Abstract
Fourier transform infrared (FT-IR) spectroscopy is a rapid, noninvasive technique with considerable potential for application in the food and related industries. We show here that this technique can be used directly on the surface of food to produce biochemically interpretable “fingerprints.” Spoilage in meat is the result of decomposition and the formation of metabolites caused by the growth and enzymatic activity of microorganisms. FT-IR was exploited to measure biochemical changes within the meat substrate, enhancing and accelerating the detection of microbial spoilage. Chicken breasts were purchased from a national retailer, comminuted for 10 s, and left to spoil at room temperature for 24 h. Every hour, FT-IR measurements were taken directly from the meat surface using attenuated total reflectance, and the total viable counts were obtained by classical plating methods. Quantitative interpretation of FT-IR spectra was possible using partial least-squares regression and allowed accurate estimates of bacterial loads to be calculated directly from the meat surface in 60 s. Genetic programming was used to derive rules showing that at levels of 107 bacteria·g−1 the main biochemical indicator of spoilage was the onset of proteolysis. Thus, using FT-IR we were able to acquire a metabolic snapshot and quantify, noninvasively, the microbial loads of food samples accurately and rapidly in 60 s, directly from the sample surface. We believe this approach will aid in the Hazard Analysis Critical Control Point process for the assessment of the microbiological safety of food at the production, processing, manufacturing, packaging, and storage levels.
The last decade has seen an exponential increase in the consumer demand for poultry and poultry products, fueled in part by dietary health considerations. Fears over microbiological food safety issues, especially the incidence of Salmonella spp. (23, 49) and Campylobacter spp. (11, 39), in conjunction with consumer demand for a product of consistently high quality, have focused attention on a particular area of the food production industry, namely, the requirement for a rapid (less than a few minutes) and accurate detection system for microbiologically spoiled or contaminated meat (3).
At present, no such technology exists in the food industry within the Hazard Analysis Critical Control Point system for the microbiological safety and quality of meat and poultry products (20, 26, 48).
Muscle foods, such as meat and poultry, are described as spoiled if organoleptic changes make them unacceptable to the consumer. These organoleptic characteristics may include changes in appearance (discoloration), the development of off odors, slime formation, changes in taste, or any other characteristic which makes the food undesirable for consumption (25, 26). While endogenous enzymatic activity within muscle tissue postmortem can contribute to changes during storage (1, 25, 32, 44), it is generally accepted that detectable organoleptic spoilage is a result of decomposition and the formation of metabolites caused by the growth of microorganisms (28, 40). The organoleptic changes which take place will also vary according to the species of microflora present, the characteristics of the meat, processing methods, product composition, and the environment in which the food is stored (25).
Provided that the atmosphere is moist, a consortium of bacteria is responsible for spoilage of meat stored at between −1 and 25°C. It is agreed that spoilage organisms belong primarily to the genus Pseudomonas, and these have been observed to attach more rapidly to meat surfaces than other spoilage bacteria (25). The other major members of the spoilage flora of meat stored aerobically include Moraxella spp., Psychrobacter spp., and Acinetobacter spp. While the gram-negative motile and nonmotile aerobic rods and coccobacilli generally dominate the spoilage microflora of meat, the initial population may also contain various levels of gram-positive genera, usually represented by micrococci and then lactic acid bacteria and Bronchothrix thermosphacta (13).
To date, in excess of 40 methods have been proposed to measure and to detect bacterial spoilage in meats (13, 26). These include enumeration methods based on microscopy, ATP bioluminescence, and the measurement of electrical phenomena (10, 45), as well as detection methods based on either immunological or nucleic acid-based procedures (42). The major drawback with the range of protocols available is that they are time-consuming and labor-intensive and give retrospective information. However, in a modern food-processing environment, monitoring procedures need to give results in real time so that corrective action can be taken as soon as possible.
The ideal method for the on-line microbiological analysis of meat would be rapid, noninvasive, reagentless, and relatively inexpensive, and these requirements can be met via the application of a spectroscopic approach, in combination with any appropriate data deconvolution strategy based on statistics or machine learning. Such statistical methods include partial least-squares (PLS) regression (36), while a popular and powerful series of machine learning strategies (37) are based on methods of evolutionary computing (4), such as genetic algorithms (GAs) (4, 19, 24) and genetic programming (GP) (6, 33). Fourier transform infrared (FT-IR) spectroscopy involves the observation of vibrations of molecules that are excited by an infrared beam, and an infrared absorbance spectrum represents a “fingerprint” which is characteristic of any chemical or biochemical substance (18, 43). This technique is also very rapid (taking seconds) and has been shown to be a valuable tool for the characterization of axenically cultured bacteria (22, 34, 38, 47), including single-gene knockout strains (41).
With the prior knowledge that organoleptic spoilage is the result of decomposition and the formation of metabolites caused by the growth of microorganisms, this information can be exploited through spectroscopic analysis. Therefore, rather than measuring exclusively the presence of bacteria per se on the meat surface, vibrational spectroscopy can also be used to measure biochemical changes within the meat substrate, enhancing and accelerating the detection of microbial spoilage. The objective of the present study was to establish this technology through a series of experiments undertaken on chicken breast muscle at room temperature and analyzed by horizontal attenuated total reflectance (HATR) FT-IR spectroscopy with PLS analysis. Our research demonstrates the utility of a novel analytical approach based on FT-IR that can enhance and accelerate the detection of microbial spoilage, providing rapid, accurate, and quantitative results. PLS methods are not always transparent as to which variables are used (2), whereas certain other methods use input variables explicitly and directly in the construction of the relationship between the spectra observed and the property of interest. GA and GP analyses fall into this category and are used here to suggest that proteolysis is the key indicator for the onset of spoilage.
MATERIALS AND METHODS
Sample preparation.
Fillets of prepacked fresh chicken breast meat were purchased from national retail outlets on the morning of each experiment. No prepreparation of the meat, such as removal of fat or connective tissue, washing, or inoculation with bacteria, was undertaken. In order to accelerate the spoilage process, the meat was first weighed and divided aseptically into 30-g subsamples and comminuted for 10 s in a Moulinex type 505 180-W coffee mill (Moulinex UK Ltd., Birmingham, United Kingdom). The bowl of the coffee mill was washed and dried with a paper towel between samples. The sample was removed from the coffee mill and placed in the upturned lid of a 90-mm-diameter petri dish and pressed manually to a thickness of ∼5 mm, using the inverted base of a petri dish as the press. A sterile upturned petri dish base was used to cover the prepared sample, and once 25 samples had been obtained, they were randomized, numbered, and stored on the bench top at ambient temperature (typically 22 ± 1°C).
HATR FT-IR spectroscopy.
FT-IR analysis was undertaken using a ZnSe HATR accessory (Spectroscopy Central Ltd., Warrington, United Kingdom) on a model IFS28 infrared spectrometer (Bruker Ltd., Coventry, United Kingdom) equipped with a deuterated triglycine sulfate detector. The ZnSe HATR crystal was capable of 10 external reflections, with the evanescent field (5) effecting a depth of 1.01 μm (Spectroscopy Central Ltd.). At 1-h intervals, six replicates were individually excised with a scalpel from a petri dish sample and placed in contact with the ZnSe crystal, and a spectrum for each replicate was collected. The replicates measured approximately 60 by 10 by 5 mm and were inverted so that the aerobic upper surface of the comminuted meat was placed in intimate contact with the HATR crystal.
The crystal surface was cleaned with distilled water and a soft tissue following collection of each spectrum and washed thoroughly with acetone, rinsed with distilled water, and dried with a soft tissue at the end of each sampling interval. The IBM-compatible personal computer (PC) used to control the IFS28 spectrometer was also programmed (using OPUS version 2.1 software, running under OS/2 Warp, provided by the manufacturers) to collect spectra over the wavenumber range 4,000 to 600 cm−1. The reference spectra were acquired from the cleaned blank crystal prior to the presentation of each sample replicate. All spectra were collected in reflectance mode with a resolution of 16 cm−1, and to improve the signal-to-noise ratio, 256 scans were coadded and averaged. The collection time for each sample spectrum was 60 s, and a total of 450 spectra were collected over the series of three experiments, all of which were undertaken within a 42-day period. At each 1-h sampling interval, a 1-g subsample of meat was also taken and vortexed for 60 s in 9 ml of 0.9% physiological saline, and the pH was recorded. A dilution series was undertaken, and plates of Lab M blood agar base (IDG PLC, Lancashire, United Kingdom) were lawned in triplicate with 50 μl of homogenate and incubated for 48 h at 25°C, and the total viable counts (TVC) were recorded as CFU.
Supervised analysis.
ASCII data were exported from the Opus software used to control the FT-IR instrument and imported into Matlab version 5.3 (The MathWorks, Inc., Natick, Mass.), which runs under Microsoft Windows NT on an IBM-compatible PC. To minimize problems arising from unavoidable baseline shifts, the spectra were scaled so that the smallest absorbance was set to 0 and the highest was set to +1 (47).
When the desired responses (targets) associated with each of the inputs (spectra) are known, then the system may be supervised. The goal of supervised learning is to find a model that correctly associates the inputs with the targets; this is usually achieved by minimizing the error between the target and the model's response (output) (16, 36). For quantitative interpretation of the FT-IR spectra, the multivariate linear regression method of PLS was applied as detailed previously (27) following computations given elsewhere (36). The input (x) data sets for the supervised-learning method contained the full HATR FT-IR spectra (441 absorbances representing a band of 16 wavenumbers) and known log10(TVC) values (y data) from the first two spoilage experiments, and these were partitioned into training and cross validation sets according to our established modeling practices (31, 46). During calibration of the model, the root mean squared (RMS) error between the true and desired levels for the cross validation data was calculated, and the lowest RMS error from this was used to find the optimal calibration which would give the best general predictive model. Following calibration, the PLS model was challenged with the independent test set of data from the entirely separate third and final experiment.
Evolutionary computation.
Although PLS is an excellent method for the quantitative analysis of biological systems (36), the information as to which wave numbers in the infrared spectrum are important is not readily available. The use of PLS, therefore, is often perceived as a “black box” approach to modeling spectra and so has limited use for the deconvolution of hyperspectral data in chemical or biochemical terms. Therefore, in this study, evolutionary computation methods based on GAs and GP were employed to aid in the deconvolution of these hyperspectra.
GA.
A GA is an optimization method based on the principles of Darwinian selection (24), where, over a series of generations, a population of parameter sets evolves until an optimal, or near-optimal, solution to a given problem is found.
First, a population of n objects (chromosomes) is created, each chromosome containing a string of numbers or binary digits representing the parameters of the problem to be optimized. The population is randomized so that n sets of “unique” parameter values can be evaluated and assigned a fitness value (usually a single numerical value). Once all n fitness values have been assigned, the next generation of chromosomes is created. In order for this new generation to be fitter than the last, principles analogous to those of sexual and asexual reproduction are applied. Using a stochastic selection method, based on parent fitness, two chromosomes are chosen to reproduce, swapping sections of their respective sequences (the probability of a particular parent chromosome being selected for sexual reproduction is proportional to its fitness, so chromosomes with a respectively high fitness value will have a greater chance of selection). This process creates two new child chromosomes inheriting characteristics of their parents. The child chromosomes are then subjected to mutation, where the value of each parameter may be randomly changed. The probability of this change is normally very small. The process of selection followed by reproduction followed by mutation is then repeated until n new chromosomes are created (i.e., a new population to replace the old). The fitness value is then evaluated for each of the new chromosomes, and the whole process repeats itself. The algorithm continues until a stopping criterion is reached. For example, the criterion may be that a given optimal fitness value is met, a certain number of generations has passed, or the chromosomes have converged to similar parameter values.
The GA-multiple linear regression (MLR) wavelength selection methodology (9) uses a GA to determine the subset of n wavenumbers, taken from the total spectral data set, which, when applied to an MLR model, will optimally discriminate between the HATR spectra and the known log10(TVC). All calculations were performed with in-house software which runs under Microsoft Windows NT on an IBM-compatible PC, and full details of GA-MLR are given elsewhere (9). Briefly, optimization is achieved by monitoring the RMS error of prediction for each model. The GA uses two-point crossover with mutation (19), operating on a population of binary-encoded chromosomes, each chromosome representing n candidate wavelengths (9); in this study, n was 2, 3, and 5. The selection of parent chromosomes for the next generation is carried out using a rank-based scheme (D. Whitley, presented at the proceedings of the Third International Conference on Genetic Algorithms, San Mateo, Calif., 1989). No two identical candidates were allowed in a given population.
GP.
A GP is an application of the GA approach to derive mathematical equations, logical rules, or program functions automatically (17, 21, 33). Rather than representing the solution to the problem as a string of parameters, as in a conventional GA, a GP usually (as here) uses a tree structure. The leaves of the tree, or terminals, represent input variables or numerical constants. Their values are passed to nodes, at the junctions of branches in the tree, which perform some numerical or program operation before passing on the result further towards the root of the tree. Mutations are performed by selecting a parent and modifying the value or variable returned by a terminal or changing the operation performed by a node. Crossovers are performed by selecting two parents and swapping subtrees at randomly selected nodes within their trees. The new individuals so generated replace less fit members of the population chosen probabilistically on the basis of their unfitness.
The GP employed the genomic computing software Gmax-bio (Aber Genomic Computing, Aberystwyth, United Kingdom), which runs under Microsoft Windows NT on an IBM-compatible PC. An introduction to Gmax-bio is given elsewhere (29), and the default parameter settings for population size (1,000), mutation, and recombination rates were used throughout. The operators that were used were +, −, /, *, log10(x), 10x, >, <, and Tanh(x). The fitness calculation used is F = 1/(0.01 + S/B), where the values of S and B are determined by the FITNESS setting. In this expression, S is a statistic derived from the model which ranges between 0 and infinity and B is a normalizing quantity. The value of B is chosen such that a perfect model yields an F of 100 and a model which performs no better than random chance yields an F of 1.
RESULTS AND DISCUSSION
The comminution of samples in order to accelerate the spoilage process was successful, as the final mean log10(TVC) of 9.02 (Table 1) was an order of magnitude above the 108 CFU g−1 generally accepted as the point at which organoleptic spoilage becomes readily detectable (13, 15). Using 108 CFU g−1 as the indicator for postspoilage, the average spoilage time over the series of experiments was 13.6 h (Table 1). The spoilage of the samples within 24 h at room temperature was anticipated, as comminution ruptures cell walls, releasing a source of nutrients; increases the surface-area-to-volume ratio; and distributes bacteria that would normally be restricted to the surface throughout the meat substrate. The initial mean pH range of fresh samples during the three experiments (5.7 to 5.9) was within those described previously in the literature (13, 15). The use of pH as an indicator of spoilage or remaining shelf life in meats would be insufficient, as the pH fluctuates prior to spoilage, only rising significantly when levels of bacteria reach ∼108 CFU g−1. At this level, sensory spoilage is readily detectable, and this is partly a consequence of the increase in pH and the production of malodorous substances, such as ammonia, dimethylsulfide, and diacetyl, by the catabolic action of the resident microflora (13, 25).
TABLE 1.
Data matrix of results from three spoilage experimentsa
| Expt | Initial pH | Final pH | Initial log10(TVC) | Final log10(TVC) | Room temp (°C) | Spoilage (h) |
|---|---|---|---|---|---|---|
| 1 | 6.02 | 6.79 | 6.86 | 9.20 | 21.5 | 14 |
| 2 | 5.52 | 6.05 | 6.62 | 8.64 | 23.1 | 17 |
| 3 | 5.94 | 6.83 | 6.77 | 9.04 | 23.3 | 10 |
| Mean (SD) | 5.87 | 6.67 | 6.76 | 9.02 | 22.6 (0.99) | 13.6 (3.5) |
The onset of spoilage is taken as the point when total bacterial numbers reach 108 CFUg−1.
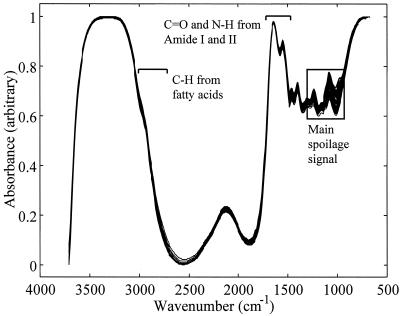
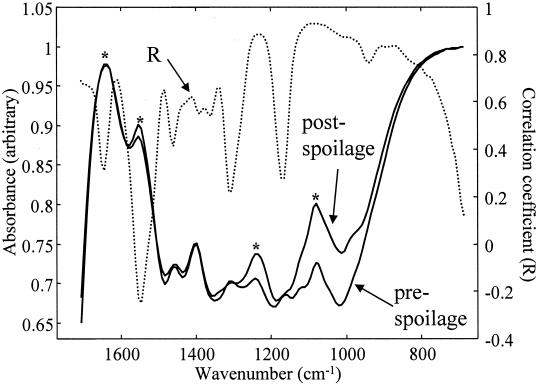
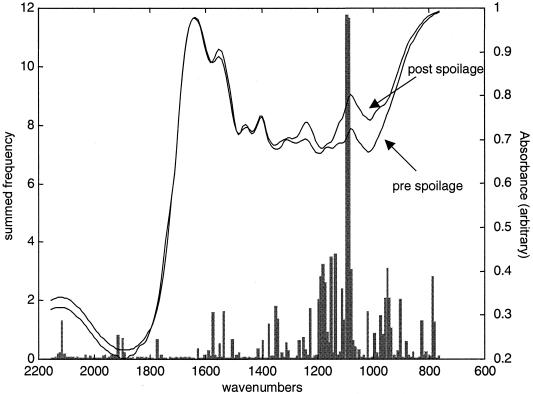
The 150 FT-IR spectra from experiment 1 are shown in Fig. 1 and illustrate the reproducibility of both HATR as a sampling method and the experimental protocol that was undertaken over a period of 6 weeks. Typical FT-IR spectral data from the 1,750- to 700-cm−1 wave number range from measurements of meat at the pre- and postspoilage stages are shown in Fig. 2. These spectra are from chicken breast meat carrying ∼7 · 106 and ∼2 · 109 CFU g−1, respectively, and are both data rich and not biased to any particular group of chemicals associated with a particular group of metabolites. Furthermore, the spectra are complex and multidimensional in nature, so they do not easily lend themselves to simple visual interpretation; this is compounded by the fact that the data set for all three experiments is substantial, with a total of 450 spectra, each containing 441 wave numbers. However, with the advent of modern machine learning approaches, the opportunity now exists to analyze such complex high-dimensional spectral patterns (7, 46) and to extract an answer to a question of biological interest with much lower dimensionality, i.e., “What is the bacterial load on the meat surface?”
FIG. 1.
All 150 HATR absorbance spectra from both fresh and spoiled meats from experiment 1. These illustrate the reproducibility of both HATR FT-IR spectroscopy and the preparation method employed throughout the series of experiments. The amide I and amide II vibrations from proteins and the CHx vibrations from fatty acids are indicated. The box indicates where the most variance in these spectra occur and hence where spoilage signals are likely to be seen.
FIG. 2.
Typical FT-IR absorbance spectra from pre- and postspoilage chicken. Also shown is the Pearson correlation coefficient (R) between the FT-IR absorbances (in experiments 1 and 2) and the log10(TVC). The asterisks indicate peaks that are attributable to amide I (1,640 cm−1), amide II (1,550 cm−1), and amine (1,240 and 1,088 cm−1) vibrations.
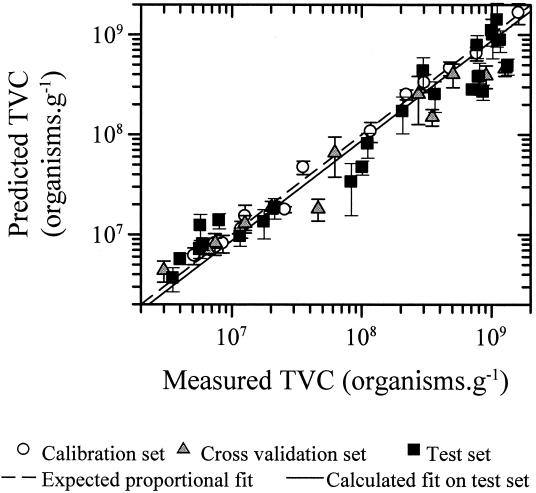
Therefore, as described above, the supervised-learning method of PLS regression was calibrated and cross validated with the FT-IR spectral data and the known log10(TVC) values from experiments 1 and 2 (Table 2 shows the details and TVC levels) before being challenged by the independent and “unseen” test set of data from experiment 3. The plots of the estimates versus the known log10(TVC) (Fig. 3) show that the FT-IR and PLS predictions were virtually indistinguishable from the expected log proportional fit [i.e., log(y) = log(x)] and so show that this approach can be used to accurately assess the spoilage status of meat. As can be seen in Fig. 3 and Table 2, the lowest level of spoilage encountered was ∼2 · 106 CFU g−1, and this necessarily restricts the detection limit. The TVC for chicken immediately postslaughter is 103 cm−2, rising to 104 to 105 cm−2 after packaging (26). That PLS gave accurate results at 2 · 106 CFU g−1 suggests that it will be possible to reach lower levels, and this will be the subject of further study with freshly killed chickens. From Fig. 3, it is evident that the spectra obtained by direct FT-IR analysis of meat do contain biochemical information that allows correlation with the spoilage status of the chicken, for data used to produce the PLS model and, more importantly, for data from a completely new experiment. The obvious question that needs to be addressed is that of which biochemical species the FT-IR is measuring that are related to the spoilage status of the chicken.
TABLE 2.
Log10(TVC) of bacteria acquired from comminuted meat samples from three experimentsa
| Time (h) | Log10(TVC) for expt:
|
Arithmetic mean | ||
|---|---|---|---|---|
| 1 | 2 | 3 | ||
| 0 | 6.87c | 6.62c | 6.77t | 6.77 |
| 1 | 6.87v | 6.35v | 6.54t | 6.64 |
| 2 | 6.93c | 6.38c | 6.60t | 6.70 |
| 3 | 6.48v | 6.61v | 6.75t | 6.63 |
| 4 | 6.71c | 6.64c | 6.76t | 6.71 |
| 5 | 6.82v | 6.50v | 7.06t | 6.85 |
| 6 | 7.07c | 6.75c | 6.90t | 6.93 |
| 7 | 7.31v | 6.53v | 7.25t | 7.14 |
| 8 | 7.10c | 6.67c | 7.32t | 7.11 |
| 9 | 7.10v | 6.76v | 7.91t | 7.52 |
| 10 | 7.41c | 6.75c | 8.00t | 7.64 |
| 11 | 7.66v | 6.65v | 8.04t | 7.73 |
| 12 | 7.55c | 6.92c | 8.31t | 7.92 |
| 13 | 7.79v | 7.07v | 8.47t | 8.09 |
| 14 | 8.07c | 6.89c | 8.56t | 8.21 |
| 15 | 8.54v | 7.42v | 8.85t | 8.56 |
| 16 | 8.34c | 7.84c | 8.89t | 8.55 |
| 17 | 8.44v | 8.36v | 8.93t | 8.66 |
| 18 | 8.48c | 8.28c | 8.88t | 8.62 |
| 19 | 8.70v | 8.49v | 8.89t | 8.72 |
| 20 | 8.68c | 8.55c | 9.00t | 8.78 |
| 21 | 8.96v | 8.49v | 9.12t | 8.93 |
| 22 | 8.88c | 8.62c | 9.00t | 8.86 |
| 23 | 9.10v | 8.56v | 9.07t | 8.97 |
| 24 | 9.21c | 8.65c | 9.04t | 9.02 |
All measurements were taken in triplicate after incubation at 25°C for 48 h and were used, in conjunction with FT-IR spectra, to calibrate (c), cross validate (v), and test (t) the PLS model.
FIG. 3.
Estimates from PLS versus the true log10(TVC). The data points are the averages of the six measurements, and the error bars show standard deviations. The RMS errors in these measurements are 0.15, 0.23, and 0.27 log units for the calibration, cross validation, and independent test sets, respectively.
The Pearson correlation coefficients between the absorbances at each wave number in the FT-IR spectra from experiments 1 and 2 and the log10(TVC) were calculated and are also plotted in Fig. 2. It can be seen that most peaks from 1,500 to 700 cm−1 are positively correlated with spoilage, but no single peak appears uniquely dominant; this necessarily means that it is difficult to pinpoint the cause of microbial spoilage to a single (or a small group of) biochemical species using this correlation approach. Therefore, GAs and GPs were evolved to discriminate qualitatively between meat carrying <107 and ≥107 bacteria (as TVC) per cm2.
GA-MLR was applied so as to extract subsets of two, three, and five wave numbers that could discriminate between fresh (<107 bacteria/cm2) and spoiled (≥107 bacteria/cm2) chicken. Because the starting population for each GA run was random, 60 GA-MLR runs were performed, and the following subsets were found to be optimal for selecting just two or three wave numbers, respectively: (1,096, 1,227 cm−1) and (1,312, 1,235, 1,088 cm−1). When the algorithm was used to look for five wave numbers, it was found that the degree of discrimination did not improve compared with selecting subsets of three, and no consistent areas of the FT-IR spectra were found to be dominant in the GA expressions; however, vibrations at 1,096 and 1,305 cm−1 were found within the best subsets.
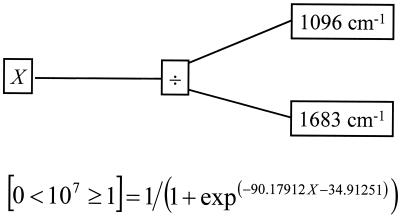
GP analyses (i) using the same 107-bacteria/cm−1 threshold as above and (ii) evolved to predict the log10(TVC) levels produced trees which could easily discriminate between fresh and spoiled chicken and quantify the level of spoilage, respectively; a typical GP parse tree is shown in Fig. 4. As with the GAs, the initial populations were produced randomly; therefore, 10 separate GPs were evolved. For the threshold GP analysis, the number of times each input (wave number) was used for the 10 evolved populations was calculated and plotted against the wave number of the infrared light (Fig. 5). Figure 5 clearly shows that the dominant area of the spectra for discriminating between fresh (<107 bacteria/cm2) and spoiled (≥107 bacteria/cm2) chicken was 1,088 to 1,096 cm−1; moreover, these wave numbers were also selected by the GA-MLR method. The functional group vibration in the region 1,088 to 1,096 cm−1 is ascribable to C-N stretching, most plausibly from amines (14, 35).
FIG. 4.
Typical GP tree evolved to discriminate between chicken carrying <107 and ≥107 bacterial counts. The use of the logistic function Pr{Y} = 1/(1 + exp(−Model Expression)), defines a maximum-likelihood (Pr{Y}) decision boundary for an output being either false (0) or true (1).
FIG. 5.
Frequency plot of the number of times an input was used in 10 independent GPs, evolved to discriminate between chicken carrying <107 and ≥107 bacterial counts (encoded as 0 and 1, respectively).
The most intense peaks that appear in fresh meat are the amide I (C=O vibration at 1,640 cm−1) and amide II (N—H deformation at 1,550 cm−1) bands from proteins and peptides, and from the Pearson correlation coefficients, the amide II band is the only vibration that is negatively correlated with spoilage (Fig. 2). This strongly suggests that the protein content of the meat was decreasing during spoilage. By contrast, the peaks at 1,240 and 1,088 cm−1, which are both ascribable to C-N stretching from amines from free amino acids, are positively correlated. Indeed, the rule in Fig. 4 shows that spoilage can be ascribed simply to the ratio of 1,096 to 1,683 cm−1 from these vibrations from amines and amides, respectively.
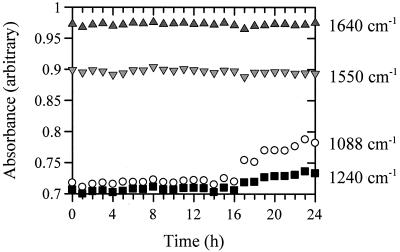
Plots of the absorbances of these vibrations versus the time for the second experiment are shown in Fig. 6. It is clear that the amide I and II bands are constant, although the amide II band does decrease very slightly after 16 h while the peaks at 1,240 and 1,088 cm−1 start to increase significantly after 16 h. It is noteworthy that the onset of spoilage, as characterized by a TVC of >108 g−1, was at 17 h, and this was the point at which the absorbance due to free amines started to increase. This was also found to be the case for experiments 1 and 3 (data not shown). These correlations, and the fact that the GAs and GPs both pick the region 1,088 to 1,096 cm−1 as the most significant area of the FT-IR spectra for the prediction of spoilage of chicken which is attributable to free amino acids, makes it clear that the most significant metabolic process that occurs at spoilage is the start of proteolysis. This is indeed highly likely, since it is known that spoilage in meat is most frequently associated with the postglucose utilization of amino acids by aerobic microorganisms, such as pseudomonads, and the onset of the enzymatic degradation of proteins and peptides, leading to the production of free amino acids (8, 12, 40).
FIG. 6.
Plots of selected IR vibrations versus time for experiment 2. The selected vibrations are amide I (C=O vibration at 1,640 cm−1) and amide II (N—H deformation at 1,550 cm−1) bands and C-N stretching from amines at 1,240 and 1,088 cm−1. Note the increase in the absorbance of 1,088 cm−1 at 17 h, which corresponds to the point at which the onset of spoilage occurs (Table 1).
In conclusion, FT-IR spectroscopy, in combination with appropriate machine learning methods, presents itself as a novel method for the quantitative detection of food spoilage. Using FT-IR, we were able to acquire a metabolic snapshot (30) and quantify, noninvasively, the microbial loads of food samples accurately and rapidly (within 60 s) directly from the sample surface. We believe that this approach has considerable potential for further development and will aid both the food safety regulatory bodies and the Hazard Analysis Critical Control Point system. In particular, we will conduct future studies testing our method for quantifying the numbers of spoilage organisms on muscle foods at the production, processing, packaging, and storage levels.
Acknowledgments
We are indebted to the Agri-Food and Engineering and Biological Systems Committees of the United Kingdom BBSRC for supporting this work.
REFERENCES
- 1.Alomirah, H. F., B. F. Gibbs, and Y. Konishi. 1998. Identification of proteolytic products as indicators of quality in whole and ground meat. J. Food Qual. 21:299-316. [Google Scholar]
- 2.Alsberg, B. K., R. Goodacre, J. J. Rowland, and D. B. Kell. 1997. Classification of pyrolysis mass spectra by fuzzy multivariate rule induction—comparison with regression, K-nearest neighbour, neural and decision-tree methods. Anal. Chim. Acta 348:389-407. [Google Scholar]
- 3.Archer, D. L. 1996. The validation of rapid methods in food microbiology. Food Control 7:3-4. [Google Scholar]
- 4.Bäck, T., D. B. Fogel, and Z. Michalewicz. 1997. Handbook of evolutionary computation. Oxford University Press, Oxford, United Kingdom.
- 5.Banwell, C. N., and E. M. McCash. 1994. Fundamentals of molecular spectroscopy, 4th ed. McGraw-Hill, London, United Kingdom.
- 6.Banzhaf, W., P. Nordin, R. E. Keller, and F. D. Francone. 1998. Genetic programming: an introduction. Morgan Kaufmann, San Francisco, Calif.
- 7.Beavis, R. C., S. M. Colby, R. Goodacre, P. B. Harrington, J. P. Reilly, S. Sokolow, and C. W. Wilkerson. 2000. Artificial intelligence and expert systems in mass spectrometry, p. 11558-11597. In R. A. Meyers (ed.), Encyclopedia of analytical chemistry. John Wiley & Sons, Chichester, United Kingdom.
- 8.Braun, P., K. Fehlhaber, C. Klug, and K. Kopp. 1999. Investigations into the activity of enzymes produced by spoilage-causing bacteria: a possible basis for improved shelf-life estimation. Food Microbiol. 16:531-540. [Google Scholar]
- 9.Broadhurst, D., R. Goodacre, A. Jones, J. J. Rowland, and D. B. Kell. 1997. Genetic algorithms as a method for variable selection in PLS regression, with application to pyrolysis mass spectra. Anal. Chim. Acta 348:71-86. [Google Scholar]
- 10.Champiat, D., N. Matas, B. Monfort, and H. Fraass. 2001. Applications of biochemiluminescence to HACCP. Luminescence 16:193-198. [DOI] [PubMed] [Google Scholar]
- 11.Chan, K. F., H. L. Tran, R. Y. Kanenaka, and S. Kathariou. 2001. Survival of clinical and poultry-derived isolates of Campylobacter jejuni at a low temperature (4°C). Appl. Environ. Microbiol. 67:4186-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dainty, R. H. 1996. Chemical/biochemical detection of spoilage. Int. J. Food Microbiol. 33:19-33. [DOI] [PubMed] [Google Scholar]
- 13.Davies, A., and R. Board (ed.). 1998. The microbiology of meat and poultry. Blackie Academic & Professional, London, United Kingdom.
- 14.Degen, I. A. 1997. Tables of characteristic group frequencies for the interpretation of infrared and Raman spectra. Acolyte Publications, Harrow, United Kingdom.
- 15.Doyle, M. P., L. R. Beuchat, and T. J. Montville. 1997. Food microbiology: fundamentals and frontiers. ASM Press, Washington, D.C.
- 16.Duda, R. O., P. E. Hart, and D. G. Stork. 2001. Pattern classification, 2nd ed. Wiley, New York, N.Y.
- 17.Gilbert, R. J., R. Goodacre, A. M. Woodward, and D. B. Kell. 1997. Genetic programming: a novel method for the quantitative analysis of pyrolysis mass spectral data. Anal. Chem. 69:4381-4389. [DOI] [PubMed] [Google Scholar]
- 18.Gillie, J. K., J. Hochlowski, and G. A. Arbuckle-Keil. 2000. Infrared spectroscopy. Anal. Chem. 72:71-79. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg, D. E. 1989. Genetic algorithms in search, optimization and machine learning. Addison-Wesley, Reading, Mass.
- 20.Gonzalez-Miret, M. L., M. T. Coello, S. Alonso, and F. J. Heredia. 2001. Validation of parameters in HACCP verification using univariate and multivariate statistics. Application to the final phases of poultry meat production. Food Control 12:261-268. [Google Scholar]
- 21.Goodacre, R., B. Shann, R. J. Gilbert, É. M. Timmins, A. C. McGovern, B. K. Alsberg, D. B. Kell, and N. A. Logan. 2000. The detection of the dipicolinic acid biomarker in Bacillus spores using Curie-point pyrolysis mass spectrometry and Fourier transform infrared spectroscopy. Anal. Chem. 72:119-127. [DOI] [PubMed] [Google Scholar]
- 22.Goodacre, R., É. M. Timmins, R. Burton, N. Kaderbhai, A. M. Woodward, D. B. Kell, and P. J. Rooney. 1998. Rapid identification of urinary tract infection bacteria using hyperspectral, whole organism fingerprinting and artificial neural networks. Microbiology 144:1157-1170. [DOI] [PubMed] [Google Scholar]
- 23.Holder, J. S., J. E. L. Corry, and M. H. Hinton. 1997. Microbial status of chicken portions and portioning equipment. Br. Poultry Sci. 38:505-511. [DOI] [PubMed] [Google Scholar]
- 24.Holland, J. H. 1992. Adaptation in natural and artificial systems: an introductory analysis with applications to biology, control, and artificial intelligence. MIT Press, Cambridge, Mass.
- 25.Jackson, T. C., G. R. Acuff, and J. S. Dickson. 1997. Meat, poultry, and seafood, p. 83-100. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers. ASM Press, Washington, D.C.
- 26.Jay, J. M. 1996. Modern food microbiology, 5th ed. Chapman & Hall, London, United Kingdom.
- 27.Jones, A., A. D. Shaw, G. J. Salter, G. Bianchi, and D. B. Kell. 1998. The exploitation of chemometric methods in the analysis of spectroscopic data: application to olive oils, p. 317-376. In R. J. Hamilton (ed.), Lipid analysis of oils and fats. Chapman & Hall, London, United Kingdom.
- 28.Kakouri, A., and G. J. E. Nychas. 1994. Storage of poultry meat under modified atmospheres or vacuum packs: possible role of microbial metabolites as indicators of spoilage. J. Appl. Bacteriol. 76:163-172. [DOI] [PubMed] [Google Scholar]
- 29.Kell, D. B., R. M. Darby, and J. Draper. 2001. Genomic computing. Explanatory analysis of plant expression profiling data using machine learning. Plant Physiol. 126:943-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kell, D. B., and P. Mendes. 2000. Snapshots of systems: metabolic control analysis and biotechnology in the post-genomic era, p. 3-25. In A. Cornish-Bowden and M. L. Cárdenas (ed.), Technological and medical implications of metabolic control analysis. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 31.Kell, D. B., and B. Sonnleitner. 1995. GMP—good modelling practice: an essential component of good manufacturing practice. Trends Biotechnol. 13:481-492. [Google Scholar]
- 32.Koohmaraie, M. 1994. Muscle proteinases and meat aging. Meat Sci. 36:93-104. [DOI] [PubMed] [Google Scholar]
- 33.Koza, J. R. 1992. Genetic programming: on the programming of computers by means of natural selection. MIT Press, Cambridge, Mass.
- 34.Lang, P. L., and S. C. Sang. 1997. The in situ infrared microspectroscopy of bacterial colonies on agar plates. Cell. Mol. Biol. 44:231-238. [PubMed] [Google Scholar]
- 35.Lin-Vien, D., N. B. Colthup, W. G. Fateley, and J. G. Grasselli. 1991. The handbook of infrared and Raman characteristic frequencies of organic molecules. Academic Press, Boston, Mass.
- 36.Martens, H., and T. Næs. 1989. Multivariate calibration. John Wiley, Chichester, United Kingdom.
- 37.Mitchell, T. M. 1997. Machine learning. McGraw Hill, New York, N.Y.
- 38.Naumann, D., D. Helm, and H. Labischinski. 1991. Microbiological characterizations by FT-IR spectroscopy. Nature 351:81-82. [DOI] [PubMed] [Google Scholar]
- 39.Newell, D. G., J. E. Shreeve, M. Toszeghy, G. Domingue, S. Bull, T. Humphrey, and G. Mead. 2001. Changes in the carriage of Campylobacter strains by poultry carcasses during processing in abattoirs. Appl. Environ. Microbiol. 67:2636-2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nychas, G. J. E., and C. C. Tassou. 1997. Spoilage processes and proteolysis in chicken as detected by HPLC. J. Sci. Food Agric. 74:199-208. [Google Scholar]
- 41.Oliver, S. G., M. K. Winson, D. B. Kell, and F. Baganz. 1998. Systematic functional analysis of the yeast genome. Trends Biotechnol. 16:373-378. [DOI] [PubMed] [Google Scholar]
- 42.Scheu, P. M., K. Berghof, and U. Stahl. 1998. Detection of pathogenic and spoilage microorganisms in food with the polymerase chain reaction. Food Microbiol. 15:13-31. [Google Scholar]
- 43.Schmitt, J., and H.-C. Flemming. 1998. FTIR-spectroscopy in microbial and material analysis. Int. Biodeterior. Biodegradation 41:1-11. [Google Scholar]
- 44.Schreurs, F. J. G. 2000. Post-mortem changes in chicken muscle. Worlds Poult. Sci. J. 56:319-346. [Google Scholar]
- 45.Seymour, I. J., M. B. Cole, and P. J. Coote. 1994. A substrate-mediated assay of bacterial proton efflux/influx to predict the degree of spoilage of beef mince stored at chill temperatures. J. Appl. Bacteriol. 76:608-615. [DOI] [PubMed] [Google Scholar]
- 46.Shaw, A. D., M. K. Winson, A. M. Woodward, A. C. McGovern, H. M. Davey, N. Kaderbhai, D. Broadhurst, R. J. Gilbert, J. Taylor, É. M. Timmins, B. K. Alsberg, J. J. Rowland, R. Goodacre, and D. B. Kell. 1999. Rapid analysis of high-dimensional bioprocesses using multivariate spectroscopies and advanced chemometrics. Adv. Biochem. Eng. Biotechnol. 66:83-114. [DOI] [PubMed] [Google Scholar]
- 47.Timmins, É. M., S. A. Howell, B. K. Alsberg, W. C. Noble, and R. Goodacre. 1998. Rapid differentiation of closely related Candida species and strains by pyrolysis mass spectrometry and Fourier transform infrared spectroscopy. J. Clin. Microbiol. 36:367-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tompkin, R. B. 1990. The use of HACCP in the production of meat and poultry products. J. Food Prot. 53:795-803. [DOI] [PubMed] [Google Scholar]
- 49.Whyte, P., J. D. Collins, K. McGill, C. Monahan, and H. O'Mahony. 2001. Quantitative investigation of the effects of chemical decontamination procedures on the microbiological status of broiler carcasses during processing. J. Food Prot. 64:179-183. [DOI] [PubMed] [Google Scholar]