Abstract
A SYBR Green LightCycler PCR assay using a single primer pair allowed simultaneous detection of stx1 and/or stx2 of Escherichia coli O157:H7. A distinct sequence of the Shiga-like toxin genes was amplified to yield products of 227 and/or 224 bp, respectively. The two products were distinguished by melting point curve analysis.
All enterohemorrhagic Escherichia coli (EHEC) strains cause serious disease in humans and possess at least one Shiga-like toxin (stx1 or stx2) gene. The detection of Shiga-like toxins is very useful for the identification of EHEC, but PCR methods for the detection of these genes have had some problems. The stx primers reported by Karch and Meyer (5) were unable to distinguish stx1 and stx2 by agarose gel electrophoresis (3, 5).
Traditional PCR methods require amplification in a thermocycler and product separation by gel electrophoresis followed by hybridization with a probe. This is a time-consuming and laborious process. However, the products of the PCR can also be detected by using a DNA binding dye, such as SYBR Green, or through the use of fluorescent probes. The non-sequence-specific SYBR Green assay is less expensive than the fluorescent-probe-based assays utilizing Taqman probes or molecular beacons. Real-time PCR assays can be automated and are sensitive and rapid. They can also quantify PCR products with greater reproducibility while eliminating the need for post-PCR processing, thus preventing carryover contamination (2, 6, 7, 8).
The present study employs amplification of two different targets with a single primer pair that amplifies stx1 and/or stx2 genes in a real-time SYBR Green fluorescent PCR assay using the LightCycler instrument. The products formed are identified based on melting point (Tm) curve analysis.
The EHEC strains tested in the present work are listed in Table 1. E. coli O157:H7 was enriched in Trypticase soy broth, incubated overnight at 37°C, spread on plates with sorbitol-MacConkey agar (SMAC), and incubated again overnight at 37°C. A loopful of colony was suspended in 100 μl of sterile distilled water in a 1.5-ml Eppendorf tube. The tube was vortexed for 5 to 10 s, floated in boiling water for 10 min, chilled on ice for 5 min, and then centrifuged at 12,000 × g for 2 min to remove debris. The supernatant (1 μl) was added directly to the PCR mixture.
TABLE 1.
Specificity of SYBR Green PCR assay to detect stx1 and/or stx2 of various EHEC serotypes based on Tm valuesa
| E. coli strain (strain number or CRIFSb Culture collection) | Sorbitol | Shiga toxin phenotype |
Tm (°C) of:
|
|
|---|---|---|---|---|
| stx1 | stx2 | |||
| O157:H7 (ATCC 43890) | − | Stx1 | 82.49 | |
| O157:H7 (ATCC 43889) | − | Stx2 | 84.03 | |
| O157:H7 (C9490) | − | Stx1 and Stx2 | 82.03 | 84.05 |
| O157:H7 (ATCC 43888) | − | |||
| O157:H7 JG1 | − | Stx1 and Stx2 | 81.74 | 83.88 |
| O157:H7 JG2 | − | Stx1 and Stx2 | 81.70 | 83.91 |
| O157:H7 JG3 | − | Stx2 | 83.46 | |
| O157:H7 JG4 | − | Stx2 | 83.53 | |
| O157:H7 JG5 | − | Stx1 and Stx2 | 81.67 | 83.86 |
| O157:H7 JG6 | − | Stx1 and Stx2 | 81.62 | 83.73 |
| O157:H7 JG7 | − | Stx1 and Stx2 | 81.45 | 83.60 |
| O157:H7 JG8 | − | Stx1 and Stx2 | 81.49 | 83.65 |
| O157:H7 JG9 | − | Stx1 and Stx2 | 81.54 | 83.66 |
| O157:H7 JG10 | − | Stx2 | 83.31 | |
| O157:H7 JG11 | − | Stx1 and Stx2 | 81.65 | 84.0 |
| O157:H7 JG12 | − | Stx1 and Stx2 | 81.42 | 83.76 |
| O157:H7 JG13 | − | Stx1 and Stx2 | 81.40 | 83.78 |
| O157:H7 JG14 | − | Stx1 and Stx2 | 81.47 | 83.77 |
| O157:H7 JG15 | − | Stx1 and Stx2 | 81.48 | 83.78 |
| O157:NM JG16 | − | Stx2 | 83.34 | |
| O157:H7 JG17 | − | Stx1 and Stx2 | 82.5 | 84.55 |
| O157:H7 JG18 | − | Stx1 and Stx2 | 82.5 | 84.55 |
| O157:H7 JG19 | − | Stx1 and Stx2 | 82.5 | 84.4 |
| O157:H7 JG20 | − | Stx1 and Stx2 | 82.61 | 84.0 |
| O2:H6 JG21 | + | |||
| O2:H8 JG22 | + | |||
| O103:H2 JG23 | + | Stx1 | 81.91 | |
| O26:H11 JG24 | + | Stx1 | 81.84 | |
| O91:H21 JG25 | + | Stx2 | 84.16 | |
| O45:H2 JG26 | + | Stx1 | 82.13 | |
| O111:H8 JG27 | + | Stx1 | 82.01 | |
| O26:H11 JG28 | + | Stx1 | 82.10 | |
| O91:H21 JG29 | + | Stx2 | 83.97 | |
| O103:H2 JG30 | + | Stx1 | 81.90 | |
| O111:H8 JG31 | + | Stx1 | 81.89 | |
| O111:H− JG32 | + | Stx1 and Stx2 | 81.28 | 83.73 |
| O113:H21 JG33 | + | Stx1 and Stx2 | 81.27 | 83.70 |
| ATCC 15597 | + | |||
The presence of stx1 was further confirmed with the PCR primer pair designed in the present study. The primers used for amplifying the stx1 gene of E. coli O157:117 were JMS1F (5′-GTCACAGTAACAAACCGTAACA-3′ [positions 7143 to 7164]) and JMS1R (5′-TCGTTGACTACTTCTTATCTGGA-3′ [positions 7237 to 7215]). After amplification, Tm analysis with a Tm value of 82.46°C was confirmed by SYBR Green PCR assay, and the expected size of the stx1 PCR product, 95 bp, was visualized in 2.5% agarose gel.
CRIFS, Canadian Research Institute for Food Safety.
To determine the detection limit of the assay, brain heart infusion broth (1 ml) was inoculated with a single colony isolated from the SMAC plates and incubated overnight at 37°C, and serial 10-fold dilutions in 1% peptone were prepared, giving counts in the range of 100 to 1010 CFU/ml. Viable counts were obtained by culturing each dilution (50 μl) on tryptose soy agar plates with overnight incubation at 37°C. DNA was extracted from 1 ml of the diluted culture using the method described above.
A single primer pair to amplify both stx1 and stx2 (Table 2) was designed, and its performance was standardized to optimize Mg2+ and primer concentrations. PCR was performed in glass capillaries using the LightCycler FastStart SYBR Green kit (Roche Diagnostics, Laval, Quebec, Canada) as described by the manufacturer. All runs included a negative DNA control without template and a positive control. Optimized reaction mixtures for both the stx1- and stx2-specific SYBR Green assays included 1× master mix, a 250 nM concentration of each primer, 4 mM Mg2+, and 1 μl of template DNA in a 10-μl PCR mixture. The cycling profile for the multiplex assay consisted of 95°C for 10 min, followed by 45 cycles of denaturation at 95°C for 15 s, annealing at 55°C for 10 s, and elongation at 72°C for 15 s. Fluorescence signals were measured once in each cycle at the end of the extension step.
TABLE 2.
Primers for detection of E. coli O157:H7
| Target and primer | Sequence (5′→3′) | Position |
|---|---|---|
| stx1a | ||
| JMS1F | GTCACAGTAACAAACCGTAACA | 7143-7164 |
| JMS1R | TCGTTGACTACTTCTTATCTGGA | 7237-7215 |
| stx2b | ||
| JMS2F | CGACC CCTCT TGAAC ATA | 6261-6278 |
| JMS2R | GATAG ACATC AAGCC CTCGT | 6368-6349 |
Product size, 95 bp.
Product size, 108 bp.
After PCR amplification, Tm curve analysis was performed. The PCR products were cooled to 65°C and then slowly heated to 95°C at a rate of 0.2°C/s. Fluorescence signals obtained were continuously monitored to confirm amplification specificity. Tm peaks of the products were calculated based on initial fluorescence curves (F/T) by plotting the negative derivative of fluorescence over temperature versus temperature (−dF/dT versus T).
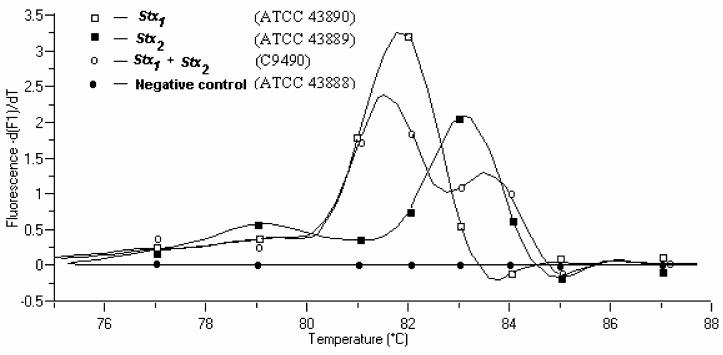
Amplicons corresponding to the regions for stx1 (227 bp with a Tm value of 81.8°C) and/or stx2 (224 bp with a Tm of 83.8°C) were observed in various E. coli O157:H7 isolates (Fig. 1), and the assay gave the expected results with the control strains: ATCC 43890, ATCC 43889, C9490, and ATCC 43888 (Table 1). The curve analysis of the two Tm peaks, corresponding to the two PCR products in the same reaction tube, could be distinguished from one another. Fluorescence from SYBR Green I binding to primer-dimers did not pose a problem due to the low Tm, which was several degrees (>5°C) less than that of the specific product (Fig. 1). This demonstrates that target sequences from these two genes could be simultaneously amplified and detected in a duplex PCR assay using a single primer pair. It was not possible to separate PCR products when they were analyzed by agarose gel electrophoresis due to the difference of only three bases between the PCR products (Table 1).
FIG. 1.
Tm curve analysis of SYBR Green assay using real-time PCR. Tm was 81.8 and 83.8°C for stx1 (ATCC 43890) and stx2 (ATCC 43889), respectively. Sample 3 contained both stx1 and stx2 (C9490), and the Tm could be resolved between the two products. Sample 4 was a negative control (ATCC 43888).
Of the 24 E. coli O157:H7 strains tested, 17 were positive for both stx1 and stx2, one was positive for stx1, 4 were positive only for stx2, and one was negative for both the Shiga toxin genes. Other isolates positive for both stx1 and stx2 were one O111:H− strain and one O113:H21 strain. Isolates of one O157:H7 strain, two O103:H2 strains, two O26:H11 strains, two O111:H8 strains, and one O45:H2 strain were positive only for stx1 and negative for stx2; whereas four O157:H7 strains, two O91:H21 strains, and one O157:NM strain were positive for the stx2 gene only (Table 1). One O157:H7 strain, one O2:H6 strain, one O2:H8 strain, and one E. coli strain of unknown serotype were negative for both stx1 and stx2.
Non-EHEC strains were negative for both stx1 and stx2. In addition, the primers did not produce any signal from other bacteria tested, including Listeria monocytogenes, Listeria grayii, Listeria ivanovii, Salmonella enterica serovar Typhimurium var. Copenhagen PT 10 SA, S. enterica serovar Enteritidis, Shigella sonnei, Yersinia enterocolitica, and Proteus vulgaris.
A single primer was chosen to amplify both stx1 and stx2 gene sequences to detect E. coli O157:H7, since strains of human origin would be predicted to possess these two virulence genes based on the prevalence among human isolates. To have a tool for a fast and reliable identification of pure cultures, the PCR assay was combined with a simple and rapid DNA isolation procedure. The direct isolation of bacterial DNA with subsequent PCR is considered to be a valuable tool for monitoring E. coli O157:H7 in mixed microbial populations. The sensitivity of the PCR assay when coupled with a simple DNA extraction method involving boiling, chilling, and centrifugation was comparable with that for chromosomal DNA targets obtained using commercial kits (data not shown).
The specificity of a single primer pair for amplification of Shiga-like-toxin genes by PCR has been well documented in other studies (3, 5). In the SYBR Green PCR assay with a single primer pair, two PCR products were generated (stx1 and stx2) with different Tm values, and these could be resolved in the LightCycler in the same reaction tube based on Tm curve analysis. While there was no problem in fluorescence data acquisition during amplification, problems were encountered during data acquisition for Tm curve analysis at the end of the amplification, especially when more than 10 samples in glass capillaries were present. Since only one detector has to acquire fluorescence data from nearly 32 samples at a difference of 0.1°C/s, when large numbers of samples are processed, the calculation of Tm curves is variable, as are the area under the specific product peak and total area under the Tm curve. However, there was no problem if the number of samples analyzed was restricted to 10 or less.
HotStart PCR provided better performance and specificity. A positive reaction was obtained using the LightCycler from inoculated and control water samples containing E. coli O157:H7 at concentrations down to 8.4 × 103 CFU/ml. No signals were obtained with all control samples that were not inoculated with E. coli O157:H7. It was possible to routinely quantify as few as eight cells per reaction. Sensitivity was increased 10-fold, either by concentration of DNA in the sample or by increasing the sample volume from 1 to 10 μl in a 100-μl reaction volume. At present 10- to 20-μl samples can be evaluated in other real-time PCR machines that are available commercially.
A number of other studies have been carried out to detect E. coli O157:H7 by real-time PCR. The sensitivity of the fluorogenic PCR assay reported by Sharma et al. (8) for EHEC O157:H7 was 2.37 × 104 CFU/ml, but the actual number of cells required to produce a positive signal was 50 for the multiplex PCR and 17 to 170 for the non-multiplex PCR assays. A real-time PCR with molecular beacons was capable of detecting E. coli O157:H7 when >102 CFU/ml was present in the samples, and as few as 1 CFU/ml in raw milk and apple juice was detected after 6 h of enrichment (2). McKillip and Drake (6) used a molecular beacon probe designed to hybridize with a region of the stx2 gene to detect E. coli O157:H7 in artificially contaminated skim milk. The degree of fluorescence increased above background levels by cycle 8, 14, or 14 in reaction mixtures containing DNA from 107, 105, or 103 CFU/ml, respectively. The detection limit of the eaeA-targeted 5′ nuclease (Taqman) detection assay was ≥103 CFU/ml for E. coli O157:H7 present in pure culture and using the QIAamp tissue kit for DNA extraction (7). In the present study the same sensitivity was achieved without the use of a DNA extraction kit by mere boiling and chilling. Use of immunomagnetic beads may further improve the sensitivity by 100- to 1,000-fold by concentrating cells from large volumes of the water or milk sample (1, 4).
Acknowledgments
This study was supported by Dairy Farmers of Ontario and Natural Sciences and Engineering Research Council of Canada.
REFERENCES
- 1.Chapman, P. A., A. T. C. Malo, C. A. Siddons, and M. Harkin. 1997. Use of commercial enzyme immunoassays and immunomagnetic separation systems for detecting Escherichia coli O157 in bovine fecal samples. Appl. Environ. Microbiol. 63:2549-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fortin, N. Y., A. Mulchandani, and W. Chen. 2001. Use of real-time polymerase chain reaction and molecular beacons for the detection of Escherichia coli O157:H7. Anal. Biochem. 289:281-288. [DOI] [PubMed] [Google Scholar]
- 3.Fratamico, P. M., S. K. Sackitey, M. Wiedmann, and M. Y. Deng. 1995. Detection of Escherichia coli O157:H7 by multiplex PCR. J. Clin. Microbiol. 33:2188-2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant, I. R., H. J. Ball, and M. T. Rowe. 1998. Isolation of Mycobacterium paratuberculosis from milk by immunomagnetic separation. Appl. Environ. Microbiol. 64:3153-3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karch, H., and T. Meyer. 1989. Single primer for amplifying segments of distinct Shiga-like toxin genes by polymerase chain reaction. J. Clin. Microbiol. 27:2751-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKillip, J. L., and M. Drake. 2000. Molecular beacon polymerase chain reaction detection of Escherichia coli O157:H7 in milk. J. Food Prot. 63:855-859. [DOI] [PubMed] [Google Scholar]
- 7.Oberst, R. D., M. P. Hays, L. K. Bohra, R. K. Phebus, C. T. Yamashiro, C. Paszko-Kolva, S. J. Flood, J. M. Sargeant, and J. R. Gillespie. 1998. PCR-based DNA amplification and presumptive detection of Escherichia coli O157:H7 with an internal fluorogenic probe and the 5′ nuclease (TaqMan) assay. Appl. Environ. Microbiol. 64:3389-3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma, V. K., E. A. Dean-Nystrom, and T. A. Casey. 1999. Semi-automated fluorogenic PCR assays (TaqMan) for rapid detection of Escherichia coli O157:H7 and other shiga toxigenic E. coli. Mol. Cell. Probes 13:291-302. [DOI] [PubMed] [Google Scholar]