Abstract
Spermatogenesis is a highly regulated process that requires precise chromatin remodeling, which includes the incorporation of testis-specific histone variants. While several of these variants have been characterized, the role of H2B.W2, a member of the H2BW family, remains largely unclear. Here, we showed that H2B.W2 expression occurs mainly in spermatocytes, slightly later than its paralog H2B.W1. Cryo-electron microscopy analysis of H2B.W2-containing nucleosomes reveals a more relaxed conformation compared to canonical nucleosomes caused by weakened interactions between the outer DNA turn and the histone core. We pinpointed the N-terminal tail and α2 helix of H2B.W2 as critical regions for nucleosome destabilization. Furthermore, we identify G73 within the L1 loop as a key residue involved in disrupting higher-order chromatin structure. Our findings suggest that H2B.W2-mediated nucleosome and chromatin destabilization may play a role in regulating gene expression during spermatogenesis, with potential implications for sperm development and function.
Graphical Abstract
Graphical Abstract.
Introduction
Spermatogenesis is the process in which mature spermatozoa develop from primordial germ cells in the testis. During this process, chromatin in the germ cells and their progenies undergoes a series of critical changes, including histone acetylation and phosphorylation, the incorporation of testis-specific histone variants, and the transition of histones to protamines as round spermatids develop into elongated spermatozoa [1]. The incorporation of testis-specific histone variants plays important roles in spermatogenesis by regulating chromatin structure and gene expression.
Histone proteins are fundamental components of the chromatin structure. In addition to the canonical H2A, H2B, H3, and H4 histones, there is a diverse group of histone variants. Many of these variants, including H2A.B (H2A.Bbd), H2B.C1 (TH2B), H2B.W1 (H2BFWT), H3.4 (H3T), and H3.5, are specifically expressed in the testis. Multiple testis-specific histone variants are known to be key regulators of gene expression and chromatin structure during specific spermatogenic events. For example, H3.4 is essential for entry into meiosis, and knock-out mice lacking H3.4 fail to enter meiosis [2]. H2B.C1 is first expressed in spermatocytes and replaces the bulk of the canonical H2B in mature spermatozoa [3]. Disruption of both H2ac1 (TH2A) and H2bc1, causes spermatogenesis defects in mice [3, 4].
In addition to H3.4 and H2B.C1, H2B.W1, which is an H2BW family histone variant only found in primates, is only expressed in the mid-to-late spermatogonia during spermatogenesis [5]. It has an extended N-terminal tail, which prevents it from participating in mitotic chromosome assembly [6, 7]. GFP-H2B.W1 was localized at telomeric region in the Chinese hamster lung V79 cell line [7]. Single nucleotide polymorphisms of the H2BW1 at −9C > T (which causes a frameshift termination) or 368A > G (which alters H100 > R) are associated with male infertility in humans [8, 9]. The H2B.W1 nucleosome is destabilized, and the instability of the nucleosome is further exacerbated by the H100R mutation [5], which is link to male infertility.
In contrast to the primate-specific H2B.W1, H2B.W2 (also known as H2B.W histone 2 and H2BFM) is found in multiple mammalian lineages and exhibits a distinct tissue expression, being present in both testis and ovary [10]. Such expression profile implies a unique role for H2B.W2 in germ cell development beyond spermatogenesis. Notably, the H2BW2 gene, located on chromosome X [NC_000023.11 (104 039 956 .104042454)] in the human genome, encodes a protein with low sequence conservation across species. H2B.W2 shares only 35% and 71% amino acid identity with canonical H2B and H2B.W1, respectively. This divergence in primary amino acid sequence further supports the idea that H2B.W2 may have distinct functions compared to its homologs. However, the structural and functional properties of H2B.W2, specifically its role in nucleosome stability and chromatin compaction during spermatogenesis, remain largely unexplored.
Here, we characterized the human H2B.W2 nucleosome structure and examined its effect on chromatin conformation. We showed that the bulk of H2B.W2 protein is expressed from late spermatogonia to leptotene spermatocytes, which is slightly later than its closest homolog H2B.W1. Our structural and biophysical analyses revealed that H2B.W2 destabilizes the nucleosome core particle (NCP) by reducing the energy barrier for the unwrapping of the outer turn nucleosomal DNA by ∼35%. We identified the extended N-terminal tail, and D85 in the α2 helix region, as critical for the H2B.W2-induced nucleosome destabilization. Furthermore, we found that the G73 residue in the L1 loop is crucial for the higher-order chromatin compaction. Given the presence of the series of tightly regulated chromatin remodeling processes in spermatogenesis, our findings provide a foundation for understanding the unique role of H2B.W2 in spermatogenesis and its potential implications for proper sperm development.
Materials and methods
Human testis immunohistochemistry and immunofluorescence staining
Formalin-fixed paraffin-embedded tissue block from the nontumor region of patients with testicular cancer purchased from (Origene Technology Inc, USA) were sectioned to a thickness of 8 μm. The sectioned samples were then deparaffinized, rehydrated, and stained with a rabbit anti-hsH2B.W2 antibody (prepared in-house, at 1:200 dilution, based on a peptide epitope, 40–54, CRGSRRRHANRRGDS, Shanghai Youke Biotechnology), a rabbit anti-hsH2B.C1 antibody (prepared in-house and used at 1:1000 dilution, 2–17, EVSSKGATISKKGFKK, Shanghai Youke Biotechnology), a rabbit anti-H2B.W1 antibody (Abcam, ab185682, at 1:35 dilution), and a rabbit anti-H4 antibody (Abcam, ab7311, at 1:1000 dilution), respectively. A rabbit specific HRP/DAB (ABC) detection IHC kit (Abcam, ab64261) was used for visualization according to the manufacturer’s protocol. Modified Mayer’s hematoxylin (Abcam, ab245880) was used to counterstain the nuclei and the images of the stained sections were captured using an Axio Scan.Z1 slide scanner (ZEISS).
For immunofluorescence staining, the sections were costained with γ-H2A.X and H2B.W2 antibodies. A monoclonal mouse antiphospho-histone H2A.X (Ser139) antibody, clone JBW301 (Sigma–Aldrich, 05-636, at 1:100 dilution) was added to the anti-H2B.W2 antibody solution (at 1:100 dilution). After washing in PBST, the sections were incubated with a mixture of Alexa Fluor™ 647 goat-anti-rabbit antibodies (Invitrogen, A-21245, at 1:250 dilution) and Alexa Fluor™488 donkey-anti-mouse (Invitrogen, A-21202, at 1:250 dilution). The cell nuclei were counterstained with freshly diluted Hoechst 33 342 (at 5 μg/μl). Images of the immunofluorescence-stained cells were captured using an LSM 980 (ZEISS) confocal microscope.
Nucleosome and tri-nucleosome reconstitution
The histone octamer was prepared by mixing an equal molar amounts of each human canonical histone H2A (histone H2A type 1-B/E, NP_003504.2), H2B (histone H2B type 1-K, NP_542160.1), H3 (histone H3.1, NP_001363866.1), and H4 (histone H4, NP_001029249.1) or human H2B.W2s (wild-type or modified H2B.W2s), in 40 μl of unfolding buffer [6-M guanidinium chloride, 20-mM Tris–HCl, pH 7.5, 4-mM dithiothreitol (DTT)] with incubation at room temperature for 1 h, followed by 4 h dialysis against water and overnight equilibrium dialysis in octamer buffer [2-M NaCl, 50-mM Tris–HCl, pH 7.5, 0.5-mM ethylenediaminetetraacetic acid (EDTA), 5-mM β-mercaptoethanol] at 4°C. H2B, H2B.W2, domain swapping H2B.W2, or point mutated H2B.W2 octamers were mixed with 147 bp [for the cryo-electron microscopy (cryo-EM) structure], 184 bp (for the MNase assay), 258 bp (for the salt stability assay), or 507 bp [for tri-nucleosome fluorescence resonance energy transfer (FRET) assay] DNA containing the 601 Widom sequence at an optimized molar ratio in the 2-M reconstitution buffer (2-M NaCl, 10-mM Tris–HCl, pH 7.5, 0.5-mM EDTA, 5-mM β-mercaptoethanol). All DNA was amplified by polymerase chain reaction (PCR) from plasmids carrying the respective nucleosome positioning sequences using primers listed in Supplementary Table S1 and purified with a QIAquick PCR purification kit (Qiagen). The mixture was then dialyzed overnight by gradually decreasing the salt concentration to 0 M. Nucleosome samples were then verified using a 5% polyacrylamide native 1× Tris-borate-EDTA (TBE) polyacrylamide gel electrophoresis (PAGE).
Cryo-EM sample preparation
H2B.W2 nucleosome samples were loaded with a 147-bp 601 DNA template (Supplementary Table S1), and then cross-linked in the 0-M reconstitution buffer (10-mM Tris–HCl, pH 7.5, 0.5-mM EDTA), 0.15% glutaraldehyde (Sigma) at 4°C for 2.5 h. The nucleosome samples were concentrated and purified using a Mini Prep Cell system (Bio-Rad), after which they were ready to be prepared for cryo-EM preparation. First, the quality of the sample was checked by negative staining using uranyl acetate in a 3.05-mm diameter carbon film copper specimen grid. Images of the stained particles were captured by scanning the specimen grid through a Talos120 kV transmission electron microscope (TEM) microscope (HKUST Biological Cryo-EM Center). Those nucleosomes that passed the quality control were then loaded into a freshly glow discharged holey carbon golden grid (Quantifoil™ R 2/2 on 300 gold mesh) (2.5 μg of nucleosome diluted in 40-mM NaCl reconstitution buffer in 3 μl was loaded in the grid) with blotting force at 0 and 100% humidity for 4.5 s with Vitrobot IV (Thermo Scientific). Micrographs of the nucleosome particles were collected with Krios G3i cryo-TEM microscope (Thermo Fisher Scientific) and a K3 camera (Gatan) in the HKUST Biological Cryo-EM Center with 81 000 times magnification at 300 kV. Electron exposure setting was 50 e−/Å2 and each pixel is 1.06 Å. Other details of the microscope setting details are listed in Supplementary Table S2.
Image processing, model building, and refinement
Movies were imported into cryoSPARC (version: V4.1.2). Patch motion correction (multi), followed by patch CTF estimation (multi), was performed with default settings. Bad images were removed with the “manually curate exposure” function. One hundred micrographs were used for blob-picker to create a template for the template-picker. An extraction box size was set at 200 pixels for the extract-from-micrographs function. After 2D selection, ab initio reconstruction was performed for 3D classification. Nonuniform refinement was used for mapping.
The template model (PDB: 3LZ0) was fitted with the density map using Chimera (Version 1.16) with the highest model-vs-map correlation coefficient. The orientation of the 601 DNA sequence was further confirmed based on previous studies, which showed that the right end of the 601 DNA is more flexible [11, 12]. Then the model was further rebuilt with WinCoot following the developer’s instructions. The model was further refined and validated with the Phenix (version 1.20–4459) real-space refinement function with multiple circles and comprehensive validation (cryo-EM) function.
Micrococcal Nuclease assay
The nucleosomes with the 184-bp Widom 601 sequence DNA (2.0 μg for DNA) (Supplementary Table S1) were mixed with 0.02 unit/μl Micrococcal Nuclease (MNase; Worthington) in 10 μl reaction solution (50-mM Tris–HCl, pH 7.5, 40-mM NaCl, 2.5-mM CaCl2, and 1.9-mM DTT) and incubated at 37°C for 0, 3, 9, 15, and 21 min. After the incubation, the reactions were quenched by adding 10 μl proteinase digestion buffer [20-mM EDTA, 0.5-mg/ml Proteinase K (Invitrogen 100 005 393), 20-mM Tris–HCl, pH 7.5, and 0.25% sodium dodecyl sulphate]. The resulting products were analyzed by nondenaturing 12% PAGE in 0.5 × TBE buffer (45-mM Tris base, 45-mM boric acid, and 1-mM EDTA).
Salt stability assay
The reconstituted and purified nucleosomes were concentrated to 200 ng/μl by Amicon Ultra centrifugal filters (50 kDa MWCO, Millipore). The concentrated nucleosomes (2 μl) were mixed with salt buffer (2 μl) comprising 10-mM Tris–HCl, pH 7.5, and 0.5-mM EDTA containing 0, 0.8, 1.6, or 2.4 M NaCl at a 1:1 ratio v/v, and then incubated for 5 min at room temperature. The samples were then dialyzed against the 0-M nucleosome reconstitution buffer for 15 min on the surface of a 47-mm MF-Millipore membrane (Millipore, VSWP04700) to remove excess salt. Then the samples were loaded in a 5% native PAGE and run at the 1 × TBE buffer. After DNA staining with SYBR Safe DNA staining (Thermo Fisher, S33102), gel images were captured using a Gel Dox XR + Gel Image (Bio-Rad), and band intensities were quantified using Image Lab (Bio-Rad) as total band volumes, which are defined as the sums of the intensity values of all pixels belonging to the band area. Relative nucleosome band volumes were calculated by dividing the total band volumes of each salt condition, by the total band volumes of the respective 0-mM NaCl conditions in each salt stability gel.
Single-molecule optical tweezers assay
Single-molecule optical tweezers measurements were conducted for quantifying the nucleosomes stability [13, 14]. Purified H2B.W2 nucleosome with 208 bp DNA containing 601 nucleosome position sequence was ligated with 0.8 and 0.5 kb lambda handle DNA, obtained from PCR with biotin or digoxigenin labeled primer (BGI). A nucleosomal DNA tether is formed between the streptavidin (SA) bead and anti-digoxigenin (AD) bead held by the optical trap and micropipette, respectively (Fig. 5A) [13, 14]. To display the nucleosome outer unfolding, inner unfolding, and refolding forces in Fig. 5B, nucleosome was incubated with the buffer containing 40-mM NaCl and 20-mM Tris–HCl, pH 7.5, and pulled at the speed of 200 nm/s. To measure the inner unfolding and refolding forces, the nucleosome was incubated the buffer containing 300-mM NaCl and 20-mM Tris–HCl, pH 7.5, and pulled at the speed of 100 nm/s. These data were collected at 200 Hz and decimated to 40 Hz to extract the forces and extensions.
Figure 5.
The H2B.W2 nucleosome destabilizes the interaction between DNA and H2A–H2B.W2 by ∼35% in single molecule optical tweezers assays. (A) Schematic to show the optical tweezers setup used in the nucleosome studies. SA represents a streptavidin-coated bead and AD represents an antidigoxigenin coated bead. (B) Comparison of two typical force-extension curves obtained for canonical H2B (blue) and H2B.W2 (red) nucleosomes. (C) The representative wrapping and unwrapping events of the canonical H2B and H2B.W2 nucleosomes in the outer hopping assay. (D) Quantification of the unwrapping and rewrapping rates under different forces. The dashed lines represent lines of best fit. Data indicates the mean ± standard error of the mean (SEM).
The wrapped rate and unwrapped rate (kw and ku) in the nucleosome outer region were extracted from being held at a constant position in a low salt buffer (5-mM NaCl, 10-mM Tris–HCl, pH 7.5) at 2–5 pN force range. The data were collected at 1 kHz and were decimated to ∼250 Hz. The kw and the ku were extracted from the inverse of the lifetimes of the nucleosome unwrapped state tu-w and wrapped state tw-u, respectively. The equilibrium force (Feq) and equilibrium rate (keq) were defined from the same length of time between tw-u and tu-w. The free energy ΔG0 of the nucleosome outer DNA wrap was calculated based on the following formula [15–17]:
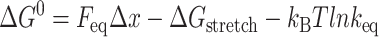 |
Here, Feq represents the equilibrium force at which the wrapping rate (kw) equals the unwrapping rate (ku). Δx refers to the average extension change of the outer unwrap in the nucleosome pulling assay. ΔGstretch denotes the energy needed to stretch the free DNA template to the Feq. The equilibrium rate (keq) corresponds to kw or ku at Feq. kBT represents the product of the Boltzmann constant (kB) and the temperature (T).
FRET assay
To investigate the impact of H2B.W2 on chromatin compaction, we reconstituted tri-nucleosome arrays using ATTO488 and ATTO594 double-end labeled N3-Widom 601–507-bp DNA templates [18]. These templates are specifically designed for tri-nucleosome reconstitution and contain three copies of the Widom 601 positioning sequence separated two 23-bp linkers, facilitating uniform nucleosome assembly (Supplementary Table S1). Following reconstitution, the arrays were dialyzed against 0-M FRET buffer (10-mM Tris–HCl, pH 7.5, 0.1-mM EDTA, 5-mM β-ME) for 4 h at 4°C and then concentrated to 200 ng/μl using Amicon Ultra-0.5 centrifugal filter units (Merck). Nucleosomes are then diluted with ice-cold 0-M FRET buffer to a concentration of 50 nM, and adding MgCl2 to varying concentrations (0, 0.2, 0.4, 0.6, 0.8, 1.0, 1.6, 2.0, and 3.2 mM). After the incubation on ice for 5 min, 40 μl aliquots of each reaction mixture were transferred to a black, flat-bottom 384-well high-throughput plate (SPL Life Sciences). The plate was incubated at 37°C for 15 min, and fluorescence signals were measured using a FlexStation 3 multimode microplate reader (Molecular Devices) (excitation/emission wavelengths: ATTO488, 488/515 nm; ATTO594, 594/610 nm). The FRET proximity ratio (P) was calculated using the following equation from Gansen et al. [19]:
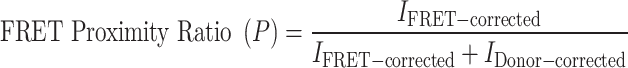 |
I FRET-corrected: fluorescence intensity of the sample in the FRET channel after subtraction of fluorescence intensity of the buffer only in the FRET channel.
I Donor-corrected: fluorescence intensity of the sample in the donor channel after subtraction of fluorescence intensity of the buffer only in the donor channel.
Fluorescence recovery after photobleaching (FRAP).
FRAP analysis was conducted on transiently transfected HeLa cell lines expressing histone-E-GFP fusion proteins according to a method from [5]. Briefly, HeLa cells were transfected with H2B-GFP or H2B.W2-GFP for 48 h, after which half of the nucleus were irradiated using a 488 nm laser for bleaching. Bleaching and fluorescence image acquisition were performed using an LSM710 DUO confocal microscope (Zeiss) with a 63×/1.4 Oil M27 objective lens using a 488-nm laser set to full power. Identical bleaching power and protocol were applied to the H2B.W2-GFP and H2B-GFP nuclei. Fluorescence images were recorded before bleaching and traced at 1-min interval after bleaching for a total duration of 40 min. Raw mean fluorescence intensities of the bleaching target (ITarget-raw) and un-bleached control (IControl-raw) areas on the nucleus and the noncell background (IBG-raw) were measured using Fiji (v2.14.0). Backgrounds were subtracted from the raw intensities and the acquisition bleaching normalization factor at each timepoint [NF(t)] was calculated for each nucleus using Equation (1).
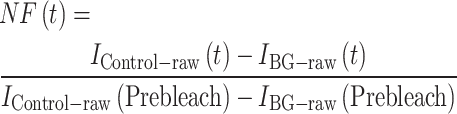 |
(1) |
The normalized relative FRAP recovery [RFRAP(t)] was computed using Equation (2), with the pre-bleaching datapoint defined as 1.
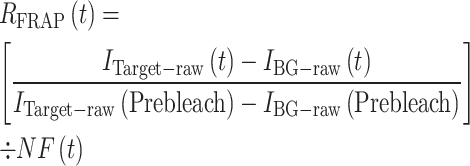 |
(2) |
To extract kinetics parameters from the observed FRAP recoveries, normalized relative FRAP recovery traces aligned at Y = 0 were fitted to a one-phase exponential recovery model (Equation 3) using GraphPad Prism (v9.5.1). The mobile fraction (%) was taken as Y∞ × 100, where K was taken as the recovery constant (min−1), and the recovery half-time T1/2 (min) was calculated using Equation (4).
 |
(3) |
 |
(4) |
Results
H2B.W2 Expression is restricted to early spermatogenesis
Understanding the temporal and spatial expression patterns of histone variants provides valuable insights into their potential functions during spermatogenesis. Given that the sequence of H2B.W2 is quite different from canonical H2B and another H2BW family histone variant, H2B.W1 (Supplementary Fig. S1), we investigated the expression of H2B.W2 in human testis using immunohistochemistry staining with a home-raised anti-H2B.W2 antibody (Fig. 1A, and Supplementary Figs S1 and S2). The results showed that H2B.W2 is only expressed in the early stages of spermatogenesis and slightly later than H2B.W1. In contrast, H2B.C1 is deposited on the chromatin of male germ cells during late spermatogenesis (Fig. 1A). To further determine the H2B.W2 expression pattern, we performed a co-staining of H2B.W2 with phosphorylated H2A.X (γ-H2A.X) (Fig. 1B and Supplementary Fig. S3). γ-H2A.X patterns can be used to identify spermatogenesis stages with decent accuracy [20, 21]. Several small double-strand DNA breaks (DSBs) and the γ-H2A.X signal appears in the middle of spermatogonia (ii, iii). The green γ-H2A.X signal is primarily limited in the sex bodies in pachytene spermatocytes (iv), and the signals largely disappeared in a round spermatid (v) [20]. Based on the γ-H2A.X signal pattern, we found that H2B.W2 is mainly present in spermatocytes and is markedly decreased in round spermatids (Fig. 1B and Supplementary Fig. S3). Furthermore, our analysis of a previously published single-cell RNA sequencing dataset (GEO: GSE106487) further supports these results (Fig. 1C) [22]. This dataset revealed the distinct expression patterns of several histone variants. H2B.C1 is expressed in the later stages of spermatogenesis, while H2B.W1 is primarily expressed in spermatogonia and leptotene spermatocytes. In contrast, H2B.W2 is expressed slightly later than H2B.W1 in differentiated spermatogonia and spermatocytes [22] (Fig. 1C and Supplementary Fig. S4). Interestingly, H2B.W2 is also expressed in around 40% of H2B.W1 expressing cells, and the expression of H2B.W2 also partially overlaps with that of H2A.X, H3.3, and another testis-specific histone variant, H3.4. However, this expression is slightly earlier than that of H1T and H3.5 (Fig. 1C). Our results indicate that H2B.W2 expression begins in late spermatogonia, then peaked in leptotene and zygotene spermatocytes, and decreases in round spermatids (Fig. 1D).
Figure 1.
H2B.W2 is mainly expressed in spermatocytes, which is later than another histone variant H2B.W1. (A) Sections of human testis were immunostained with an (a) anti-H4, (b) anti-H2B.C1, (c) anti-H2B.W1, or (d) anti-H2B.W2 antibody. The region bounded by the black rectangle in (d) is shown at higher magnification in (d’), where H2B.W2-positive spermatocytes are indicated by black arrows. Scale bars are 50 μm (a-d) and 10 μm (d’). (B) (Upper panel) Representative immunofluorescence staining examples of human testis sections with anti-H2B.W2 (red) and anti-phospho-histone H2A.X (γ-H2A.X; green) antibodies. The DNA was counter-stained with Hoechst 33 342 (blue). Scale bars = 50 μm. (Lower panel) Enlarged images of representative cells at different stages of spermatogenesis, which were extracted from the merged image shown in the upper row panel or merged images of samples treated with the same immunofluorescence staining. These images show: an early spermatogonia located at the outermost part of seminiferous tubule without any γ-H2A.X signal in its oval-shaped nucleus (i); a mid-late spermatogonia with very weak γ-H2A.X signal and weak H2B.W2 signal that appear as numerous small foci (ii); an early spermatocyte with strong and globally distributed γ-H2A.X signal and strong H2B.W2 signal (iii); a pachytene spermatocyte with strong γ-H2A.X-staining in the sex bodies and weaker H2B.W2 signal (iv); a round spermatid without γ-H2A.X staining and weak H2B.W2 signal (v); an elongating spermatid with condensed nuclei and almost disappeared H2B.W2 signal (vi); an elongated spermatid with conically-shaped condensed nuclei (vii). H2B.W2 (red), γ-H2A.X (green), and DNA (blue). Scale bars = 2 μm. (C) Uniform Manifold Approximation and Projection plots of 2854 human testicular cells (left panel). This plot is the same as the figure from Ding et al. (2024) [5]. The expression heat map of stage marker genes and histone variants according to different stages (right panel). The single-cell RNA sequencing data of human testicular cells were rebuilt from a data set previously published by Wang M. et al. [22]. H2B variants are highlighted in red. (D) Schematic view of H2B variant expression in spermatogenesis.
H2B.W2 Destabilizes nucleosome structure
While this specific expression pattern suggests a potential role for H2B.W2 during meiosis, its precise function at the molecular level remains unclear, especially on the nucleosome level. To gain further insights into the structural basis of H2B.W2 function, we determined the cryo-EM structure of the H2B.W2 NCP (PDB: 9JC6) using 601 DNA (Supplementary Figs S5 and S6) and compared it with the structures of H2B (PDB: 8JBX) and H2B.W1 (PDB: 8CJJ) NCPs [5]. A total of 264 491 particles were selected from micrographs of H2B.W2 NCPs and classified into several distinct groups (Supplementary Fig. S6). From these, we chose 127 551 particles of homogeneous H2B.W2 NCPs, resulting in a final cryo-EM map with a resolution of 3.34 Å (Fig. 2A, Supplementary Table S2, and Supplementary Fig. S6).
Figure 2.
Cryo-EM structure of nucleosomes containing human H2B.W2. (A) Cryo-EM sharped density map of the H2B.W2 nucleosome at 3.34 Å resolution with contoured at 1.6σ and (B–D) atomic models of (B) H2B.W2-NCP, (C) H2B-NCP, and (D) H2B.W1-NCP, in disc (middle) and gyre (left and right) views. The H2B.W1 model was updated from previous version [5] with lower contour level set at 2.0σ. The same applies to all H2B.W1-related figures in this manuscript. The right most panel shows that distance changes between H2BS123-H2B’S123 in the H2B-NCP, H2B.W2L145-H2B.W2’L145 in the H2B.W2-NCP, and H2B.W1L144-H2B.W1’L144 in the H2B.W1-NCP.
Our structural analysis indicated that the H2B.W2-NCP is similar to the H2B-NCP in overall structure (Fig. 2A–C and Supplementary Fig. S7A), as well as the H2B.W2 and H2B structures (Supplementary Fig. S7C). However, we observed distinct conformational and structural rearrangements, including a flexible DNA end and an increased width of the nucleosome (Fig. 2B and C, and Supplementary Fig. S7A and B). The H2B.W2 model orientation was chosen based on the observed asymmetry in DNA density according to prior studies that found the “Right” end of 601 DNA to be less well-resolved than the “Left” end [11, 12, 23]. Specifically, in the H2B-NCP, we could resolve 147 bp of nucleosomal DNA, but the last 13 bp at the 3′ end of the 601 sequence of the H2B.W2 nucleosomal DNA remained unresolved (density map was contoured at 1.6σ for model building). We observed that the 3′end of the 601 DNA (i.e. 601-Right) is present at lower occupancy, likely due to partial unwrapping (Supplementary Fig. S6F). Furthermore, the nucleosome width—in terms of the distance from H2BS123 to H2B'S123, or from H2B.W2L145 to H2B.W2'L145—increased by 3.9 Å, from 61.0 Å in H2B-NCP to 64.9 Å in H2B.W2-NCP (Fig. 2B and C). In the H2B-NCP structure (PDB: 8JBX), H2B V118, and Y121 (chain D) pack against neighbouring residues in H2A (chain C): H2B Y121 interacts with H2A R17 and V49, while H2B V118 packs against H2A V49 and Y50 (Supplementary Fig. S8A, middle). These are replaced by smaller residues H2B.W2 A140 and T143, which lack the side-chain bulkiness to sustain equivalent interactions in H2B.W2 (Supplementary Figs S1 and S8A, left). Similar effects apply to the H2B.W1 nucleosome (PDB: 8CJJ) (Fig. 2D and Supplementary Figs S7A and B, and S8A). These changes likely reduce the inward pull on the variants’ αC helix, resulting in a more relaxed, outward-rotated conformation in H2B.W2 and H2B.W1 nucleosomes, resulting in wider nucleosome structures.
Upon rebuild of the H2B.W1 cryo-EM data using the same lowered contour threshold (i.e. density map was contoured at 2.0σ for model building) applied to the H2B.W2 reconstruction, we find that the H2B.W1 and H2B.W2 nucleosomes exhibit comparable DNA end resolution and flexibility (Fig. 2D). A similar asymmetric flexibility of the two DNA ends was also observed in the CENPA-NCP [23]. However, the H2B.W1-NCP was missing 11 bp at one end and 18 bp at the other compared to the H2B-NCP when a higher counter threshold was used (i.e. density map was contoured at 3.5σ for model building) [5]. This previously reported disorderliness at both DNA ends of the H2B.W1 nucleosome structure reflected a failure to model these regions at the higher thresholds used in the original analysis.
Even though the H2A/H2B.W2 and H2A/H2B.W1 dimers are very similar as the H2A/H2B dimer in the nucleosome structures (Fig. 3A and Supplementary Fig. S7D), there are still a few differences between these variants and H2B. H2A-R42, H2A-R77, H2B-R33, and DNA form the minor groove anchors, and these anchors are critical for nucleosome stability or the dimer–DNA interaction [24]. In the H2B.W2 nucleosome, the corresponding amino acid of H2B-R33 is H2B.W2-D53, which has an opposite electrostatic property from H2B-R33 (Supplementary Fig. S1). Together with other negative charged amino acids residues at the N-terminus of H2B.W2, they made the N-terminus of H2B.W2 more flexible, also the H2B-R33 and DNA anchor corresponding interaction cannot be observed in the H2B.W2 nucleosome. Besides, even though the DNA around H2A-R77 in the H2B.W2-NCP is resolved at the lower threshold of 2.0σ (Fig. 3B), the same region cannot be resolved when a higher contour threshold was applied (3.5σ for model building). This structural change was only observed on one side, and H2A'-R77 in the H2B.W2-NCP was inserted into a minor grove of DNA on the other side (Supplementary Fig. S8B). These effects were also observed in the H2B.W1-NCP (Fig. 3B and Supplementary Fig. S8B). Similar to the H2A.B-NCP, the last 5–10 bp of superhelical location (SHL) ±5 regions in the H2B.W2-NCP exhibit conformational fluctuations (Fig. 2B and C, and Supplementary Fig. S7B) [25]. However, unlike the H2A.B-NCP, the H2A–H2B.W2 dimer does not tilt away from the histone H3-H4 tetramer (Fig. 3A and Supplementary Fig. S7D). Again, the N-terminal tails of both H2B.W2 and H2B.W1 may contribute to these fluctuations. While H2B interacts through end of N-terminal tail, R31, R33, and K34, neither H2B.W1 nor H2B.W2 possess these positive charge residues [26, 27]. These positively charged residues, located between the two DNA gyres, neutralize the electrostatic repulsion between adjacent phosphate backbones. Their absence in H2B.W2 and H2B.W1 likely reduces this stabilizing effect, contributing to the outward DNA shift (Supplementary Fig. S8C).
Figure 3.
H2B.W2 increases the negative charges including the acidic patch region. (A) Comparison of the overall structure of H2A–H2B.W2, H2A–H2B, and H2A–H2B.W1 dimer within the nucleosome context. (B) Comparison of a copy of H2A/H2B dimer and DNA interaction among the H2B.W2 (left), H2B (middle), and H2B.W1 (right) nucleosomes and merged views. (C) Comparison of α1 and α2 helix of a copy of H2B.W2 (left), H2B (middle), and H2B.W1 (right) and DNA interaction, with the hidden of the L1 loop. H band was indicated by a blue dashed line. (D) Comparison of the surface electrostatic charges in the nucleosome acidic patch region in H2B.W2-NCP (left), H2B-NCP (middle), and H2B.W1-NCP (right). The regions bounded by the dotted green squares are those regions of the acidic patch in H2B and the variants. The potential display levels are between −10 and 10 kcal/(mol*e).
Notably, H2B.W2 has a negatively charged amino acid residue D53 and, together with H2B.W1, have several more negatively charged residues in their extended N-terminal tails (Supplementary Fig. S1). Therefore, it is conceivable that the H2B.W2-NCP and H2B.W1-NCP have fewer DNA–histone interactions, which partially underlies the pronounced outward shift of DNA at SHL ±5 (Supplementary Fig. S7B). Beyond differences in the N-terminal tail, sequence variations in helices α1 and α2 also likely contribute to this observed DNA displacement. In helix α2 of H2B-NCP (8JBX), H2B S56 and K57 (chains D and H) are adjacent to the phosphate of nucleotide −54 (in chains I and J, respectively) (Fig. 3C and Supplementary Fig. S8D). In H2B.W2, they are replaced by Q78 (bulkier) and E79 (acidic), respectively, potentially weakening local protein–DNA interactions and further contributing to the DNA shift. In helix α1 of H2B-NCP (8JBX), H2B Y42 (chains D and H) forms a hydrogen bond with the phosphate of nucleotide −53 (chains I and J, respectively). This interaction is lost in H2B.W2, where the corresponding residue is P64, which is not able to form a hydrogen bond (Fig. 3C and Supplementary Fig. S8D). A similar outward DNA shift is also observed in the H2B.W1, however the contribution of H2B.W1 R63, R77, and E78 to the shift at SHL ±5 remains unclear.
To further confirm the length of DNA protected by the H2B.W2-NCP, we performed an MNase sensitivity assay. As expected, we observed a ∼150-bp band after MNase digestion of the H2B-NCP, consistent with previous studies (Fig. 4A) [28]. In contrast, digestion of the H2B.W2-NCP initially produced a band around 140 bp, which then shifted to a primary band around 120 bp, with even shorter fragments appearing over time (Fig. 4A). This result aligns with the DNA length observed in our cryo-EM analysis of the H2B.W2-NCP. Additionally, based on the cryo-EM structure, the extended N-terminal tail may contribute to the protection of DNA fragment size. Therefore, we made an artificial construct swapping the N-terminal tail from H2B.W2 to H2B (i.e. H2B.W2-s.H2B-Nter; Supplementary Fig. S9). Although the overall protection size of the H2B.W2-sH2B-Nter-NCP was similiar to that of the wild-type H2B.W2-NCP, the 120 bp band was more persistent in the chimeric nucleosome (Supplementary Fig. S10). These findings suggest that while the extended H2B.W2 N-terminal tail contributes to the destabilization of nucleosomal DNA ends, other regions within H2B.W2 likely play a more dominant role in altering DNA–histone interactions and overall nucleosome stability.
Figure 4.
The stability of the H2B.W2 nucleosome is decreased and the H2B.W2 protects fewer DNA. (A) Illustration of MNase digestion assay (left) and representative 12% 0.5 × TBE gel image of H2B and H2B.W2 nucleosome samples after MNase digestion for 0, 3, 9 15, and 21 min (right). (B) Nucleosome stability assay of the H2B, the H2B.W2 and the domain swapping nucleosomes. Polyacrylamide gels of the nucleosome after they were treated with different ionic concentrations (i.e. 0, 0.4, 0.8, or 1.2 M) of NaCl. (C) 0.5 × TBE 6% polyacrylamide gels of the H2B.W2-G73D, H2B.W2-E79K, H2B.W2-D85N, H2B.W2-H89N, and H2B.W2-Q101R nucleosomes after they were treated with different ionic concentrations (i.e. 0, 0.4, 0.8, or 1.2 M) of NaCl. (D) Quantification of the nucleosome band in the gels from panel (B). The data (n = 3) were compared using a Student’s t-test (two-tail, equal and unequal variances) and the asterisks indicate statistically significant differences at P <.01 (**), P <.001 (***), or P <.0001 (****), ns represents no significant statistical difference. (E) Quantification of the nucleosome band in the gels from panel (C). The data (n = 3) were compared using a Student’s t-test (two-tail, equal, and unequal variances) and the asterisks indicate statistically significant differences at P <.05 (*), P <.01 (**), ns represents no significant statistical difference.
In addition to the observed differences in mononucleosome stability, our cryo-EM structure revealed alterations in the acidic patch of the H2B.W2 nucleosome. Specifically, two acidic patch amino acids in H2B (E105 and E113) are replaced with a positive residue (K127) and a neutral residue (Q135) in H2B.W2, while H2B.W1 only has one amino acid changed (Fig. 3D and Supplementary Fig. S1). The acidic patch, formed by six H2A and two H2B amino acid residues, is well known to play a role in chromatin-level interactions, influencing nucleosome–nucleosome interactions, and the chromatin remodelling complex interactions [29]. These alternations in H2B.W2 could potentially influence higher-order chromatin structure and dynamics by modulating interactions with neighboring nucleosomes or altering the binding of chromatin remodeling factors.
The negatively charged N-terminal tail, and D85 in the α2 helix underlie the H2B.W2–nucleosome destabilization
To further understand which domains of H2B.W2 contribute to its destabilizing effect, we systematically swapped each domain with its counterpart from H2B and examined their stability under high salt environments (Fig. 4B and D, and Supplementary Figs S9 and S11). These experiments revealed that, in addition to some contribution from the N-terminal tail, the α2 helix region (H2B.W2 79–105) is the major determinant of H2B.W2-NCP stability (Fig. 4B and D, and Supplementary Figs S9 and S11). This result is consistent with a recent solid-state NMR study, which found that part of the H2B α2 helix region positioned near the H4 α3 helix exhibits enhanced structural dynamics and is likely engaged in the coordination of octamer-DNA interactions [30].
However, swapping the α1 helix region did not significantly affect H2B.W2-NCP stability or had a minor effect (Supplementary Fig. S11), as other regions might compensate for the effects of the destabilization. In fact, the N-terminal tail and the α2 helix region appear to contribute more to nucleosome stability than the α1 helix region. Given that the α2 helix region of H2B.W2 has more negative charge residues compared to H2B (Supplementary Fig. S1), we hypothesized that these charges might be responsible for the H2B.W2-NCP destabilization. To test this hypothesis, we created a series of charge-altering mutants within the L1 loop and α2 helix regions (G73D, E79K, D85N, H89N, Q101R; Fig. 4C, E and Supplementary Fig. S9). Salt stability assays of these mutants showed that neutralizing the charge of D85 (H2B.W2 D85N) significantly increased nucleosome stability compared to the wild-type H2B.W2 (Fig. 4C and E).
H2B.W2 destabilizes the nucleosome by ∼35% along the interaction regions between histone H2A–H2B dimers and DNA
To quantify the nucleosome destabilization effects of H2B.W2, we performed single-molecule optical tweezers experiments. Optical tweezers experiments allow us for precise force measurement to extract the strength of histone–DNA interaction. Nucleosomes were attached to biotin- and digoxigenin-labeled DNA handles (Fig. 5A). When the applied force was increased in the nucleosome pulling assay, we observed two distinct unwrapping events: a lower-force event (outer rip) and a higher-force event (inner rip) [31] (Fig. 5B). The outer rip corresponds to the unwrapping of the outer DNA turn, releasing ∼60–70 bp of DNA, while the inner rip reflects the unwrapping of the inner DNA turn, releasing ∼70–80 bp.
To quantify the effect of H2B.W2 on the outer rips, we performed a single-molecule optical tweezers nucleosome hopping assay at 5-mM NaCl. In this assay, we held a nucleosome tether at a constant trap position under varying force ranges to observe transitions between the wrapped and unwrapped state (Fig. 5C and D). In the H2B.W2-NCP, the lifetime of the nucleosome wrapped stage tw-u was decreased, and the lifetime of the nucleosome unwrapped state tu-w was increased compared to the H2B-NCP (Fig. 5C and D). Then, we extracted the equilibrium force (Feq), the force at which tw-u equates to tu-w, was reduced from 2.97 pN in the H2B-NCP to 2.36 pN in the H2B.W2-NCP (Fig. 5C and D, and Supplementary Table S3). We then calculated the free energy cost of the outer rip at 0 pN (ΔGo) to be 20.46 kJ/mol for the H2B-NCP, whereas it was significantly reduced to 13.22 kJ/mol for the H2B.W2-NCP [5]. This 35% reduction in the energy barrier for the outer rip in the presence of H2B.W2 is similar to the 37% reduction observed in the presence of H2B.W1 [5].
Average inner rip forces were at around ∼15 pN and ∼7 pN in the H2B-NCP and H2B.W2-NCP, respectively, in presence of 300-mM NaCl (Supplementary Fig. S12). These results are consistent with our MNase and salt stability assay data, suggesting that weakened interactions between the H2A–H2B.W2 dimer and the rest of the nucleosome promote dimer dissociation. This destabilization is reflected in the significantly lower inner unwrapping force observed in H2B.W2-NCP compared to H2B-NCP. The inner unwrapping force for H2B.W2 (∼7 pN) was also lower than that observed for H2B.W1-NCP (∼10 pN) (Supplementary Fig. S12) [5], indicating that H2B.W2 has more destabilizing effect on the nucleosome than H2B.W1.
H2B.W2 disrupts chromatin compaction through G73 in the L1 loop
Beyond the mononucleosome level, chromatin structure is the primary biologically functional form under in vivo condition, and nucleosomal DNA end interactions and linker DNA length are known to influence chromatin folding [32, 33]. We investigated whether the increased DNA ends flexibility in H2B.W2 nucleosomes impacts higher-order structure. We compared the stacking dynamics of H2B and H2B.W2 tri-nucleosomes, representing the minimal unit of chromatin, across varying Mg2+ concentrations (Fig. 6A–E, and Supplementary Figs S13–S16) [18, 34]. The H2B tri-nucleosome adopted a more compact structure with increasing Mg2+ concentration (Supplementary Figs S13 and S16), which is consistent with previous reports [18, 34]. A similar trend was observed for the H2B.W2 tri-nucleosome; however, the initial FRET signal was much lower in the H2B.W2 condition, indicating a less compact chromatin conformation at low Mg2+ concentrations (Fig. 6C and E and Supplementary Figs S13 and S16). This relative openness is maintained across all Mg2+ concentrations tested, which encompassed the physiological Mg2+ concentration at around 1 mM (Fig. 6C and E, and Supplementary Fig. S16). To test the possibility of inter-array interactions, we repeated the FRET experiment using 50/50 mixtures of donor-only and acceptor-only tri-nucleosomes. Interestingly, FRET signals of the donor-only/acceptor-only mixtures and 50/50 mixtures of donor-only and acceptor-only tri-nucleosomes are much lower than those of the corresponding bi-labeled tri-nucleosomes (Supplementary Figs S14 and S15). Therefore, the signal increased observed upon Mg2+ addition is mainly caused by intra-array interactions between nucleosomes. Notably, a crystal structure of a 12-nucleosomal fiber revealed that H2BD51, the canonical H2B residue equivalent to G73 in H2B.W2, within the L1 loop interacts with H2AR71 in neighboring nucleosomes [35]. We hypothesized that H2B.W2G73 may change the interaction with H2AR71 of neighboring nucleosome, thus changing the chromatin structure. The decreased chromatin compaction of H2B.W2 tri-nucleosome is partially reverted by the H2B.W2-G73D mutation (Fig. 6C–E and Supplementary Fig. S16). These results indicate that H2B.W2 not only destabilizes individual nucleosomes but also disrupts higher-order chromatin structure, with G73 playing a key role in this destabilization at the chromatin level.
Figure 6.
H2B.W2G73 plays an important role in opening up the chromatin structure composed of H2B.W2. (A) The schematic diagram of tri-nucleosome array condensation FRET assay. (B) Representative native PAGE of the loaded H2B, H2B.W2, and H2B.W2-G73D tri-nucleosomes. (C, D) Representative emission spectra of (C) H2B.W2 tri-nucleosome, and (D) H2B.W2-G73D tri-nucleosome from 515 nm to 710 nm upon excitation at 488 nm exposed to the indicated amounts of Mg2+ ions. The spectra were normalized to their corresponding donor emission peaks at 525 nm. (E) The relationship between the relative FRET proximity ratios (P) and the Mg2+ ion concentrations for H2B.W2 and H2B.W2-G73D tri-nucleosomes. Datapoints were normalized to the corresponding mean proximity ratios at 0- and 3.2-mM Mg2+ concentrations, which were defined as having the relative proximity ratios of 0 and 1, respectively. Each datapoint represents the mean relative proximity ratio ± 1 standard deviation (n = 3 for all conditions tested). The mean relative proximity ratios at each given Mg2+ concentrations were compared using a Student’s t-test and the asterisks indicate statistically significant differences at P <.01 (**), or P <.001 (***). (F) Representative fluorescence images of the HeLa cell nuclei expressing either H2B-GFP or H2B.W2-GFP pre-bleach and at 0-, 10-, 20-, and 40-min post-bleach. White rectangles indicate areas targeted for photo-bleaching. (G) Line graph showing the relative fluorescence signal recovery after photo-bleaching over time for H2B.W2-GPF and H2B-GFP. The mean normalized relative FRAP recoveries of the target areas immediately after bleaching (at t = 0) are defined as having relative fluorescence recovery values of 0. Datapoints represent the mean ± SEM. The mean normalized FRAP recoveries of H2B.W2-GFP and H2B-GFP at each timepoint were compared individually using the Student’s t-test (two tail, unequal variance). Asterisks indicate statistically significant differences at P <.05 (*) and unmarked timepoints indicate nonsignificant differences [n = 13 for H2B.W2 and n = 9 for H2B (H2B dataset is from [5])]. (H) Table summarizing the fluorescence recovery kinetics parameters of H2B.W2-GFP and H2B-GFP derived from the FRAP experiment in panel (G) using nonlinear regression fitted to a one-phase exponential recovery model. Values represent the mean, and the corresponding 95% confidence intervals (CIs) are shown in brackets.
To further validate the destabilizing effect of H2B.W2 on nucleosomes and chromatin within a cellular context, we performed FRAP experiments using EGFP-tagged H2B or H2B.W2 (Fig. 6F and G, and Supplementary Fig. S17). As expected, EGFP-tagged H2B or H2B.W2 were both shown to be localized in the nucleus (Fig. 6F). Same as previous reports, the EGFP fluorescence recovery of canonical H2B was very slow because canonical histones are stably bound to chromatin and thus virtually immobile (Fig. 6F–H and Supplementary Fig. S17) [36–38]. In contrast, the H2B.W2-EGFP signal exhibited a comparatively greater extent of recovery, with the mobile fraction of H2B.W2-EGFP measured as 42.41% (95% CI: 36.87–54.61%), significantly higher than the 25.33% (95% CI: 22.35–32.70%) for H2B-GFP (Fig. 6G and H). At the same time, there were no significant differences in the extracted recovery constants (K) and the recovery half-times (T1/2) (Fig. 6G and H). These results suggest that H2B.W2 is indeed more mobile and diffusive compared to canonical H2B in cells, which support the presence of less stable H2B.W2–nucleosomes in the nucleus and are in good agreement with known FRAP recovery responses of other nucleosome destabilizing histone variants such as H2A.B and H3.5 [39, 40].
Discussion
Our study reveals that H2B.W2, a testis-specific histone variant, exhibits unique structural and dynamic properties that distinguish it from canonical H2B. Specifically, we found that H2B.W2 destabilizes nucleosomes, disrupts chromatin compaction, and exhibits increased mobility within the nucleus. These findings suggest a specialized role for H2B.W2 in regulating chromatin structure and accessibility during spermatogenesis. The timing of H2B.W2 expression in spermatogenesis is distinct from that of H2B.C1, which is a well-studied testis-specific variant of histone H2B. H2B.C1 is expressed in the later stages of spermatogenesis, particularly in the spermatids, and it is known to be distributed evenly across whole chromosomes [41]. The histone variant evolutionarily closest to H2B.W2 is H2B.W1; it is expressed slightly earlier than H2B.W2 but there is some overlap in the expression stages. Single-cell RNA sequence analysis showed that ∼40% of the cells that express H2B.W1, also express H2B.W2 (Fig. 1). H2B.W1-NCP and H2B.W2-NCP have similar nucleosome destabilizing effects and are expressed in the early stages of spermatogenesis. Although it is still not clear whether disruption of H2B.W2 function might affect human spermatogenesis, disruptions of H2B.W1 gene (i.e. two SNPs of the H2B.W1 at −9C > T and 368A > G) are known to be related to infertility [8, 9]. Therefore, it is possible that mutations of H2B.W2 will also be associated to infertility. Further studies would be helpful to understand how H2B.W1 and H2B.W2 function in spermatogenesis.
The significant decrease in H2B.W2-NCP stability compared to canonical nucleosomes suggests a more dynamic role for this variant. Based on the amino acid sequence alignment and the domain swapping salt stability assay, the H2B.W2 N-terminal tail and the α2 helix significantly contributed to the nucleosome destabilization. The N terminus of H2B.W2 contains much more negatively charged amino acid residues than that of H2B, and slightly more than that of H2B.W1. Previous studies have shown that the H2B N-terminus can adopt different orientations within the nucleosome, influencing DNA–histone interactions [42]. We speculate that the excess negative charge in H2B.W2 disrupts these interactions through enhanced electrostatic repulsion with DNA. This is supported by our cryo-EM structure, which reveals increased flexibility in the H2B.W2 N-terminal region and the DNA shift at SHL (±5).
The observation that H2B.W2-NCP protected a shorter DNA region, potentially indicative of hexasome formation, further supports its role in creating a more open chromatin conformation. While we cannot exclude the possibility of hexasome formation, both asymmetric unwrapping and hexasome structures represent weaker conformations that could facilitate transcription [43]. This is particularly relevant for H2B.W2, as its destabilization effects, exerted on both the NCP structures and on chromatic folding, might be crucial for upregulating genes expressed during early spermatogenesis and facilitating the dynamical chromosomal structural changes required in spermatogenesis.
In addition to its impact on nucleosome stability, H2B.W2 also influences for the chromatin compaction. G73, located in the L1 loop of H2B.W2, appears to play a role in chromatin compaction, potentially by reducing the attraction with H2A R71 of neighboring nucleosome. This suggests that while certain regions of H2B.W2 contribute to nucleosome destabilization, other parts may be involved in modulating higher-order chromatin structure. Furthermore, our FRAP assay revealed a significantly larger intranuclear mobile fraction for EGFP-tagged H2B.W2 compared to EGFP-tagged H2B, suggesting increased mobility of H2B.W2 within the nucleus. This enhanced mobility likely results from the combined effects of destabilized H2B.W2–nucleosomes and relaxed H2B.W2-containing chromatin structures. This observation aligns with recent findings highlighting the importance of positive charges in histone tails for stable nucleosome association [40]. Including the changes in the acidic patch region, these results suggest that the altered charge distribution in H2B.W2 not only destabilizes its nucleosome structure but also makes it more accessible to chromatin remodelers and histone chaperones or promote the recruitment of spermatogenesis-specific factors. Similar destabilization effects are observed in other testis-specific histone variants such as H2B.W1, H3.4, H3.5, and H2A.B, suggesting distinct functions compared to canonical histones [2, 5, 39, 44–46]. H3.5, present near transcription start sites (TSSs) in spermatogonia, may facilitate H3.3 replacement [39]. Canonical histone H3.1/H3.2 is replaced by H3.4 during spermatogonial stem cell differentiation, and H3.4 deletion arrests meiosis, causing infertility [2]. H2A.B accumulates at TSSs and gene bodies of transcribing genes [45, 46], forming an open intermediate conformation that can even replace H2A without additional factors [44]. This instability, similar to other histone variants, likely facilitate rapid chromatin remodelling during early spermatogenesis form meiosis and histone-to-protamine replacement. Given the partially overlapping expression of H3.4 and H2B.W2, investigating their potential co-occupancy within nucleosomes and the functional consequences of this potentially further destabilized nucleosome would illustrate the roles of histone variants in spermatogenesis.
This increased accessibility, coupled with the potential for hexasome formation, raises intriguing questions about the localization and function of H2B.W2 during spermatogenesis. While canonical histones typically exhibit a more widespread distribution, unique properties of H2B.W2 might target it to specific genomic regions, as hinted by the uneven and dotted staining patterns observed in H2B.W2-positive spermatocytes (Fig. 1A and B, and Supplementary Fig. S3). Interestingly, H2B.W1 has been found to localize at telomeric regions in the V79 cell line [7]. Transiently expressed H2B.W2 in HeLa cells also localized to sub-telomeric and telomeric regions (data not shown), it is possible that the function of H2B.W2 may be similar to that of H2B.W1. Since spermatogenic cells are different from cultured cell lines, both H2B.W2 and H2B.W1 may be localized at different locus in male germ cells. Investigating their localization in these cells in telomeric or elsewhere and their involvement in telomere maintenance or gene regulation is necessary for understanding the functional role of H2B.W2. Given the sequence specificity of CENP-A for pericentromeric major satellite DNA, exploring whether H2B.W2 exhibits similar sequence specificity is also interesting [47]. Moreover, destabilized nucleosomes have been implicated in homologous recombination [48, 49]. The destabilization of nucleosomes and chromatin in early spermatogenesis by H2B.W2 could potentially facilitate meiotic recombination. This is consistent with reports demonstrating the importance of chromatin remodeling during meiosis [50]. Further investigation of the precise mechanisms by which H2B.W2 destabilizes chromatin structure within the context of native genomic sequences will be essential for fully understanding the complexities of spermatogenesis and male fertility.
Supplementary Material
Acknowledgements
EM dataset was collected at the Biological Cryo-EM Center, generously supported by a donation from the Lo Kwee Seong Foundation, at HKUST.
Author contributions: April - Thi Thuy Nguyen (Investigation [equal], Methodology [equal], Writing—original draft [equal], Writing—review & editing [supporting]), Yue LIU (Investigation [supporting], Methodology [supporting]), Tingyu Jin (Investigation [supporting], Methodology [supporting], Visualization [supporting], Writing—review & editing [supporting]), Zhichun Xu (Investigation [supporting], Resources [supporting]), Yingyi Zhang (Methodology [supporting], Supervision [supporting], Validation [supporting], Visualization [supporting]), Toyotaka Ishibashi (Conceptualization [lead], Funding acquisition [equal], Methodology [supporting], Project administration [lead], Supervision [lead], Writing—original draft [lead], Writing—review & editing [lead])
Contributor Information
April T T Nguyen, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China; Present address:Centre for Translational Stem Cell Biology, Hong Kong Science and Technology Park, Shatin, New Territories, Hong Kong, HKSAR, China.
Dongbo Ding, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China; Present address:Institute of Biophysics, Chinese Academy of Science, Chaoyang, Beijing 100101, China.
Xingpeng Bai, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China.
Matthew Y H Pang, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China.
Mingxi Deng, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China.
Yue Liu, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China.
Tingyu Jin, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China.
Zhichun Xu, School of Biological Sciences, The University of Hong Kong, Pokfulam, Hong Kong, HKSAR, China.
Yingyi Zhang, Biological Cryo-EM Center, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China.
Yuanliang Zhai, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China; School of Biological Sciences, The University of Hong Kong, Pokfulam, Hong Kong, HKSAR, China.
Yan Yan, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China; Shenzhen Peking University, The Hong Kong University of Science and Technology Medical Center, Shenzhen, Guangzhou 518055, China.
Toyotaka Ishibashi, Division of Life Science, The Hong Kong University of Science and Technology, Clear Water Bay, NT, HKSAR, China; The Hong Kong University of Science and Technology Fok Ying Tung Research Institute, Nansha, Guangzhou 511462, China.
Supplementary data
Supplementary data is available at NAR online.
Conflict of interest
None declared.
Funding
This work was supported grants from the Hong Kong Innovation & Technology Commission Innovation and Technology Fund [MHP/033/20], the National Natural Science Foundation of China [32170548] awarded to T.I., Shenzhen Science and Technology Innovation Commission [JCYJ20200109140201722] to YY, and grants from the Research Grants Council of the Hong Kong SAR [C6036-21GF] awarded to Y.Z. and T.I., [C7035-23G] to Y.Z., and [16104324], [16104725], [T13-602/21N] to YY.
Data availability
The structure of the H2B.W2 nucleosome is available in the PDB (PDB ID: 9JC6), and the other data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Hammoud SS, Nix DA, Zhang H et al. Distinctive chromatin in human sperm packages genes for embryo development. Nature. 2009; 460:473–8. 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ueda J, Harada A, Urahama T et al. Testis-specific histone variant H3t gene is essential for entry into spermatogenesis. Cell Rep. 2017; 18:593–600. 10.1016/j.celrep.2016.12.065. [DOI] [PubMed] [Google Scholar]
- 3. Montellier E, Boussouar F, Rousseaux S et al. Chromatin-to-nucleoprotamine transition is controlled by the histone H2B variant TH2B. Genes Dev. 2013; 27:1680–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shinagawa T, Huynh LM, Takagi T et al. Disruption of Th2a and Th2b genes causes defects in spermatogenesis. Development. 2015; 142:1287–92. [DOI] [PubMed] [Google Scholar]
- 5. Ding D, Pang MY, Deng M et al. Testis-specific H2B. W1 disrupts nucleosome integrity by reducing DNA–histone interactions. Nucleic Acids Res. 2024; 52:11612–25. 10.1093/nar/gkae825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boulard M, Gautier T, Mbele GO et al. The NH2 tail of the novel histone variant H2BFWT exhibits properties distinct from conventional H2B with respect to the assembly of mitotic chromosomes. Mol Cell Biol. 2006; 26:1518–26. 10.1128/MCB.26.4.1518-1526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Churikov D, Siino J, Svetlova M et al. Novel human testis-specific histone H2B encoded by the interrupted gene on the X chromosome. Genomics. 2004; 84:745–56. 10.1016/j.ygeno.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 8. Lee J, Park HS, Kim HH et al. Functional polymorphism in H2BFWT-5′ UTR is associated with susceptibility to male infertility. J Cellular Molecular Medi. 2009; 13:1942–51. 10.1111/j.1582-4934.2009.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ying H-q, Scott MB, Zhou-Cun A Relationship of SNP of H2BFWT gene to male infertility in a Chinese population with idiopathic spermatogenesis impairment. Biomarkers. 2012; 17:402–6. 10.3109/1354750X.2012.677066. [DOI] [PubMed] [Google Scholar]
- 10. Papatheodorou I, Moreno P, Manning J et al. Expression Atlas update: from tissues to single cells. Nucleic Acids Res. 2020; 48:D77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ngo TT, Zhang Q, Zhou R et al. Asymmetric unwrapping of nucleosomes under tension directed by DNA local flexibility. Cell. 2015; 160:1135–44. 10.1016/j.cell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chua EY, Vasudevan D, Davey GE et al. The mechanics behind DNA sequence-dependent properties of the nucleosome. Nucleic Acids Res. 2012; 40:6338–52. 10.1093/nar/gks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jing Y, Ding D, Tian G et al. Semisynthesis of site-specifically succinylated histone reveals that succinylation regulates nucleosome unwrapping rate and DNA accessibility. Nucleic Acids Res. 2020; 48:9538–49. 10.1093/nar/gkaa663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wan YCE, Leung TCS, Ding D et al. Cancer-associated histone mutation H2BG53D disrupts DNA–histone octamer interaction and promotes oncogenic phenotypes. Sig Transduct Target Ther. 2020; 5:1–4. 10.1038/s41392-020-0131-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bustamante C, Chemla YR, Forde NR et al. Mechanical processes in biochemistry. Annu Rev Biochem. 2004; 73:705–48. 10.1146/annurev.biochem.72.121801.161542. [DOI] [PubMed] [Google Scholar]
- 16. Li W, Chen P, Yu J et al. FACT remodels the tetranucleosomal unit of chromatin fibers for gene transcription. Mol Cell. 2016; 64:120–33. 10.1016/j.molcel.2016.08.024. [DOI] [PubMed] [Google Scholar]
- 17. Liphardt J, Onoa B, Smith SB et al. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001; 292:733–7. 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 18. Poirier MG, Oh E, Tims HS et al. Dynamics and function of compact nucleosome arrays. Nat Struct Mol Biol. 2009; 16:938–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gansen A, Felekyan S, Kühnemuth R et al. High precision FRET studies reveal reversible transitions in nucleosomes between microseconds and minutes. Nat Commun. 2018; 9:4628. 10.1038/s41467-018-06758-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Blanco-Rodríguez J γH2AX marks the main events of the spermatogenic process. Microsc Res Tech. 2009; 72:823–32. 10.1002/jemt.20730. [DOI] [PubMed] [Google Scholar]
- 21. Hamer G, Roepers-Gajadien HL, van Duyn-Goedhart A et al. DNA double-strand breaks and γ-H2AX signaling in the testis. Biol Reprod. 2003; 68:628–34. 10.1095/biolreprod.102.008672. [DOI] [PubMed] [Google Scholar]
- 22. Wang M, Liu X, Chang G et al. Single-cell RNA sequencing analysis reveals sequential cell fate transition during human spermatogenesis. Cell Stem Cell. 2018; 23:599–614. 10.1016/j.stem.2018.08.007. [DOI] [PubMed] [Google Scholar]
- 23. Boopathi R, Danev R, Khoshouei M et al. Phase-plate cryo-EM structure of the Widom 601 CENP-A nucleosome core particle reveals differential flexibility of the DNA ends. Nucleic Acids Res. 2020; 48:5735–48. 10.1093/nar/gkaa246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kobayashi W, Sappler AH, Bollschweiler D et al. Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition. Nat Struct Mol Biol. 2024; 31:757–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhou M, Dai L, Li C et al. Structural basis of nucleosome dynamics modulation by histone variants H2A.B and H2A.Z.2.2. EMBO J. 2020; 40:e105907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lehmann K, Felekyan S, Kuhnemuth R et al. Dynamics of the nucleosomal histone H3 N-terminal tail revealed by high precision single-molecule FRET. Nucleic Acids Res. 2020; 48:1551–71. 10.1093/nar/gkz1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krajewski WA, Li J, Dou Y Effects of histone H2B ubiquitylation on the nucleosome structure and dynamics. Nucleic Acids Res. 2018; 46:7631–42. 10.1093/nar/gky526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nozawa K, Takizawa Y, Pierrakeas L et al. Cryo–electron microscopy structure of the H3-H4 octasome: a nucleosome-like particle without histones H2A and H2B. Proc Natl Acad Sci USA. 2022; 119:e2206542119. 10.1073/pnas.2206542119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kalashnikova AA, Porter-Goff ME, Muthurajan UM et al. The role of the nucleosome acidic patch in modulating higher order chromatin structure. J R Soc Interface. 2013; 10:20121022. 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shi X, Kannaian B, Prasanna C et al. Structural and dynamical investigation of histone H2B in well-hydrated nucleosome core particles by solid-state NMR. Commun Biol. 2023; 6:672. 10.1038/s42003-023-05050-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mihardja S, Spakowitz AJ, Zhang Y et al. Effect of force on mononucleosomal dynamics. Proc Natl Acad Sci USA. 2006; 103:15871–6. 10.1073/pnas.0607526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Luger K, Dechassa ML, Tremethick DJ New insights into nucleosome and chromatin structure: an ordered state or a disordered affair?. Nat Rev Mol Cell Biol. 2012; 13:436–47. 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Perišić O, Collepardo-Guevara R, Schlick T Modeling studies of chromatin fiber structure as a function of DNA linker length. J Mol Biol. 2010; 403:777–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Engelhardt M Condensation of chromatin in situ by cation-dependent charge shielding and aggregation. Biochem Biophys Res Commun. 2004; 324:1210–4. 10.1016/j.bbrc.2004.09.175. [DOI] [PubMed] [Google Scholar]
- 35. Adhireksan Z, Sharma D, Lee PL et al. Near-atomic resolution structures of interdigitated nucleosome fibres. Nat Commun. 2020; 11:4747. 10.1038/s41467-020-18533-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harada A, Maehara K, Ono Y et al. Histone H3.3 sub-variant H3mm7 is required for normal skeletal muscle regeneration. Nat Commun. 2018; 9:1400. 10.1038/s41467-018-03845-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kujirai T, Horikoshi N, Sato K et al. Structure and function of human histone H3.Y nucleosome. Nucleic Acids Res. 2016; 44:6127–41. 10.1093/nar/gkw202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wiedemann SM, Mildner SN, Bönisch C et al. Identification and characterization of two novel primate-specific histone H3 variants, H3. X and H3. Y. J Cell Biol. 2010; 190:777–91. 10.1083/jcb.201002043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Urahama T, Harada A, Maehara K et al. Histone H3. 5 forms an unstable nucleosome and accumulates around transcription start sites in human testis. Epigenetics Chromatin. 2016; 9:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Arimura Y, Kimura H, Oda T et al. Structural basis of a nucleosome containing histone H2A. B/H2A. Bbd that transiently associates with reorganized chromatin. Sci Rep. 2013; 3:3510. 10.1038/srep03510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Patankar A, Gajbhiye R, Surve S et al. Epigenetic landscape of testis specific histone H2B variant and its influence on sperm function. Clin Epigenet. 2021; 13:1–18. 10.1186/s13148-021-01088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ohtomo H, Kurita J-i, Sakuraba S et al. The N-terminal tails of histones H2A and H2B adopt two distinct conformations in the nucleosome with contact and reduced contact to DNA. J Mol Biol. 2021; 433:167110. 10.1016/j.jmb.2021.167110. [DOI] [PubMed] [Google Scholar]
- 43. Burgos-Bravo F, Tong AB, Li C et al. FACT weakens the nucleosomal barrier to transcription and preserves its integrity by forming a hexasome-like intermediate. Mol Cell. 2025; 85:2097–2109.e8. 10.1016/j.molcel.2025.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hirano R, Arimura Y, Kujirai T et al. Histone variant H2A.B-H2B dimers are spontaneously exchanged with canonical H2A–H2B in the nucleosome. Commun Biol. 2021; 4:191. 10.1038/s42003-021-01707-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Soboleva TA, Nekrasov M, Pahwa A et al. A unique H2A histone variant occupies the transcriptional start site of active genes. Nat Struct Mol Biol. 2012; 19:25–30. [DOI] [PubMed] [Google Scholar]
- 46. Soboleva TA, Parker BJ, Nekrasov M et al. A new link between transcriptional initiation and pre-mRNA splicing: the RNA binding histone variant H2A.B. PLoS Genet. 2017; 13:e1006633. 10.1371/journal.pgen.1006633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arora UP, Sullivan BA, Dumont BL Variation in the CENP-A sequence association landscape across diverse inbred mouse strains. Cell Rep. 2023; 42:113178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Prado F, Aguilera A Partial depletion of histone H4 increases homologous recombination-mediated genetic instability. Mol Cell Biol. 2005; 25:1526–36. 10.1128/MCB.25.4.1526-1536.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Poirier GG, de Murcia G, Jongstra-Bilen J et al. Poly(ADP-ribosyl)ation of polynucleosomes causes relaxation of chromatin structure. Proc Natl Acad Sci USA. 1982; 79:3423–7. 10.1073/pnas.79.11.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hauer MH, Seeber A, Singh V et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat Struct Mol Biol. 2017; 24:99–107. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structure of the H2B.W2 nucleosome is available in the PDB (PDB ID: 9JC6), and the other data underlying this article will be shared on reasonable request to the corresponding author.









