealth of sequence information accumulated for 16S rRNA genes from thousands of organisms and stored in the Ribosomal Database Project (25) has become a standard tool for studying microbial communities. Libraries of total genomic DNA extracted from a community of interest can be screened for rRNA genes, or libraries of PCR amplified rRNA genes or gene segments can be generated and sequenced.
Other gene targets used in microbial identification and for elucidation of phylogenetic relationships include rpoB (7), gyrB (22), pmoA (5), and cpn60, which encodes the 60-kDa chaperonin found in virtually all eubacteria and the mitochondria and chloroplasts of eukaryotes (38). A robust molecular method has been developed for the identification of microorganisms based on amplification of a portion of the cpn60 gene by universal, degenerate PCR primers (13). This method has demonstrated advantages over 16S rRNA-based methods in that for closely related organisms, there is more phylogenetic information in the protein-encoding cpn60 sequence relative to the structural RNA-encoding 16S rRNA gene (6).
The overriding limitation to sequence-based studies to date has been scale. The most thorough, direct analysis of cloned 16S rRNA gene sequences from a complex microbial community to date involved the sequencing of 284 16S rRNA gene fragments from a human fecal sample (42). A study of this scale, which resulted in the identification of 82 different 16S rRNA sequences, is not adequate to catalog the microbial diversity in feces, thought to contain at least 500 different bacterial species (28), or soil, which is estimated to contain ca. 13,000 different species (46).
The development of high-throughput technologies for genomics applications presents an opportunity to conduct large-scale, even comprehensive, studies of complex microbial communities. Here we describe a modestly scaled feasibility study of the application of genomics technology and the cpn60 molecular diagnostic method to cataloguing the diversity in a microbial community. We chose pig feces as our target population because it is a tractable microbial community, rich in microbial life (estimated to exceed 1011 organisms per g of feces [11, 28]), for which there is a wealth of descriptive literature. Results will be discussed in terms of the potential for this method in larger studies of microbial communities and the establishment of cpn60 as a universal target for studying the phylogenetic relationships of microorganisms in complex communities.
MATERIALS AND METHODS
Pigs and feces collection.
Fecal samples were obtained from the recta of 6-week-old pigs (n = 5) housed in a commercial swine facility (Prairie Swine Centre Inc., Saskatoon, Saskatchewan, Canada). A medicated (chlortetracycline [308 mg/kg], sulfamethazine [308 mg/kg], and penicillin [154 mg/kg]) wheat and soybean meal-based diet formulated to meet nutrient requirements was fed from 21 days of age (weaning). Fecal samples were pooled (a total of approximately 2 g, i.e., 0.4 g from each of the five animals) and stored at −20°C until genomic DNA was extracted.
Genomic DNA extraction.
Two methods of genomic extraction were used. In a modification of the benzylchloride extraction method (52), approximately 0.8 g of feces was thawed and dispersed in 5 ml of benzylchloride extraction buffer (100 mM Tris-HCl [pH 9.0], 40 mM EDTA). To 500 μl of the suspension was added 100 μl of 10% sodium dodecyl sulfate (SDS) and 300 μl of benzyl chloride. The remaining 4.5 ml of fecal suspension was reserved at −20°C. The sample was mixed by vortexing and incubated at 50°C for 30 min, with vortexing at 5-min intervals. Then 300 μl of 3 M sodium acetate (pH 5) was added, and the sample was mixed by inversion and incubated on ice for 15 min, followed by centrifugation at maximum speed in a microcentrifuge at 4°C for 15 min to separate the aqueous and organic phases. The supernatant was transferred to a clean tube and nucleic acids were precipitated by the addition of 400 μl of isopropanol followed by centrifugation at top speed in a microcentrifuge for 10 min at 4°C. The pellet was washed in cold 70% ethanol, dried, and resuspended in 100 μl of TE (100 mM Tris-HCl, pH 8, 1 mM EDTA).
Approximately 0.8 g of feces was dispersed in 5 ml of 25% sucrose-40 mM Tris, pH 8. To 500 μl of the suspension was added 100 μl of lysozyme (10 mg/ml in 25 mM Tris, pH 8), and the sample was incubated at 4°C for 10 min, followed by the addition of 100 μl EDTA (0.5 M, pH 8) and incubation at 4°C for 10 min. Then 1 ml of lysis buffer (62.5 mM EDTA, 50 mM Tris [pH 8], 1% [vol/vol] Triton X-100) was added, and the sample was incubated at 4°C for 15 min with periodic mixing. The lysate was extracted twice with 25:24:1 (vol/vol/vol) phenol-chloroform-isoamyl alcohol, and nucleic acids were precipitated by the addition of 85 μl of 3 M sodium acetate and 850 μl of isopropanol, followed by centrifugation at maximum speed in a microcentrifuge for 10 min at 4°C. Pellet was washed once with 70% ethanol, air dried, and resuspended in 100 μl of TE.
PCR and cloning of PCR products.
Genomic DNA extracted from feces (1 μl of either benzylchloride or phenol-chloroform-extracted DNA) was used as the template in PCRs. The PCR primers used were H279, 5′-GAI III GCI GGI GA(C/T) GGI ACI ACI AC-3′, and H280, 5′-(C/T)(G/T)I (C/T)(G/T)I TCI CC(A/G) AAI CCI GGI GC(C/T) TT-3′. Inosine (I) was used to reduce the degeneracy of the sequences (29). Primers were designed to amplify the region between codons 92 and 277 based on the Escherichia coli groEL sequence (accession number X07850). The PCRs contained 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 250 μM each of the four deoxynucleoside triphosphates, 2 U of Taq DNA polymerase, and 0.5 μg (50 pmol) of each primer.
PCRs were performed on a Stratagene Robocycler thermocycler according to the following parameters: 3 min at 95°C, 40 cycles of 1 min at 95°C, 1 min at 40°C, 1 min at 72°C, and 10 min at 72°C. PCRs included a negative control reaction containing no template DNA to ensure that no contaminating template was present in the reactions. An additional set of PCRs were done as described except that the annealing temperature was 56°C. The resulting four PCR products were agarose gel purified and ligated into vector pCR2.1-TOPO with the TOPO T-A cloning kit (Invitrogen), and transformed Escherichia coli was plated on Luria-Bertani agar (LB) containing ampicillin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The resulting libraries were named according to the template extraction method and PCR annealing temperature used in their production: B56 and B40 (benzylchloride template amplified with an annealing temperature of 56°C or 40°C, respectively); P56 and P40 (phenol-chloroform template amplified with an annealing temperature of 56°C or 40°C, respectively). Colonies (576 white colonies from each library) were picked and used to inoculate 96-well plates containing 100 μl of LB with ampicillin (50 μg/ml) per well. Culture plates were incubated overnight in humidified containers at 37°C with shaking. Glycerol (100 μl of 30% glycerol in LB) was added to each well, and plates were sealed and stored at −80°C.
Plasmid DNA isolation and DNA sequencing.
Plasmid DNA for sequencing template was isolated either by the Qiagen R.E.A.L. Prep 96 plasmid kit according to the manufacturer's protocol or by a solid-phase reversible immobilization method modified from an earlier published procedure (19) for use on an integrated automation platform (ELVIS; see http://bioinfo.pbi.nrc.ca/robotics). For the robotic plasmid preparation, recombinant clones were cultured in 1.2 ml of Terrific Broth in deep-well (2 ml) 96-channel microtiter plates, pelleted by centrifugation, and lysed by an alkaline-SDS procedure. Lysates were made up to 10% polyethylene glycol 8000 and 0.5 M NaCl prior to the addition of 200 μg of COOH-derivatized paramagnetic beads (Seradyn). The bead slurry mixture was incubated with shaking for 5 min, and the beads were subsequently fractionated over permanent magnets, washed in 50% ethanol, dried, and resuspended in double-distilled H2O. Plasmid concentration was estimated by resolving plasmid preps on 1% agarose gels.
High-throughput DNA sequencing reactions were conducted in 384-well microtiter plate format, by using 100 to 300 ng of template DNA in combination with 5′-biotinylated T7 and M13RP sequencing primers, in a 1/3 volume Big Dye sequence reaction (PE Biosystems). Reactions were assembled by the robotic system described above and thermocycled according to the supplier's recommended protocol. Sequence extension reaction products were purified by addition of 10 μg of streptavidin-paramagnetic beads (M-280; Dynal Corporation), followed by fractionation over permanent magnets. Fractionated beads were resuspended in 12 μl of 50% deionized formamide and treated at 95°C for 5 min prior to immobilization and transfer of up to 12 μl of the reaction product-containing supernatant to a fresh 384-well microtiter plate. Completed reactions were sealed and stored at −80°C prior to resolution on a PE-3700 capillary sequencing device.
Sequence data assembly and analysis.
All sequence data assembly, analysis, and storage were done by software available from the Canadian Bioinformatics Resource (http://www.cbr.nrc.ca). Raw sequencing data were assembled into contigs for each template by Pregap4 (version 1.1) and Gap4 (version 4.6) in the Staden software package (release 2000.0; J. Bonfield, K. Beal, M. Betts, M. Jordan, and R. Staden, 2000). Contig nucleotide and peptide sequences were compared to a database of approximately 1,000 cpn60 sequences by BlastP and BlastN. Sequence data, template information, and Blast results were deposited in a MySQL database for data storage and further analysis. Sequence manipulations, such as format changes and amino acid translations, were done by GCG (Wisconsin package, version 10.1 for Unix). Sequence alignments were done with ClustalW and viewed with GeneDoc.
Phylogenetic analysis was done by programs in the PHYLIP software package. Specifically, alignments were sampled for bootstrap analysis by Seqboot, distances were calculated with the PAM option of Protdist (for peptide sequences) or the maximum-likelihood option of Dnadist. Dendrograms were constructed from distance data by using neighbor-joining by neighbor. Consensus trees were calculated by Consense, and branch lengths were superimposed on consensus trees by Fitch. Completed trees were viewed by TreeView and manipulated for presentation with Microsoft Powerpoint.
RESULTS
cpn60 gene sequences amplified from piglet feces total DNA.
To provide a mixed DNA template representing a complex microbial community, total DNA was extracted from piglet feces. From this template, a region of the cpn60 gene sequence was amplified by universal, degenerate primers. Four independently amplified DNA products were produced by application of two methods for DNA extraction combined with two annealing temperatures for PCR, 40 and 56°C. The amplified products were cloned independently to produce four libraries. High-quality sequence data were obtained for 1,125 clones that were randomly selected from the four libraries (278 from B40, 332 from B56, 293 from P40, and 222 from P56). Disregarding the flanking degenerate primer sequences, the cloned cpn60 gene region was either 552, 555, or 558 nucleotides in length (184, 185, or 186 codons, respectively).
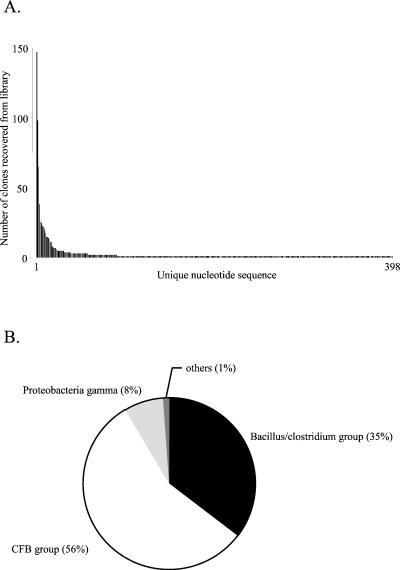
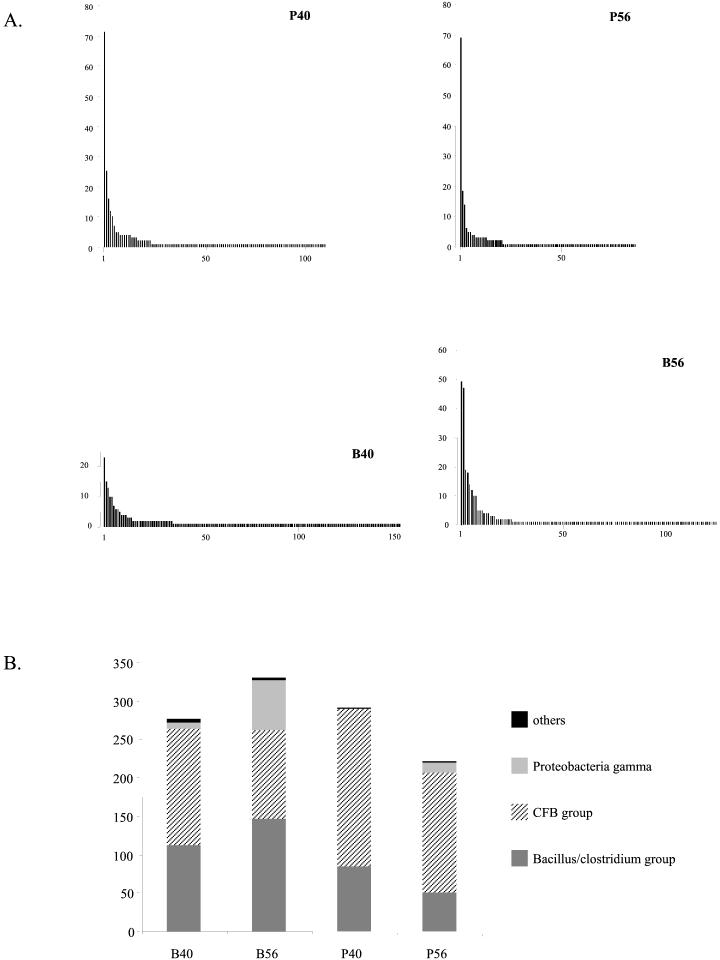
Pairwise comparisons of the 1,125 sequences by ClustalW revealed the presence of 398 unique nucleotide sequences (encoding 280 unique peptide sequences). These were deposited in GenBank as a phylogenetic study and assigned accession numbers AF436893 to AF437290. Figure 1A shows the number of times each unique nucleotide sequence was recovered from the total library. A few sequences were recovered frequently, and one sequence was recovered 148 times whereas 307 sequences were recovered only once. Only 10 sequences were recovered more than 20 times. Pairwise comparisons among the 398 unique sequences gave from 47 to 99% nucleotide sequence identity.
FIG. 1.
(A) Frequency distribution of unique nucleotide sequences recovered from the combined pig feces cpn60 libraries. (B) Taxonomic breakdown of total library contents. Assignment to a taxonomic group was based on comparisons of clone sequences to a database of cpn60 reference sequences.
Phylogenetic analysis of cpn60 sequence data.
Each DNA and peptide sequence was compared to a database of cpn60 sequences by Blast (1). The database is a curated and growing collection of approximately 1,100 eubacterial and eukaryotic cpn60 sequences harvested from public databases or generated in the laboratories of a network of collaborating researchers. The nearest database neighbors of the most frequently recovered library sequences are shown in Table 1. The estimated taxonomic breakdown of the total library contents, based on nearest-neighbor taxonomy, is illustrated in Fig. 1B and Table 2.
TABLE 1.
Nearest cpn60 database neighbors of sequences recovered from a library at least four times among 1,125 clones
| Clone | Nearest cpn60 database neighbor | GenBank accession no. | Taxonomic group | % Peptide identity (similarity) | % DNA identity | Frequencya |
|---|---|---|---|---|---|---|
| 002_a03 | Prevotella intermedia ATCC25611 | AF440234 | CFB group | 88 (94) | 78 | 148∗ |
| 001_f12 | Prevotella bivia ATCC29303 | AF440233 | CFB group | 88 (92) | 73 | 98∗ |
| 002_a11 | Anaerobiospirillum succiniciproducens ATCC700195 | AF441383 | Proteobacteria gamma | 86 (94) | 80 | 65∗ |
| 001_c11 | Prevotella bivia ATCC29303 | AF440233 | CFB group | 88 (92) | 72 | 38∗ |
| 001_e03 | Bacillus halodurans | AP001508 | Bacillus/Clostridium group | 64 (83) | 64 | 26∗ |
| 001_a02 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 71 (85) | 69 | 25∗ |
| 001_g07 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 72 (86) | 70 | 23∗ |
| 005_a04 | Prevotella bivia ATCC29303 | AF440233 | CFB group | 88 (92) | 73 | 22∗ |
| 002_c12 | Bacteroides ovatus ATCC8483 | AF440236 | CFB group | 97 (97) | 83 | 21∗ |
| 002_b08 | Clostridium difficile 79-685 | AF080547 | Bacillus/Clostridium group | 73 (88) | 63 | 19∗ |
| 003_b04 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 72 (86) | 70 | 17∗ |
| 001_a05 | Chryseobacterium gleum ATCC35910 | AF440235 | CFB group | 68 (84) | 67 | 15∗ |
| 002_g03 | Thermoanaerobacter brockii Rt8.G4 | U56021 | Bacillus/Clostridium group | 75 (89) | 68 | 15∗ |
| 002_e03 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 71 (85) | 70 | 14∗ |
| 005_b04 | Prevotella bivia ATCC29303 | AF440233 | CFB group | 88 (92) | 73 | 13∗ |
| 002_e11 | Bacteroides ovatus ATCC8483 | AF440236 | CFB group | 96 (96) | 80 | 11∗ |
| 003_a04 | Anaerobiospirillum succiniciproducens ATCC700195 | AF441383 | Proteobacteria gamma | 83 (91) | 79 | 11 |
| 005_c01 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 74 (88) | 71 | 8∗ |
| 005_e05 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 72 (88) | 72 | 8∗ |
| 001_h02 | Clostridium perfringens | X62914 | Bacillus/Clostridium group | 71 (88) | 66 | 7∗ |
| 003_f04 | Prevotella bivia ATCC29303 | AF440233 | CFB group | 88 (93) | 74 | 7∗ |
| 008_h10 | Clostridium difficile 79-685 | AF080547 | Bacillus/Clostridium group | 73 (89) | 64 | 7∗ |
| 002_c10 | Prevotella bivia ATCC29303 | AF080547 | CFB group | 87 (93) | 77 | 6∗ |
| 001_h09 | Lactobacillus amylovorous ATCC33620 | Bacillus/Clostridium group | 100 (100) | 100 | 5 | |
| 001_h11 | Bacillus halodurans | AP001508 | Bacillus/Clostridium group | 64 (83) | 64 | 5∗ |
| 003_b12 | Pediococcus pentosaceus ATCC43200 | Bacillus/Clostridium group | 95 (96) | 84 | 5∗ | |
| 003_d12 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 72 (86) | 70 | 5 |
| 005_e02 | Prevotella intermedia ATCC25611 | AF440234 | CFB group | 89 (93) | 82 | 5 |
| 005_e06 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 72 (86) | 70 | 5 |
| 006_b07 | Clostridium perfringens | X62914 | Bacillus/Clostridium group | 64 (84) | 64 | 5∗ |
| 006_f02 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 78 (89) | 71 | 5∗ |
| 001_e12 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 74 (88) | 71 | 4∗ |
| 002_d06 | Prevotella intermedia ATCC25611 | AF440234 | CFB group | 88 (94) | 78 | 4 |
| 002_d12 | Clostridium thermocellum ncib10682 | Z68137 | Bacillus/Clostridium group | 71 (86) | 68 | 4 |
| 002_g10 | Prevotella intermedia ATCC25611 | AF440234 | CFB group | 88 (94) | 78 | 4∗ |
| 011_c01 | Lactobacillus acidophilus T-13 | Bacillus/Clostridium group | 100 (100) | 100 | 4∗ | |
| 014_a12 | Clostridium perfringens | X62914 | Bacillus/Clostridium group | 72 (87) | 70 | 4 |
| 018_b06 | Lactococcus garvieae ATCC43921 | AF245674 | Bacillus/Clostridium group | 66 (86) | 63 | 4 |
∗, recovered from at least two libraries.
TABLE 2.
Summary of all library clones classified by nearest cpn60 database neighbors
| Taxonomic group | Nearest cpn60 database neighbor | GenBank accession no. | No. of unique sequences | No. of clones | % DNA identity | % peptide identity (similarity) |
|---|---|---|---|---|---|---|
| CFB group | Bacteroides forsythus ATCC43037 | AJ006516 | 21 | 25 | 69-75 | 76 (86)-88 (92) |
| Bacteroides ovatus ATCC8483 | AF440236 | 29 | 68 | 66-83 | 69 (81)-97 (97) | |
| Bacteroides vulgatus ATCC8482 | AF440238 | 10 | 10 | 63-85 | 70 (82)-94 (95) | |
| Chryseobacterium gleum ATCC35910 | AF440235 | 3 | 17 | 67-68 | 68 (84)-73 (82) | |
| Prevotella bivia ATCC29303 | AF440233 | 87 | 274 | 72-79 | 84 (89)-89 (94) | |
| Prevotella intermedia ATCC25611 | AF440234 | 44 | 210 | 72-82 | 72 (84)-93 (96) | |
| Prevotella nigrescens ATCC33563 | AF441382 | 19 | 20 | 68-81 | 67 (83)-89 (93) | |
| Bacillus/Clostridium group | Bacillus coagulans CECT 12 | AF441379 | 5 | 6 | 68-70 | 72 (87)-74 (86) |
| Bacillus firmus CECT 14 | AF441380 | 1 | 2 | 66 | 65 (82) | |
| Bacillus halodurans | AP001508 | 11 | 42 | 63-67 | 64 (84)-71 (86) | |
| Bacillus psychrophilus CECT 4073 | AF441381 | 8 | 9 | 66-70 | 72 (86)-73 (88) | |
| Bacillus sp. MS | AB028452 | 3 | 3 | 68-69 | 72 (87) | |
| Clostridium acetobutylicum | M74572 | 2 | 3 | 66-68 | 65 (84)-73 (87) | |
| Clostridium difficile 79-685 | AF080547 | 14 | 42 | 61-65 | 64 (78)-74 (89) | |
| Clostridium perfringens | X62914 | 12 | 28 | 63-70 | 63 (83)-73 (88) | |
| Clostridium thermocellum ncib10682 | Z68137 | 88 | 210 | 65-75 | 67 (83)-82 (91) | |
| Enterococcus asini ss-1501 | AF245671 | 7 | 11 | 59-100 | 71 (85)-100 (100) | |
| Globicatella sanguinis ATCC51173 | AF441384 | 1 | 1 | 62 | 63 (83) | |
| Lactobacillus acidophilus T-13 | 2 | 5 | 99-100 | 99 (99)-100 (100) | ||
| Lactobacillus amylovorous ATCC33620 | 3 | 9 | 95-100 | 97 (99)-100 (100) | ||
| Lactobacillus jensenii ATCC25258 | 1 | 1 | 64 | 66 (83) | ||
| Lactococcus garvieae ATCC43921 | AF245674 | 1 | 4 | 63 | 66 (86) | |
| Pediococcus pentosaceus ATCC43200 | 5 | 10 | 84-98 | 95 (96)-100 (100) | ||
| Thermoanaerobacter brockii Rt8.G4 | U56021 | 2 | 16 | 68 | 75 (89) | |
| Proteobacteria gamma | Anaerobiospirillum succiniciproducens ATCC700195 | AF441383 | 12 | 88 | 79-80 | 83 (91)-86 (94) |
| Proteobacteria beta | Burkholderia vietnamiensis DSM 11319 | AF104908 | 1 | 1 | 82 | 87 (93) |
| Chlamydiales | Chlamydia muridarum | NP_296764 | 1 | 2 | 55 | 50 (68) |
| Spirochetes | Borrelia burgdorferi | NC_001318 | 3 | 5 | 62-67 | 65 (82)-67 (83) |
| Treponema pallidum | AE001188 | 1 | 1 | 64 | 71 (89) | |
| Total | 398 | 1,125 |
The largest taxonomic group, represented by 55% of the total library clones and 54% of the unique nucleotide sequences, was the Cytophaga-Flexibacter-Bacteroides (CFB) group. The Bacillus/Clostridium subgroup of gram-positive bacteria represented 36% of the total library clones and 42% of the unique nucleotide sequences, and gamma-class Proteobacteria accounted for 8% of the total clones and 3% of the unique nucleotide sequences. The group labeled “others” in Fig. 1B consisted of clones whose nearest database neighbors were in the spirochete, Chlamydiales, or beta Proteobacteria families (see Table 2 for details). Sequence length was strictly correlated with taxonomic assignment. That is, all clones with nearest neighbors in the CFB group had lengths of 558 bp (186 codons), whereas all the clones with nearest neighbors in the Proteobacteria gamma group and Bacillus/Clostridium group were 555 bp (185 codons) and 552 bp (184 codons), respectively. These are identical to the lengths observed for database reference sequences from each of these groups.
The most abundant sequence in the library (recovered 148 times), represented by clone 002_a03, was 88% identical at the amino acid level (78% nucleotide identity) to Prevotella intermedia ATCC25611. Other sequences recovered at least four times from the library are identified in Table 1 along with their nearest database neighbors. In three cases, library clones showed 100% DNA sequence identity with database reference strains Lactobacillus amylovorus ATCC 33620, Lactobacillus acidophilus T13, and Enterococcus asini ss-1501. Another clone showed 100% amino acid sequence identity and 98% nucleotide sequence identity with Pediococcus pentocaceus ATCC 43200. Overall, the level of sequence identity between each of the 398 unique library sequences and its nearest database neighbor ranged from 56 to 100% DNA identity (51 to 100% peptide identity, 71 to 100% peptide similarity), with only two clones having less than 60% peptide identity to their nearest database neighbor. Table 2 shows the overall composition of recovered sequences in terms of their nearest database neighbors.
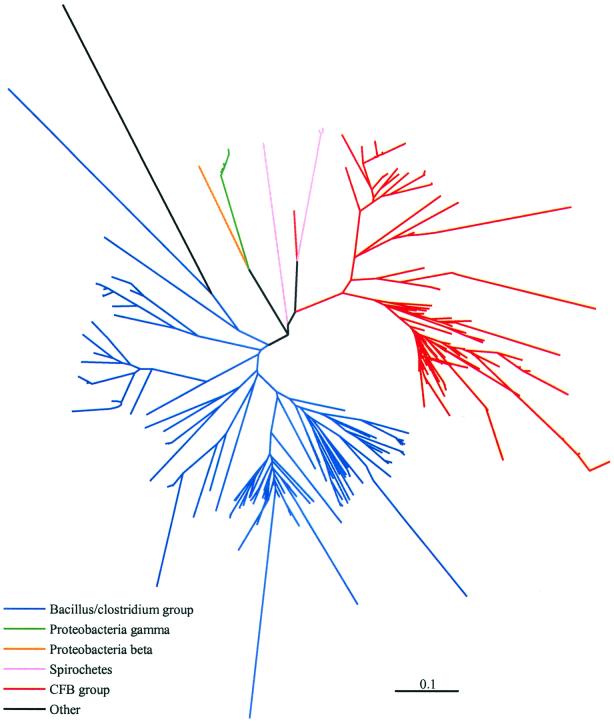
Inferred phylogenetic relationships among unique library sequences are illustrated in Fig. 2. The 280 unique peptide sequences translated from the 398 unique nucleotide sequences were subjected to a multiple sequence alignment with ClustalW. Pairwise distances between the aligned sequences were calculated by the Protdist program within PHYLIP (PAM matrix), and the tree was generated by neighbor joining. Branches were color-coded according to the taxonomic group of the nearest database neighbor of each clone sequence. Overall, the phylogenetic relationships outlined in the tree in Fig. 2 reflect the initial taxonomic estimates made based on the Blast results.
FIG. 2.
Phylogenetic relationships of 280 unique Cpn60 peptide sequences translated from 398 unique nucleotide sequences. Distance calculations were made by the Dayhoff PAM matrix, and the dendrogram was produced by neighbor joining. The scale bar represents 0.1 substitution per site. Branches are colored according to the assigned taxonomic group of the sequences (red, CFB group; green, Proteobacteria gamma; blue, Bacillus/Clostridium group; orange, Proteobacteria beta; pink, spirochetes; black, other).
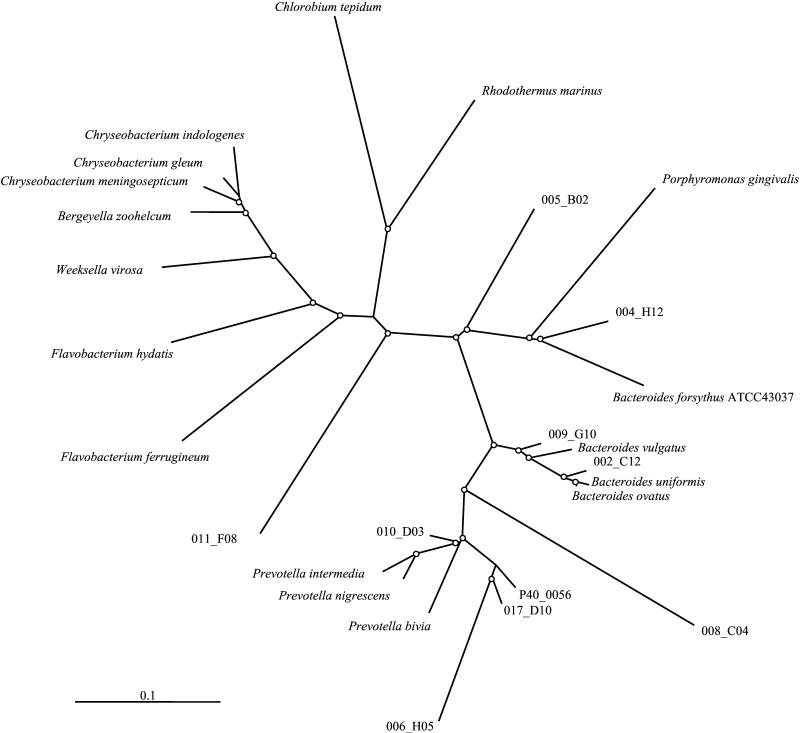
Following the gross phylogenetic analysis presented in Fig. 2, groups of cloned sequences from each of the represented taxonomic categories were selected for detailed phylogenetic tree construction, incorporating reference sequences from the cpn60 database. By this technique, clone sequences were tentatively identified to the level of taxonomic subclass, family, or genus. An example of this analysis, including 10 clone sequences with nearest database neighbors in the CFB group and reference sequences from the genera Chlorobium, Rhodothermus, Flavobacterium, Bergeyella, Chryseobacterium, Bacteroides, and Weeksella, is shown in Fig. 3.
FIG. 3.
Phylogenetic relationships of 12 clone peptide sequences assigned to the CFB group, including the two most abundant cloned sequences (represented by 001_f12 and 002_a03). The tree is a consensus of 100 neighbor-joined trees. Distance calculations were made by the Dayhoff PAM matrix, and branch lengths were imposed on the consensus tree by Fitch. Nodes with bootstrap values of >50% are indicated with white dots. Reference sequences used in the tree are Flavobacterium hydatis (GenBank accession no. AAK32145), Flavobacterium ferrugineum (AAK32146), Bergeyella zoohelcum (ATCC 43767), Chryseobacterium meningosepticum (ATCC 13253), Chryseobacterium gleum (ATCC 35910), Bacteroides forsythus (CAB43992), Bacteroides vulgatus (ATCC 8482), Bacteroides uniformis (ATCC 8492), Bacteroides ovatus (ATCC 8483), Prevotella bivia (ATCC2 9303), Prevotella intermedia (ATCC 25611), Rhodothermus marinus (strain ITI 376, AAD37976), and Chlorobium tepidum (derived from contig 3499, TIGR unfinished genome database).
Genetic diversity of sampled microorganisms.
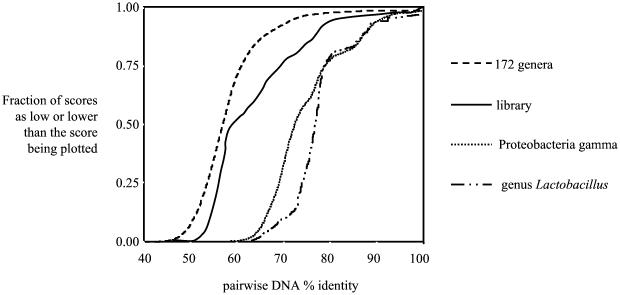
The cumulative frequency distribution was plotted for the DNA sequence identity scores from all pairwise comparisons of library clone sequences (Fig. 4). To produce plots for comparison to this cumulative frequency distribution plot of our experimental population, three other populations of cpn60 sequences were synthesized by selecting sequences from our database of cpn60 reference sequences. The first of these populations consisted of individual species from 172 different genera (including both prokaryotes and eukaryotes) represented in the database. A second population was constructed by pooling cpn60 universal target sequences from 77 species (34 genera) of Proteobacteria gamma. The third population consisted of 37 species from a single genus, Lactobacillus. The experimental pig feces library population, while less diverse than the population of 172 genera, was more diverse than the genus Lactobacillus or the Proteobacteria gamma taxon, with approximately half of the pairwise comparisons within the library having DNA identities of 60% or less.
FIG. 4.
Cumulative frequency distribution plots for the pig feces library (solid line), a population of individual species from 172 different eubacterial and eukaryotic genera, a single taxonomic subclass (77 species from 34 genera of Proteobacteria gamma), and a single genus, Lactobacillus. Plots were generated from DNA identity matrices derived from ClustalW multiple sequence alignments by GeneDoc.
Sequence accuracy and microheterogeneity.
Clusters of nearly identical clone sequences (98 to 99% nucleotide identity) ranging in size from 2 to 20 sequences (191 total sequences) were further analyzed to determine the nature of the differences between the sequences. Multiple alignments of these groups of sequences showed that a disproportionate (P < 0.001) number of the differences within the alignments were synonymous changes, occurring in the third position of codons. Examination of a total of 320 differences revealed that 61 were in the first position of codons, 63 were in the second position, and 196 were in the third position. Almost all third-position differences (191 of 196) were synonymous changes in terms of their effects on the encoded peptide sequence. No in-frame stop codons were observed in any of the 1,125 clone sequences determined. Also, 29 of the 38 sequences in Table 1 (sequences occurring at least four times) were recovered from at least two of the four libraries.
DNA extraction methods and PCR conditions used affect organisms sampled.
To assess the effects of library construction parameters on library contents, sequence data were grouped by library of origin, and clone frequencies and taxonomic distributions were analyzed for each of the four data sets (Fig. 5A). The B40, B56, P40, and P56 libraries contained 156, 125, 112, and 91 different sequences, respectively. The frequency distributions of unique sequences varied markedly between libraries. While the most prevalent clone in the P56 and P40 libraries was recovered approximately 70 times from each library (accounting for 25 to 30% of clones sequenced) and the most prevalent clone in the B56 library was recovered 49 times (15% of clones), the most abundant clone in the B40 library was recovered only 23 times (8% of clones). Figure 5B shows the taxonomic composition of each of the four libraries. The relative proportions of each taxon clearly varied between the four libraries, with the largest proportion of Proteobacteria gamma-like clones occurring in the B56 library, while this taxon was completely absent from the P40 library.
FIG. 5.
(A) Frequency distributions of unique nucleotide sequences recovered from clone libraries P56, P40, B40, and B56. (B) Taxonomic composition of libraries B40, B56, P40, and P56.
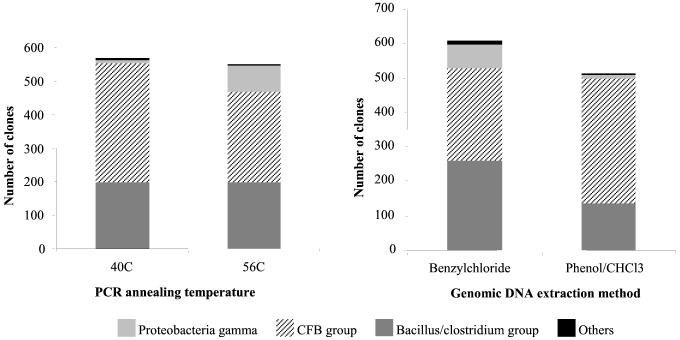
Data were also grouped for analysis according to the PCR annealing temperature or genomic DNA extraction method used in library construction. Figure 6 shows the taxonomic composition of clones produced with a PCR annealing temperature of 40°C versus 56°C and DNA template prepared by the benzylchloride versus phenol-chloroform extraction methods. While all four taxonomic subclasses (CFB group, Bacillus/Clostridium group, Proteobacteria gamma, and others) were detected in each group, the relative proportions of each taxon present varied with library construction conditions. For example, the highest proportion of Proteobacteria gamma class clones were produced with a PCR annealing temperature of 56°C and a benzylchloride-extracted template (see also Fig. 5A).
FIG. 6.
Taxonomic composition of groups of library clones pooled by PCR annealing temperature used in library construction (left panel) and genomic DNA template extraction method (right panel).
DISCUSSION
The microbial community present in the gastrointestinal tract is a complex and dynamic one, varying in composition with age, diet, stress, medication, temperature, etc., as well as varying along the length of the tract. Despite its importance, the microbial flora of the animal gut remains poorly characterized. In fact, microbial communities in general are poorly understood. The technologies developed for genomics programs present us with an unprecedented opportunity to advance our understanding of these populations. The current study, which combined high-throughput genomics technologies with an existing cpn60-based molecular diagnostic to characterize the pig feces microbial community, was undertaken as a feasibility study for the general application of this approach to other complex microbial communities.
Sequence data reveal biologically based microheterogeneity.
The degenerate PCR primers used in this study were previously demonstrated to amplify the universal target region of the cpn60 gene from a wide variety of organisms, including eubacteria, fungi, plants, and animals (6, 12-15; unpublished observations). The region of template-specific cpn60 amplified from pig feces total DNA varied in length, being either 552, 555, or 558 bp (184, 185, or 186 codons), and the complete sequence of each cloned PCR product was determined with two sequencing reactions initiated from sites within the cloning vector. Only unambiguous full-length sequences were included in our analysis.
To assess the potential impact of sequence artifacts that might have been introduced by PCR or Taq polymerase infidelity, clusters of nearly identical nucleotide sequences were examined. If the observed microheterogeneity in these sequence groups resulted from PCR-generated errors, then the sequence differences should be distributed uniformly among the first, second, and third positions within codons. We observed, however, that a significantly disproportionate number of nucleotide differences occurred in the third codon position and that virtually all of these were synonymous differences, resulting in no change in the encoded peptide sequence. Also significant was the fact that no in-frame stop codons were observed in any of the clone sequences assembled.
The genetic code includes 18 codons that, with a single nucleotide change, can be converted to a stop codon, and analysis of the 1,125 clone sequences indicated that there were 58,881 codons that were vulnerable to single-nucleotide mutation to a nonsense codon. Thus, we suggest that while PCR artifacts cannot be ruled out entirely, much of the minor sequence variation observed within clusters of related clone sequences was a reflection of real biological diversity. The sequence differences observed in many of these clusters was typical of the sorts of differences observed previously between cpn60 universal target sequences from serotypes of a single bacterial species, Streptococcus suis (6).
A major concern in 16S rRNA sequence-based studies of microbial communities is the occurrence of chimeric PCR products (35, 40, 49). The generation of chimeric PCR products through template switching is facilitated by the presence of a number of highly conserved stretches along the primary structure of ribosomal DNAs (rDNAs) and can involve closely related sequences, including multiple copies of 16S rRNA genes within a single genome. Chimeric cpn60 PCR products may be less likely than 16S rRNA chimeras, since cpn60 is present in fewer copies per genome (a single copy in most prokaryotic genomes), the amplified sequence is shorter (552 to 558 bp versus the approximately 1.6 kb amplified from bacterial 16S rRNA genes), providing fewer opportunities for template switching, and the cpn60 sequence lacks the intermittent highly conserved sequence stretches present in 16S rRNA genes. Obviously, chimera formation would be more likely, and difficult to detect if it occurred in-frame, between closely related cpn60 sequences.
The best evidence that a sequence is not a PCR artifact is that it is recovered from more than one library, since the libraries were generated from independent PCRs. In this study, 29 of the 38 most frequently recovered sequences were recovered from at least two libraries. Also, several examples of pairs of very similar sequences independently recovered from at least two libraries were found. For example, the sequences of clones 001_a02 and 001_g07 were 98% identical and were each independently recovered from three of the four libraries. The same is true of 002_a03 and 002_g10 (99% identical, each recovered from four and two libraries, respectively) and 001_f12, 005_a04, 005_b04, and 001_c11, which have pairwise identities of 98% and were each recovered from B40 and P40 libraries. These examples provide evidence of real microheterogeneity in the population. The best way to systematically detect PCR chimeras in cpn60 libraries will be to apply computational tools such as Check_Chimera (25), developed for use in 16S rRNA-based studies. Options are currently being investigated in our laboratory so that a reliable chimera-checking tool is available for use with the cpn60 sequence database.
The identification of microheterogeneity within bacterial populations is not practical by traditional culture-based methods. The ability to track sequence microheterogeneity in complex microbial communities may have implications for our ability to understand the dynamics of these populations, particularly with respect to microbial evolution and concepts such as lateral gene transfer (9). Subtle sequence variation, typical of collections of sequences from closely related organisms, is overlooked in population profiling methods such as denaturing gradient gel electrophoresis, which rely on gross sequence attributes, or cloning and sequencing methods where sequences are grouped into general operational taxonomic units based on restriction fragment length polymorphisms before individuals representative of each unit are sequenced (8).
A numbers game.
Authors of previous studies of porcine fecal microflora have reported that the most predominant bacterial species outnumbers the next most abundant species by at least an order of magnitude and that culturable organisms were retrieved at frequencies that varied over many orders of magnitude. For example, while Bacteroidaceae have been found at 1010 cells per g of feces and Bifidobacterium spp. have been found at 109 per g of feces (4), less abundant organisms such as E. coli have been reported at only 105 per g of adult pig feces (21). Based on these observations, it might be predicted that a study involving PCR amplification, cloning, and sequencing of target genes from DNA extracted from such a population would yield very little sequence diversity and that only the most abundant organisms would be represented. Instead we observed a great deal of sequence diversity.
Among 1,125 clones, 398 distinct nucleotide sequences were observed, varying in frequency from 148 to 1, only two orders of magnitude. A likely explanation for the discrepancy between what was predicted and what was observed is the C0t effect (26). In a PCR involving mixed template molecules, the amplification of the most abundant templates declines more rapidly during amplification cycles than that of less abundant templates due to the tendency for abundant templates and amplified products to reanneal rather than undergo primer-mediated amplification. Thus, over the course of the PCR, there is normalization, so that abundant templates become underrepresented and rare templates become overrepresented. While this phenomenon presents a serious challenge to experimental design strategies aimed at quantitating templates in the original sample, it works in favor of sampling maximum diversity.
The extent of sequence diversity found in the clone library is illustrated in Fig. 4 in a cumulative frequency distribution plot of the 79,003 pairwise DNA sequence comparisons derived from the 398 unique clone nucleotide sequences. When compared to populations constructed from a single genus, a taxonomic subclass and single representatives of 172 eukaryotic and eubacterial genera, the pig feces library falls between the taxonomic subclass distribution and the 172-genera population. The figure also shows that the pig feces library population plot is not a smooth curve like the 172-genera plot, indicating the presence of clusters of various sizes of closely related sequences in the population as opposed to the uniform heterogeneity of the artificially constructed 172-genera population.
Comparison of experimental population to known fecal organisms.
Although some limited descriptive analysis of microbial populations resident in various gut compartments of pigs has been conducted (34, 39), the majority of studies of gut microflora have used feces as starting material. It is well established that the composition of fecal microbial populations varies widely from birth to adulthood (21, 23, 27, 44) as well as with diet changes (4) and various disease states (24, 43). Although there is tremendous variation in the proportions of gross taxonomic groups of organisms reported due to the different methods of isolation and characterization, there is some agreement on the types of organisms that constitute the normal porcine fecal flora.
As the fecal flora changes in composition from birth to maturity, there is an increase in the proportion of anaerobes and facultative anaerobes. Culture-based studies suggest that with maturity, CFB group organisms become dominant, particularly Bacteroides, with much smaller proportions of coliforms and Lactobacillus species and highly variable populations of Clostridium species being present (44). Greater phylogenetic and taxonomic detail is available for human fecal populations, which have been the focus of more molecular characterization and are thought to be somewhat similar to microbial populations in pig feces (44). In their analysis of PCR-amplified and cloned 16S rRNA sequences from human feces, Suau et al. (42) found that 95% of the 284 cloned sequences were related to CFB group organisms (particularly Bacteroides species) and Clostridium species. The taxonomic subclasses identified in this study, CFB group, Bacillus/Clostridium group, and gamma class Proteobacteria, are consistent with these previous studies, as are the proportions identified (55% CFB group, 36% Bacillus/Clostridium group, and 8% gamma class Proteobacteria).
The primary goal of the current study was not to quantitate the constituent members of the population but rather to identify and characterize taxonomically diverse organisms within the population. However, despite the potential normalizing effects of the PCR conditions, including the C0t effect, the frequencies of sequences recovered in the library were most likely influenced by the frequencies of the source organisms in the population.
In addition to the three major taxonomic subclasses detected, our sequencing efforts also revealed relatively rare clone sequences assigned to the spirochete group and the beta class Proteobacteria. Our observation of clone sequences with similarity to the spirochete family is not unexpected because nonpathogenic spiral rods have been reported in microscopic observations of feces from healthy pigs (37) and have been cultured from similar sources (4). The observation of a cloned sequence with 82% nucleotide identity and 87% peptide identity (93% similarity) to Burkholderia vietnamiensis, a member of the beta class of Proteobacteria, is interesting because members of this bacterial family have not been reported in studies of fecal flora from animals. However, members of the genus Burkholderia are known to include soil and rhizosphere bacteria as well as plant and human pathogens (10), so perhaps it is not surprising that genomic DNA from this group of organisms would be present in pig feces.
Interestingly, we recovered no sequences with similarity to bifidobacteria. Bifidobacterium spp. are reportedly a major constituent of the fecal flora of monogastrics such as pigs (4) and humans (18), where they have been detected in culture-based studies at frequencies of 109 CFU per g of feces. It seems unlikely that the absence of bifidobacterial PCR products was due to failure of the PCR primers to anneal to these templates, since previous work done in our laboratory and available sequence data from a number of bifidobacterial cpn60 genes demonstrate that primer-binding sites are preserved in these organisms. It seems more likely that the genomic DNA preparation methods used failed to capture bifidobacterial DNA due to a lack of mechanical force sufficient to break open these bacteria or that the relatively high G+C content of bifidobacterial sequences prevented efficient PCR amplification of these targets.
Failures to amplify and clone 16S rRNA sequences from Bifidobacterium spp. have also been reported in studies of human fecal flora (42, 51). Recently, we addressed this issue by isolating total genomic DNA from a similar pig fecal sample by a bead-beating method and conducting PCR with Bifidobacterium-specific primers, designed to amplify a 180-bp region within the cpn60 target. The resulting PCR products were cloned, and when a small number of them were sequenced, they were found to be 99% identical at the DNA level to Bifidobacterium animalis. This result suggests that bifidobacteria are indeed present in pig feces and either that their absence in the library sample is the result of a failure to isolate the genomic DNA template by chemical methods or that the bifidobacterial template DNA is so rare in the template pool that it was not represented in the sample of 1,125 clones.
Library construction methods affected library contents.
To assess the effects of library construction parameters on our results, we created four libraries by genomic DNA templates prepared by one of two methods and conducted the PCRs at either of two annealing temperatures. Figures 5 and 6 clearly illustrate that these parameters did indeed have a pronounced effect on the contents of the resulting library. The Proteobacteria gamma group of templates seem particularly affected by template preparation method and PCR annealing temperature, indicated by a higher proportion of these sequences in libraries constructed from benzylchloride-extracted template DNA and higher annealing temperature. Thus, while accounting for approximately 20% of clones in the B56 library, Proteobacteria gamma species were completely absent from the P40 library.
Potential challenges for approaches such as ours include the possibility for systematic biases in the representativeness of templates within total DNA extracts compared to that of the original microbial population and biases introduced by the degenerate primers and PCR conditions with respect to amplification from specific DNA templates. The small size of our initial fecal sample (approximately 2 g of feces from five pigs) may also be a factor in the representativeness of the genomic DNA extracts, since feces are likely not homogeneous and areas of concentration of some bacterial species may exist. The methods described here offer a way of addressing these issues systematically. Currently, we have a powerful comparative method that could be used to compare microbial populations from similar sources.
cpn60 sequence database.
The clone with weak sequence similarity to Chlamydia muridarum (clone 007_D05) is indicative of the current limitations to sequence identification (Table 2). The ability to assign cloned sequences to a taxonomic subclass or beyond that to the level of genus or species is necessarily limited by the availability of relevant reference sequence data. The tree shown in Fig. 3 is a good illustration of both the strengths and weaknesses in our ability to identify clone sequences. While in some cases identification to the level of genus or even species is possible, there are other cases where limited reference data make it possible to identify sequences only to the level of taxonomic subclass. Currently, our database contains approximately 1,100 reference sequences. We intend to continue to expand the database and release it to the public domain, where we hope it will become a valuable resource complementary to the existing 16S rRNA resource, the Ribosomal Database Project, for microbial population studies as well as for identification of organisms and for phylogenetics.
The results presented here confirm the feasibility of conducting a similar study on a large scale, and the large number of distinct sequences, 307, that were recovered only once in 1,125 sequenced clones indicates that a larger study will identify many more sequences from phylogenetically diverse organisms. Genomics-inspired technologies such as robotic colony picking, template preparation, and sequencing and automated data assembly and analysis can be employed to produce potentially comprehensive profiles of important microbial communities. The libraries of sequence data produced will be tools for developing methods to quantitate organisms within a population, for the detection of pathogens or specific organisms of interest, to monitor changes in populations over time or treatment, and for creating specific probes for techniques, such as fluorescence in situ hybridization.
Acknowledgments
We are grateful to David Carter and Albana Zeko for automated DNA template and sequencing reaction preparation, to Jason Marshall for technical assistance, to Noor Syed for design of Bifidobacterium-specific primers, to the PBI sequencing laboratory for automated sequencing, and to the Canadian Bioinformatics Resource for provision of bioinformatics tools.
We are grateful to the Canadian Biotechnology Strategy and the Saskatchewan Agriculture Development Fund for funding.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai, Q., A. Gattinger, and L. Zelles. 2000. Characterization of microbial consortia in paddy rice soil by phospholipid analysis. Microb. Ecol. 39:273-281. [PubMed] [Google Scholar]
- 4.Benno, Y., K. Endo, K. Suzuki, T. Mitsuoka, and S. Namioka. 1985. Use of nonprotein nitrogen in pigs: effects of dietary urea on the intestinal microflora. Am. J. Vet. Res. 46:959-962. [PubMed] [Google Scholar]
- 5.Bourne, D. G., I. R. McDonald, and J. C. Murrell. 2001. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils. Appl. Environ. Microbiol. 67:3802-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brousseau, R., J. E. Hill, G. Prefontaine, S. H. Goh, J. Harel, and S. M. Hemmingsen. 2001. Streptococcus suis serotypes characterized by analysis of chaperonin 60 gene sequences. Appl. Environ. Microbiol. 67:4828-4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahllof, I., H. Baillie, and S. Kjelleberg. 2000. rpoB-based microbial community analysis avoids limitations inherent in 16S rRNA gene intraspecies heterogeneity. Appl. Environ. Microbiol. 66:3376-3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diez, B., C. Pedros-Alio, and R. Massana. 2001. Study of genetic diversity of eukaryotic picoplankton in different oceanic regions by small-subunit rRNA gene cloning and sequencing. Appl. Environ. Microbiol. 67:2932-2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doolittle, W. F. 1999. Lateral genomics. Trends Cell Biol. 9:M5-M8. [PubMed] [Google Scholar]
- 10.Estrada-De Los, S. P., R. Bustillos-Cristales, and J. Caballero-Mellado. 2001. Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67:2790-2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finegold, S. M., H. R. Attebery, and V. L. Sutter. 1974. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am. J. Clin. Nutr. 27:1456-1469. [DOI] [PubMed] [Google Scholar]
- 12.Goh, S. H., D. Driedger, S. Gillett, D. E. Low, S. M. Hemmingsen, M. Amos, D. Chan, M. Lovgren, B. M. Willey, C. Shaw, and J. A. Smith. 1998. Streptococcus iniae, a human and animal pathogen: specific identification by the chaperonin-60 gene identification method. J. Clin. Microbiol. 36:2164-2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goh, S. H., R. R. Facklam, M. Chang, J. E. Hill, G. J. Tyrrell, E. C. Burns, D. Chan, C. He, T. Rahim, C. Shaw, and S. M. Hemmingsen. 2000. Identification of Enterococcus species and phenotypically similar Lactococcus and Vagococcus species by reverse checkerboard hybridization to chaperonin 60 gene sequences. J. Clin. Microbiol. 38:3953-3959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goh, S. H., S. Potter, J. O. Wood, S. M. Hemmingsen, R. P. Reynolds, and A. W. Chow. 1996. HSP60 gene sequences as universal targets for microbial species identification: studies with coagulase-negative staphylococci. J. Clin. Microbiol. 34:818-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goh, S. H., Z. Santucci, W. E. Kloos, M. Faltyn, C. G. George, D. Driedger, and S. M. Hemmingsen. 1997. Identification of Staphylococcus species and subspecies by the chaperonin-60 gene identification method and reverse checkerboard hybridization. J. Clin. Microbiol. 35:3116-3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gordon, D. A., J. Priscu, and S. Giovannoni. 2000. Origin and phylogeny of microbes living in permanent Antarctic lake ice. Microb. Ecol. 39:197-202. [DOI] [PubMed] [Google Scholar]
- 17.Greenblatt, C. L., A. Davis, B. G. Clement, C. L. Kitts, T. Cox, and R. J. Cano. 1999. Diversity of microorganisms isolated from amber. Microb. Ecol. 38:58-68. [DOI] [PubMed] [Google Scholar]
- 18.Hartemink, R., and F. M. Rombouts. 1999. Comparison of media for the detection of bifidobacteria, lactobacilli and total anaerobes from faecal samples. J. Microbiol. Methods 36:181-192. [DOI] [PubMed] [Google Scholar]
- 19.Hawkins, T. L., T. O'Connor-Morin, A. Roy, and C. Santillan. 1994. DNA purification and isolation by a solid-phase. Nucleic Acids Res. 22:4543-4544. [DOI] [PMC free article] [PubMed]
- 20.Hooper, L. V., and J. I. Gordon. 2001. Commensal host-bacterial relationships in the gut. Science 292:1115-1118. [DOI] [PubMed] [Google Scholar]
- 21.Jensen-Waern, M., L. Melin, R. Lindberg, A. Johannisson, L. Petersson, and P. Wallgren. 1998. Dietary zinc oxide in weaned pigs—effects on performance, tissue concentrations, morphology, neutrophil functions and faecal microflora. Res. Vet. Sci. 64:225-231. [DOI] [PubMed] [Google Scholar]
- 22.Kasai, H., K. Watanabe, E. Gasteiger, A. Bairoch, K. Isono, S. Yamamoto, and S. Harayama. 1998. Construction of the gyrB database for the identification and classification of bacteria. Genome Inform. Ser. Workshop Genome Inform. 9:13-21. [PubMed]
- 23.Katouli, M., A. Lund, P. Wallgren, I. Kuhn, O. Soderlind, and R. Mollby. 1997. Metabolic fingerprinting and fermentative capacity of the intestinal flora of pigs during pre- and postweaning periods. J. Appl. Microbiol. 83:147-154. [DOI] [PubMed] [Google Scholar]
- 24.Katouli, M., L. Melin, M. Jensen-Waern, P. Wallgren, and R. Mollby. 1999. The effect of zinc oxide supplementation on the stability of the intestinal flora with special reference to composition of coliforms in weaned pigs. J. Appl. Microbiol. 87:564-573. [DOI] [PubMed] [Google Scholar]
- 25.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed]
- 26.Mathieu-Daude, F., J. Welsh, T. Vogt, and M. McClelland. 1996. DNA rehybridization during PCR: the ‘Cot effect’ and its consequences. Nucleic Acids Res. 24:2080-2086. [DOI] [PMC free article] [PubMed]
- 27.Melin, L., M. Jensen-Waern, A. Johannisson, M. Ederoth, M. Katouli, and P. Wallgren. 1997. Development of selected faecal microfloras and of phagocytic and killing capacity of neutrophils in young pigs. Vet. Microbiol. 54:287-300. [DOI] [PubMed] [Google Scholar]
- 28.Moore, W. E., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohtsuka, E., S. Matsuki, M. Ikehara, Y. Takahashi, and K. Matsubara. 1985. An alternative approach to deoxyoligonucleotides as hybridization probes by insertion of deoxyinosine at ambiguous codon positions. J. Biol. Chem. 260:2605-2608. [PubMed] [Google Scholar]
- 30.Olsen, G. J., D. J. Lane, S. J. Giovannoni, N. R. Pace, and D. A. Stahl. 1986. Microbial ecology and evolution: a ribosomal RNA approach. Annu. Rev. Microbiol. 40:337-365. [DOI] [PubMed] [Google Scholar]
- 31.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 32.Pace, N. R., D. A. Stahl, D. J. Lane, and G. J. Olsen. 1986. The analysis of natural microbial populations by ribosomal RNA sequences. Adv. Microb. Evol. 9:1-55. [Google Scholar]
- 33.Paster, B. J., F. E. Dewhirst, S. M. Cooke, V. Fussing, L. K. Poulsen, and J. A. Breznak. 1996. Phylogeny of not-yet-cultured spirochetes from termite guts. Appl. Environ. Microbiol. 62:347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiu, X., L. Wu, H. Huang, P. E. McDonel, A. V. Palumbo, J. M. Tiedje, and J. Zhou. 2001. Evaluation of PCR-generated chimeras, mutations, and heteroduplexes with 16S rRNA gene-based cloning. Appl. Environ. Microbiol. 67:880-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranjard, L., F. Poly, and S. Nazaret. 2000. Monitoring complex bacterial communities by culture-independent molecular techniques: application to soil environment. Res. Microbiol. 151:167-177. [DOI] [PubMed] [Google Scholar]
- 37.Salanitro, J. P., I. G. Blake, and P. A. Muirhead. 1977. Isolation and identification of fecal bacteria from adult swine. Appl. Environ. Microbiol. 33:79-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sigler, P. B., Z. Xu, H. S. Rye, S. G. Burston, W. A. Fenton, and A. L. Horwich. 1998. Structure and function in GroEL-mediated protein folding. Annu. Rev. Biochem. 67:581-608. [DOI] [PubMed] [Google Scholar]
- 39.Simpson, J. M., V. J. McCracken, B. A. White, H. R. Gaskins, and R. I. Mackie. 1999. Application of denaturant gradient gel electrophoresis for the analysis of the porcine gastrointestinal microbiota. J. Microbiol. Methods 36:167-179. [DOI] [PubMed] [Google Scholar]
- 40.Speksnijder, A. G., G. A. Kowalchuk, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Staley, J. T., and A. Konopka. 1985. Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39:321-346. [DOI] [PubMed] [Google Scholar]
- 42.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svendsen, J., N. Bille, N. C. Nielsen, J. L. Larsen, and H. J. Riising. 1975. Preweaning mortality in pigs. 4. Diseases of the gastrointestinal tract in pigs. Nord. Vet. Med. 27:85-101. [PubMed] [Google Scholar]
- 44.Swords, W. E., C. C. Wu, F. R. Champlin, and R. K. Buddington. 1993. Postnatal changes in selected bacterial groups of the pig colonic microflora. Biol. Neonate 63:191-200. [DOI] [PubMed] [Google Scholar]
- 45.Theron, J., and T. E. Cloete. 2000. Molecular techniques for determining microbial diversity and community structure in natural environments. Crit. Rev. Microbiol. 26:37-57. [DOI] [PubMed] [Google Scholar]
- 46.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van Leeuwenhoek, A. 1684. Microscopical observations about animals in the scurf of the teeth. Phil. Trans. R. Soc. London 14:568-574. [Google Scholar]
- 48.Vaughan, E. E., F. Schut, H. G. H. J. Heilig, E. G. Zoetendal, W. M. De Vos, and A. D. L. Akkermans. 2000. A molecular view of the intestinal ecosystem. Curr. Issues Intest. Microbiol. 1:1-12. [PubMed] [Google Scholar]
- 49.Wang, G. C., and Y. Wang. 1997. Frequency of formation of chimeric molecules as a consequence of PCR coamplification of 16S rRNA genes from mixed bacterial genomes. Appl. Environ. Microbiol. 63:4645-4650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weidner, S., W. Arnold, E. Stackebrandt, and A. Puhler. 2000. Phylogenetic analysis of bacterial communities associated with leaves of the seagrass Halophila stipulacea by a culture-independent small-subunit rRNA gene approach. Microb. Ecol. 39:22-31. [DOI] [PubMed] [Google Scholar]
- 51.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu, H., F. Qu, and L. H. Zhu. 1993. Isolation of genomic DNAs from plants, fungi and bacteria by benzyl chloride. Nucleic Acids Res. 21:5279-5280. [DOI] [PMC free article] [PubMed]