Abstract
Comparisons of the activities and diversities of ammonia-oxidizing bacteria (AOB) in the root environment of different cultivars of rice (Oryza sativa L.) indicated marked differences despite identical environmental conditions during growth. Gross nitrification rates obtained by the 15N dilution technique were significantly higher in a modern variety, IR63087-1-17, than in two traditional varieties. Phylogenetic analysis based on the ammonium monooxygenase gene (amoA) identified strains related to Nitrosospira multiformis and Nitrosomonas europaea as the predominant AOB in our experimental rice system. A method was developed to determine the abundance of AOB on root biofilm samples using fluorescently tagged oligonucleotide probes targeting 16S rRNA. The levels of abundance detected suggested an enrichment of AOB on rice roots. We identified 40 to 69% of AOB on roots of IR63087-1-17 as Nitrosomonas spp., while this subpopulation constituted 7 to 23% of AOB on roots of the other cultivars. These results were generally supported by denaturing gradient gel electrophoresis of the amoA gene and analysis of libraries of cloned amoA. In hydroponic culture, oxygen concentration profiles around secondary roots differed significantly among the tested rice varieties, of which IR63087-1-17 showed maximum leakage of oxygen. The results suggest that varietal differences in the composition and activity of root-associated AOB populations may result from microscale differences in O2 availability.
The supply of nitrogen in paddy fields is recognized as a major limitation toward increased productivity of rice (Oryza sativa L.) (17, 18). Since rice paddies are typically maintained under flooded conditions, ammonium constitutes the major form of mineral N in the bulk soil (14), which is a form to which rice is especially adapted (47, 56). However, considerable amounts of nitrate formed through nitrification do accumulate in the oxygen-rich surface layer of irrigated paddy soils as well as in well-drained upland rice fields and in rain-fed environments during the dry season (14). Compared to ammonium, which is usually bound to the soil matrix, nitrate is mobile within soils, making it susceptible to loss due to leaching and denitrification (21, 53). Nitrification is therefore critical to the supply of N in rice fields as well as to the balance of ammonium and nitrate.
The initial, rate-limiting step of nitrification is carried out by the chemolithoautotrophic ammonia-oxidizing bacteria (AOB) of the β subdivision of the class Proteobacteria (β-Proteobacteria) (38). All AOB are obligately aerobic and generally recalcitrant to cultivation in artificial culture (38). Therefore, molecular methods have been instrumental to our current understanding of their ecology. Based on 16S rRNA sequence analysis, all known AOB of the β-Proteobacteria occurring in terrestrial environments belong to a monophyletic assemblage represented by members of only two genera: Nitrosomonas spp. and Nitrosospira spp. (24, 52). Analysis of the functional ammonium monooxygenase gene (amoA) yields similar, although not identical, evolutionary relationships (39). Studies conducted in various systems suggest that in a variety of soils, Nitrosospira spp. are predominant (23, 27, 31, 43, 49), while environments with high ammonium loads, such as sewage sludge and wastewater, favor Nitrosomonas spp. (32, 35, 43). At present, very little is known about how the population structure of these bacteria affects nitrification in rice soils or how AOB populations are influenced by rice plants.
Under flooded conditions, rice soils become anoxic almost immediately beneath the soil-water interface (42). However, the immediate surface of rice roots offers a supply of oxygen through leakage from aerenchymatous tissue (7, 42). It is recognized that rice roots release oxygen at rates sufficient to support nonspecific aerobic microbial processes (9, 13). The possibility that rice roots may support significant rates of nitrification has also been suggested (10, 33). However, very little information is available regarding the microbiology of AOB on or near the root surface of rice plants. The purpose of this study was to study the abundance, composition, and activity of populations of AOB occurring in the rice root environment.
To detect and characterize the AOB from the roots and rhizosphere, we used PCR coupled with denaturing gradient gel electrophoresis (DGGE) targeting the amoA gene. This approach, combined with clone screening and sequencing (generally but not exclusively targeting the 16S rRNA gene), has been used successfully in various systems, including agronomic soils (16, 29, 31, 36, 43, 50). For quantification of AOB, we employed fluorescence in situ hybridization (FISH) using rRNA-targeted oligonucleotide probes. The method has been reliably used for identification and quantification of AOB (22, 32, 35, 55). Aside from allowing direct visualization of bacteria in the environment, the method also has the added advantage of detecting active cells, since the target molecule is the rRNA. However, the above studies have focused mainly on nutrient-rich environments where the abundance and ribosome content of cells are relatively high. The application of FISH in nutrient-poor environments such as soil and roots is more problematic due to lower cell numbers and the presence of interfering autofluorescence from roots and soil particles. In this study, however, we found that the biofilm coating the root surface can be detached by moderate sonication, concentrated, and then probed using FISH. Since the abundances detected were within the detection limits of FISH, we are able to present, for the first time, population estimates of AOB on the rice root surface.
In this study, all experiments were conducted under artificial conditions set to simulate rain-fed paddy fields that favor nitrification (alternating flood and dry periods). In order to account for the possible effect of genotypic variation, all experiments were performed using different rice cultivars. The varieties were chosen based on their drought tolerance and represent a modern hybrid rice variety (IR63087-1-17) and two improved traditional rice varieties (Kao Dawk Mali 105 [KDML 105] and Mahsuri). To complement results of molecular analysis, gross nitrification rates associated with the three varieties and unplanted soil were estimated by the 15N isotopic dilution technique. Finally, oxygen concentration profiles from the roots of two contrast varieties grown in hydroponic culture were measured to determine the possible relationship between the composition and activity of AOB populations in the root environment and oxygen release from rice roots.
MATERIALS AND METHODS
Plant growth conditions.
Three drought-tolerant rain-fed lowland rice cultivars (Oryza sativa L. cv. KDML 105, Mahsuri, and IR63087-1-17) were pregerminated prior to transplanting into pots. Soil was collected from a rain-fed lowland area in Tarlac Province, the Philippines. The soil is a Luisita series, coarse-loamy, mixed, nonacid, isohyperthermic typic tropaquept with the following characteristics: pH, 6.2; C, 0.94%; N, 0.093%; C/N ratio, 11.9; sand, 39.2%; silt, 43.2%; clay, 17.6%. Fourteen days after transplanting, the pots were maintained flooded (water depth 1 to 2 cm above the soil plane) for 4 weeks. Thereafter, water was withdrawn to simulate drought conditions by monitoring the plants daily and applying water when approximately 5% of the plant leaves had begun to roll. Nitrogen in the form of urea was applied to all plants after the flooded phase at amounts equivalent to a field application rate of 50 kg of N ha−1. The plants were grown in a phytotron maintaining a 12-h-12-h photoperiod. Day and night cycles reached maximum and minimum temperatures of 30 and 25°C, respectively. During the day cycle, plants received 1,450 microeinsteins of light m−2 s−1. Humidity was maintained at 70% relative humidity. Pots containing no plants were included as nonrhizosphere controls.
A total of three pot experiments were conducted using the same conditions as described above. Two experiments were conducted in plastic pots (inside diameter, 16 cm; soil height, 13 cm) and sampled for PCR and FISH analysis, respectively. The 15N dilution experiment was conducted in polyvinyl chloride cylinders (8.3 cm [inner diameter] by 16 cm [height]; stoppered at the bottom) with holes drilled at the sides for application of labeled N. A second set of samples for PCR analysis was also collected from this experiment. All experiments were set up in a completely randomized design with three replicates.
Sampling for PCR and FISH.
Soil samples for DNA extraction were obtained after the 3rd week of induced drought. Rhizosphere samples were obtained by scraping off the soil adhering to the root surface. Roots that had been scraped free of clumps of adhering soil comprised our source for root bacterial DNA. All samples were immediately stored at −20°C until DNA extraction.
Root samples for FISH (also collected after the 3rd week of induced drought) were first gently washed in phosphate-buffered saline (PBS) (130 mM sodium chloride, 10 mM sodium phosphate buffer; pH 7.2) and then fixed in 4% paraformaldehyde in PBS. The roots were immersed in this solution for 3 to 4 days to allow the soil loosely adhering to the root surface to settle. Thereafter, roots were transferred to a 1:1 mixture of ethanol and PBS and then stored at −20°C for long-term storage.
DNA extraction, PCR, and DGGE.
DNA from rhizosphere and roots was extracted using a FastDNA SPIN Kit for Soil (Bio 101, Inc. Vista, Calif.) following the manufacturer's instructions. Prior to extraction, root samples were snap frozen in liquid nitrogen and then ground by mortar and pestle. Extracts from 400 mg of fresh soil and 100 mg of crushed root materials comprising rhizosphere and root DNA extractions, respectively, were finally resuspended in 50 μl of sterile DNase- and RNase-free water (Sigma, St. Louis, Mo.). Two microliters of a 1:10 dilution of the DNA extracts was used as the template for subsequent PCRs using amoA-1F and amoA-2R primers (43) with modifications suggested by Stephen et al. (50). PCRs were performed in a total volume of 50 μl using Ex Taq DNA polymerase (Takara Shuzo, Shiga, Japan) in the supplied buffer and a Robocycler 96 thermocycler (Stratagene, La Jolla, Calif.). Amplification proceeded for 35 cycles using previously published cycling parameters (43). PCR products were resolved by electrophoresis on 1.5 to 2% agarose gels and visualized by UV transillumination after staining with SYBR Green I (FMC Bioproducts, Rockland, Maine). The amount of PCR product was estimated from the gel by using image analysis software (ImageJ 1.26 [freely available at http://rsb.info.nih.gov/ij]) with appropriate DNA size and concentration standard (stable DNA ladder; Sigma-Genosys, Ishikari, Japan). For DGGE, a GC clamp (5′-CCGCCGCGCGGCGGGCGGGGCGGGGGCACGGGG-3′) (34) was attached to the forward primer. PCR amplification for DGGE analysis of environmental samples used 1.5 to 2 ng of initial amoA PCR product as the template followed by 12 cycles using the primers with the GC clamp and employing the cycling parameters described above (43).
DGGE of amoA PCR products was performed as described by Muyzer et al. (34) using a D-Code system (Bio-Rad, Hercules, Calif.). Polyacrylamide gradient gels (6% acrylamide, 2% glycerol, and 37.5:1 acrylamide-bisacrylamide in 1× Tris-acetate-EDTA; 60 to 100% denaturant; 1 mm thick by 16 cm by 16 cm) were poured to achieve a parallel gradient with the aid of a model 475 gradient delivery system (Bio-Rad) according to the manufacturer's instructions. A 100% denaturant was defined as 6% acrylamide containing 7 M urea and 40% deionized formamide. An estimated 50 ng of PCR product was loaded into each lane. Environmental samples were run at 120 V for 7.5 h at 50°C and were visualized by UV transillumination after staining with SYBR Green I.
Construction and analysis of amoA gene fragment libraries.
Purified amoA PCR products retrieved from Mahsuri roots and IR63087-1-17 roots and rhizosphere were cloned using the pPCR-Script Amp cloning kit (Stratagene). Ligation and transformation steps were performed according to the manufacturer's instructions. All plasmids bearing inserts of the correct size (491 bp) were used as template (approximately 0.1 ng of plasmid DNA) for PCR employing the same thermocycling conditions as described for environmental samples, except that cycling was allowed to proceed for only 20 cycles. From each library, a total of 22 to 38 clones bearing an amoA insert were screened by DGGE under the same conditions as described for environmental samples, except that run times were extended to 12 h in order to better resolve the more stable DNA duplexes that tend to migrate to the bottom half of the gel. Under these conditions, certain less-stable DNA duplexes that migrate within the top half of the gel may become fully denatured. Therefore, clones representing these sequences were screened using a 7.5-h run time. Fifteen unique amoA sequences were resolved based on differences in mobility in DGGE gels. Representative clones from each DGGE type were selected for sequencing and phylogenetic analysis.
Sequencing and phylogenetic analysis.
Representative clones from each DGGE type were sequenced using the ABI-PRISM Dye Terminator Cycle Sequencing Core Kit (Applied Biosystems, Perkin-Elmer, Foster City, Calif.). Sequencing products were analyzed on an Applied Biosystems model 310 DNA sequencer following the manufacturer's recommendations. Phylogenetic analyses based on either 450-nucleotide (excluding primers) or deduced 150-amino-acid sequences were performed using algorithms in PHYLIP version 3.57c (J. Felsenstein, University of Washington, Seattle). Evolutionary distances between amino acid sequences were generated using the point-accepted mutation-Dayhoff substitution model implemented in PROTDIST, while distances between nucleotide sequences were generated by a maximum-likelihood method using DNADIST. Distance dendrograms based on the Fitch-Margoliash method were generated using the FITCH algorithm. The topologies of the resulting trees were consistent with those presented below. Robustness of derived groupings was tested by bootstrap using 100 replications.
15N dilution assays.
Gross nitrification assays were based on methods described by Barraclough (11). Each treatment (rice cultivar, including unplanted control) received either K15NO3 or 15NH4Cl (10 atom%) at a rate of 30 μg of N (cm3 of soil)−1. The 15NH4+ treatment was included in order to estimate gross mineralization rates as well. N solutions were injected into each polyvinyl chloride cylinder (described above) at the end of the 3-week drought phase to achieve a homogeneous distribution of the label throughout the rhizosphere. Unlabeled controls were included to account for background 15N. Samples of 10 g for inorganic N extraction were obtained immediately after injection and at 3 and 7 days after 15N application. Samples were shaken in 100-ml aliquots of 2 M KCl, after which the extracted ammonium and nitrate were captured onto acidified Whatman GF/D filter paper disks using a diffusion technique developed by Brooks et al. (15). After each diffusion step, filter paper disks were dried at 40°C for 1 h and then placed into Sn capsules (Europa Scientific, Cheshire, United Kingdom) and analyzed for total N and 15N by automated nitrogen carbon analysis-mass spectrometry (Europa Scientific ANCA-SL Stable Isotope Analysis System). No significant dilution of the labeled NO3− was observed during the first 3 days of incubation. Therefore, nitrification rates were determined between 3 and 7 days of incubation as described by Barraclough (11).
FISH.
Root samples that had been fixed and washed as described above were embedded in Tissue-Tek OCT compound (Miles, Elkhart, Ind.), cryosectioned, mounted, and processed in the same manner as described for wastewater biofilms (35). Subsequent microscopy found that the detection and quantification of AOB on the root surface was difficult due to the patchy distribution of bacteria and intense autofluorescence from roots. Therefore, a method was devised to separate the root surface biofilm from the roots. The method presented below was optimized during preliminary experiments that compared sonication and bead beating as possible methods for stripping surface biofilms from roots.
Root samples totaling 10 cm were resuspended in 2 ml of fresh ethanol-PBS (1:1) solution. The surface biofilm was stripped by sonication (Branson Sonifier; Branson Ultrasonics, Danbury, Conn.) using an output control setting of 2 for 4 min with brief pauses every minute to prevent sample overheating. Under these conditions, most of the surface biofilm was stripped off but not totally disintegrated. The efficiency of stripping was monitored by checking the sonicated roots under the epifluorescent microscope after staining with 4′,6-diamidino-2-phenylindole.
After sonication, the stripped biofilm was concentrated by centrifugation at low speed (ca. 2,000 × g) and resuspended in 500 μl of ethanol-PBS. Fifteen microliters of biofilm suspension was spotted on gelatin (0.1% gelatin, 0.01% chromium potassium sulfate)-coated hybridization slides (Cel-line/Erie Scientific, Portsmouth, N.H.), air-dried overnight, and subsequently dehydrated through a series of ethanol washes (50, 80, and 98%; 3 min each). All hybridization procedures were performed as described by Amann (5) using previously published hybridization and washing stringencies (4, 32). The following oligonucleotide probes were used: Eub338, specific for the domain Bacteria (4); Nso190, specific for AOB of the β-Proteobacteria (32); and Nsm156, specific for the Nitrosomonas lineage (32). Probe Eub338 was labeled with 5-carboxyfluorescein (FAM; Sigma-Genosys), while probes Nso190 and Nsm156 were labeled with 6-carboxy-x-rhodamine (6-ROX; Sigma-Genosys). Eub338 was used as a positive control for confirming that cells stained with either Nso190 or Nsm156 were true bacteria. Since this required double hybridizations, the first hybridization was carried out on the probe with higher thermal stability (Nso190 or Nsm156) followed by the second probe (Eub338). Negative control probes, consisting of the complementary sequences of each probe conjugated to the same dyes, were used to determine background counts due to nonspecific binding of DNA or the dye. The same hybridization and washing stringencies were used for the corresponding negative control probes.
Confocal microscopy and image analysis.
Hybridized samples were analyzed by confocal laser scanning microscopy (CLSM) using an LSM 510 microscope (Carl Zeiss, Jena, Germany) equipped with Ar and HeNe lasers (488 and 543 nm, respectively). For quantification of cells hybridizing to each probe, images having an optical section thickness of 1.2 μm were acquired by random movement of the microscope's stage controls. Cell numbers per unit area were derived using the image analysis software, ImageJ. Firstly, the highest background value of fluorescence intensity in each image was determined manually. This was set as the minimum threshold value to eliminate blank spaces and background fluorescence. Then, the image analysis program was used to count the number of cells corresponding to a specified range of particle sizes and intensity threshold values. The area of the biofilm was similarly measured by setting the appropriate threshold and size range values. Since the images have a known section thickness (1.2 μm), all cell counts could be expressed per unit volume, assuming that each section contains a single layer of cells. For each set of root biofilm samples, at least 15 randomly acquired projection images from three replicate samples were analyzed. Only cells displaying fluorescence from both specific (Nso190 and Nsm156) and general (Eub338) probes were counted as either authentic AOB or Nitrosomonas. All counts were corrected by subtracting counts obtained from the negative control probes.
Oxygen measurements from roots of Mahsuri and IR63087-1-17.
Plants were grown for 3 weeks in liquid culture solution having the following composition: (NH4)2SO4, 50 μM; K2SO4, 1 mM; MgSO4, 2 mM; CaCl2, 1 mM; NaH2PO4, 300 μM; Fe-EDTA, 100 μM; MnCl2, 9 μM; (Na)6Mo7O24, 25 μM; H3BO3, 20 μM; ZnSO4, 1.5 μM; CuSO4, 1.5 μM. The pH of the medium was adjusted to 6.5 with NaOH and replenished every 2 days. In order to immobilize the roots during microelectrode measurements, the plants were transplanted to 1% agar blocks containing the same components as the growth medium prior to microelectrode measurements. Clark-type oxygen microelectrodes with tip diameters of about 10 μm were prepared and calibrated as described by Revsbech and Jorgensen (41). Oxygen profiles were obtained using methods described by de Beer and van den Heuvel (20). Concentration profiles were obtained by using a motor-driven micromanipulator (model ACV-104-HP; Chuo Precision Industrial, Tokyo, Japan) at intervals of 100 to 200 μm from the surface of selected plant roots. Microprofiles were determined three times on different roots at points 2 to 3 cm from the base of three plants.
Data analysis.
Data for 15N analysis (N pools, 15N excess abundance; n = 3) for each treatment within each N source were compared by analysis of variance and least significant difference comparison of means using Statistix for Windows (version 2.0; Analytical Software, Tallahasee, Fla.). The cell densities from FISH were tested for evenness of distribution by applying Wilk-Shapiro tests and constructing histograms. Both tests showed nonnormal distribution in all cases. Therefore, differences between means were verified by Kruskal-Wallis analysis of variance using Statistix for Windows.
Nucleotide sequence accession numbers.
The environmental amoA sequences generated in this study have been deposited in the GenBank nucleotide sequence database under accession numbers AY050674 through AY050688.
RESULTS AND DISCUSSION
The PCR-DGGE-cloning approach.
The combination of PCR and DGGE targeting the 16S rRNA gene has been widely used to detect and characterize AOB populations from various environments (16, 28-30, 51). More recently, the approach has also been demonstrated to be feasible when targeting the amoA gene (36). In this study, DGGE was used to screen libraries of amoA clones and to profile the AOB populations based on amoA retrieved from the root and soil environments.
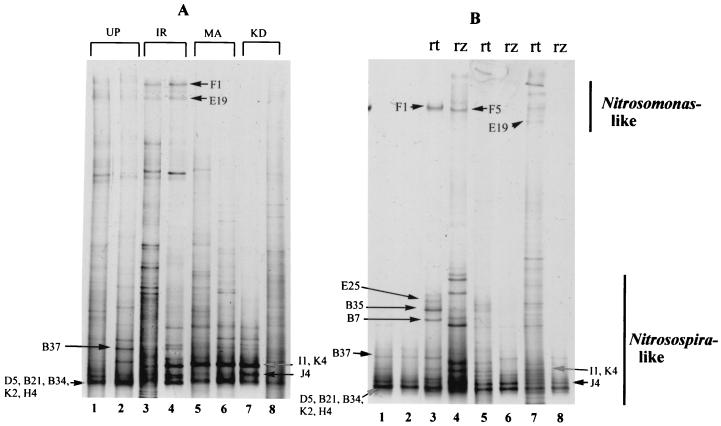
Bands corresponding to Nitrosomonas-like and Nitrosospira-like sequences migrated to the top and bottom halves, respectively, of DGGE gels (Fig. 1). This agrees with results of Oved et al. (36), although our environmental profiles were generally more complex. This might be explained by the two-step amplification procedure that was performed to link amoA with the GC clamp as described above. During preliminary experiments, we found that amplification of environmental samples using the primer pair with the GC clamp was sometimes less efficient compared to that with the original primers. Furthermore, the introduction of a second PCR step allowed us to load more-uniform amounts of DNA in the DGGE gels, since we could use a measured amount of template (1.5 to 2 ng) for the second PCR and limit the additional cycles in order to minimize errors in amplification. This strategy probably increased the yield of nonspecific amplification products, although the detection of less-predominant authentic sequences was also probably increased. Since amoA library clones could be used as reference bands, we confined our analysis to bands with corresponding clones. Based on comparisons between individual amoA clones and the environmental profiles, we were able to determine that the two most prominent bands in Fig. 1 may actually correspond to up to two and five different amoA clones, respectively.
FIG. 1.
(A) DGGE of amoA retrieved from the rhizosphere of three rice varieties. Band designations represent cloned amoA sequences displaying similar migration rates in the DGGE gel. Abbreviations: UP, unplanted; IR, IR63087-1-17; MA, Mahsuri; KD, KDML 105. (B) DGGE of amoA PCR products retrieved from roots and rhizosphere soil. Band designations represent cloned amoA sequences displaying similar migration rates in the DGGE gel. Arrow D5, B21, B34, K2, H4 and arrow I1, K4 denote bands that may be composed of different amoA sequences represented by designated clones. Lanes 1 and 2, unplanted soil; lanes 3 and 4, IR63087-1-17; lanes 5 and 6, Mahsuri; lanes 7 and 8, KDML 105. Abbreviations: rt, root; rz, rhizosphere.
The environmental profiles show that certain AOB strains are more prominent in the presence of rice roots. In particular, bands represented by Nitrosospira clones I1, K4, and J4 appear prominently only in the planted treatments (Fig. 1A). The levels of detection of amoA by PCR and Nitrosomonas by DGGE are compared to their respective detection levels by using FISH in Table 1. In general, amoA was detected in all (roots, rhizosphere, and unplanted soil) samples. DGGE bands corresponding to Nitrosomonas, however, were consistently detected only in the root environment of IR63087-1-17, particularly in the root samples. This was supported by FISH estimates, which detected around three times as many Nitrosomonas cells on the roots of IR63087-1-17 as on the roots of the traditional varieties.
TABLE 1.
Detection of AOB and Nitrosomonas spp. in root, rhizosphere, and soil by PCR, DGGE, and FISHa
| Sample | AOB (Nitrosomonas) detected by:
|
||
|---|---|---|---|
| amoA PCRb | DGGEb | FISHc (107 cells [cm3 of roots]−1) | |
| KDML roots | +++ | (+) | 5.0 ± 0.8 (1.2 ± 0.4) |
| Mahsuri roots | +++ | (−) | 6.0 ± 1.0 (1.3 ± 0.4) |
| IR63087 roots | +++ | (+++) | 6.8 ± 1.2 (4.0 ± 0.3) |
| KDML rhizosphere | +++ | (+) | ND |
| Mahsuri rhizosphere | +++ | (−) | ND |
| IR63087 rhizosphere | +++ | (++) | ND |
| Unplanted soil | +++ | (+) | ND |
PCR was used to amplify the ammonium monooxygenase gene (amoA) which was subsequently resolved by DGGE. Cell densities were detected by FISH using group-specific oligonucleotide probes.
Symbols: +, ++, and +++, presence of amoA or (Nitrosomonas) PCR product in one, two, or three replicates, respectively; −, no product was detected in all replicates. Qualitative results from three replicates are presented.
Means (n = 15) of cell densities ± 95% confidence limits (all symbols and values in parentheses refer to Nitrosomonas).
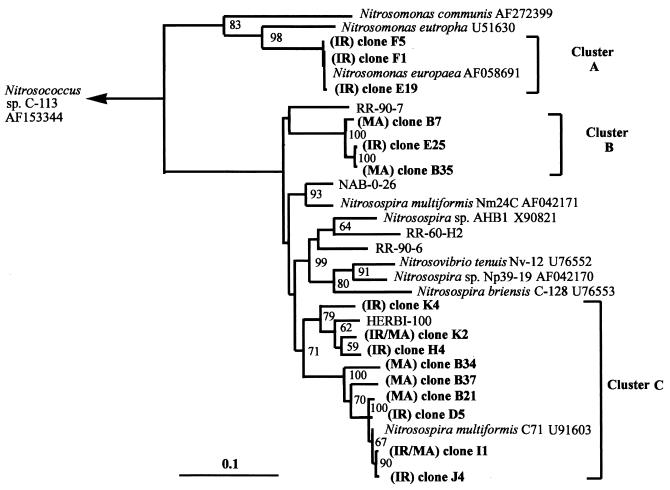
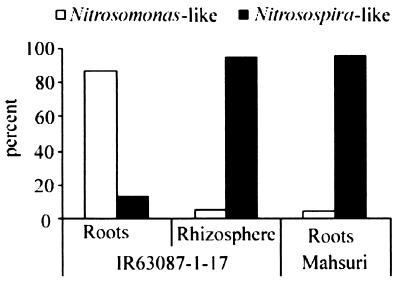
Phylogenetic analysis of all the sequenced amoA clones related one major cluster (cluster A) of sequences to Nitrosomonas europaea and two clusters (clusters B and C) with two different strains of Nitrosospira multiformis (Fig. 2). There was rough agreement between clusters generated by phylogenetic analysis and clone mobility in DGGE gels, indicating that a limited amount of phylogenetic information could be derived from DGGE analysis. The similarities of the retrieved sequences to the DNA and amino acid sequences of the cultured sequences were greater than 80 and 85%, respectively. Based on this criterion (39), all the sequences described in this study are likely to be different strains of either N. europaea or N. multiformis. Based on clone frequency, Nitrosomonas sequences were retrieved at a higher frequency from IR63087-1-17 roots, while Nitrosospira-like amoA sequences were predominant in all other cases (Fig. 3). While these frequencies are based only on single libraries consisting of a small number of clones (20 to 38) from each DNA source, the contrast in AOB population structure associated with roots is partially supported by FISH data (below).
FIG. 2.
Fitch-Margoliash tree of amoA sequences recovered from rice root environment. The sequences span a common stretch of 450 nucleotide bases corresponding to the fragment amplified by the amoA primer pair (minus primer sequences). Sequences in boldface type were generated during this study. Clones were selected from three clone libraries by DGGE. Prefixes indicate the cultivar source of amoA as follows: IR, IR63087-1-17; MA, Mahsuri. Numbers at bifurcations represent percentage bootstrap values (100 repetitions). The scale bar represents 10% sequence divergence.
FIG. 3.
Frequencies of clones containing Nitrosomonas-like and Nitrosospira-like amoA sequences. The clone libraries were generated by PCR of the amoA gene retrieved from IR63087-1-17 roots (22 clones) and rhizosphere (38 clones) and Mahsuri roots (24 clones).
The FISH approach.
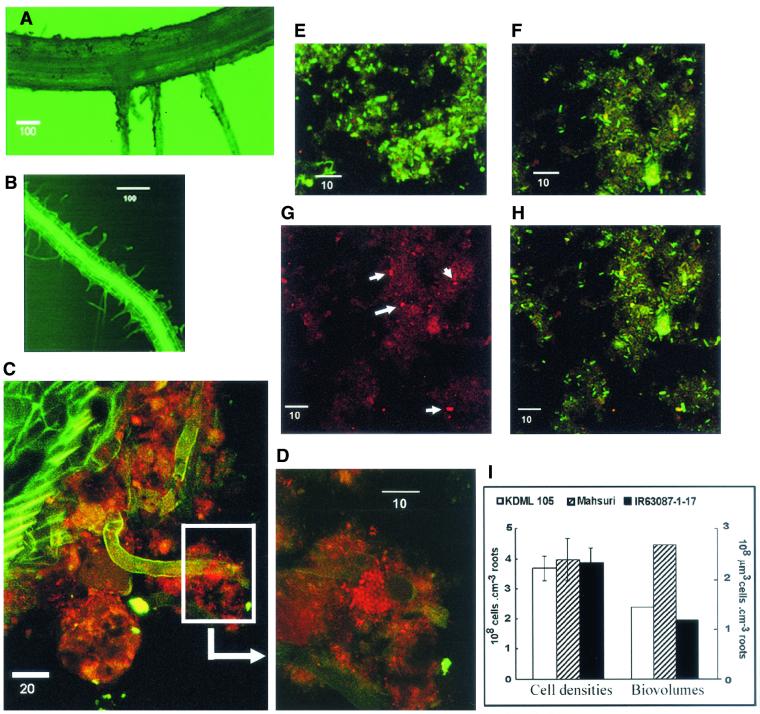
A thin (usually <100-μm-thick) biofilm was observed on the surface of all root samples examined. The roots themselves were strongly autofluorescent, and using epifluorescence microscopy, it was possible to see the biofilm as a dark coating on the root surface (Fig. 4A). On whole root sections, it was possible to detect individual cells and cell clusters stained by Nso190 (Fig. 4C and D). However, the abundance of AOB was relatively low, their distribution was uneven, and root autofluorescence was strong, making quantification impossible on whole root sections. These problems were solved by applying moderate sonication to root samples and probing only the detached biofilm, which could be concentrated on a small surface area (diameter, 8 mm) on glass slides. After sonication, fine root hairs that are typically embedded within the biofilm were revealed (Fig. 4B). This suggests that the increase in root surface area attributed to root hairs may be highly relevant in relation to processes occurring within the biofilm
FIG. 4.
CLSM images of rice roots and root surface biofilm samples. (A) Rice root before sonication; (B) rice root after sonication; (C) biofilm situated on root surface; (D) close-up of boxed region in C. (E to H) Root biofilm samples detached from root surface by sonication: Mahsuri biofilm (E), IR63087-1-17 biofilm (F), and biofilm showing stained AOB (G). (H) Same image as in panel G, but showing all bacterial cells stained by probes Eub338 (green) and Nsm156 (red). Colocalization of probes produces a yellow color. (I) Total cell abundance expressed as cell densities and biovolumes. Error bars indicate 95% confidence intervals. Scale bars in CLSM images are in units of micrometers.
Differences were detected in the general bacterial constituents of the root biofilms of different cultivars. The cells on Mahsuri biofilms were generally larger, with clearer outlines and brighter fluorescence compared to indigenous populations on the other two varieties (Fig. 4E and F). No significant differences were detected in the total abundance of cells on the root biofilms of the three varieties. However, because cells on Mahsuri biofilms were generally larger, the total bacterial biovolume was nearly twice that present in the other varieties (Fig. 4I). These observations suggest that root exudates from Mahsuri favor the development of more heterotrophic copiotrophs.
Detection and quantification of AOB and Nitrosomonas spp.
Under conditions favorable to nitrification, AOB generally occur as large aggregates within thick (>100-μm-thick) biofilms and are frequently associated with nitrite-oxidizing bacteria (22, 35, 46). In whole root sections, we were able to detect clusters of AOB (Fig. 4C and D), although individual cells dispersed unevenly within the biofilm were also frequently observed. It was difficult to make a more detailed analysis of their distribution in situ because of root autofluorescence and their scant and uneven distribution relative to the total bacterial community. While our in situ observations were limited, they do suggest that a mixed community of AOB is present on rice roots, based on cell aggregation characteristics. After sonication, the AOB were mostly dispersed; hence, we should assume that dispersal of aggregates and some cell loss could have occurred during sonication. This may have led to a certain degree of underestimation in our counts. The AOB cells had diameters between 0.5 and 1.0 μm and were usually short or curved rods and spheres (Fig. 4D and G).
We detected no significant differences in the abundances of AOB among the three rice cultivars (Table 1). However, significantly higher (P < 0.05) levels of Nitrosomonas cells were detected in the root biofilms of IR63087-1-17 (Table 1). In this study, we considered the Nitrosomonas density subtracted from the total AOB cell density to reflect the abundance of Nitrosospira spp., since all terrestrial AOB are comprised of either Nitrosomonas or Nitrosospira spp. based on 16S rRNA sequences (24, 52). The percentage of Nitrosomonas associated with IR63087-1-17 ranged from 40 to 69% of the total AOB (based on the ratio of ranges between 95% confidence intervals). In the case of KDML 105 and Mahsuri, the Nitrosomonas cells ranged from 7 to 23% of the total AOB, and therefore, the predominant AOB present on roots of these varieties are Nitrosospira spp. The presence of a higher proportion of Nitrosomonas cells on roots of IR63087-1-17 corresponded with the highest gross nitrification rates measured among the three varieties and unplanted soil (Table 2). Population estimates of Nitrosomonas on root biofilms were positively correlated (R2 = 0.93) with gross nitrification rate estimates. The latter lacked enough sensitivity to provide reliable estimates for Mahsuri and unplanted soil. This may be attributed to low soil ammonium N pools (5 to 8 μg of NH4+-N [cm3 of soil]−1) relative to the added 15NO3− (30 μg NO3−-N [cm3 of soil]−1). The relative differences between estimates, however, should be valid and indicate that significantly lower nitrification rates are associated with Mahsuri and unplanted soil than with the other treatments.
TABLE 2.
Gross nitrification and mineralization ratesa
| Sample | Gross nitrification rate (μg of N · g of soil−1 day−1) | Gross mineralization rate (μg of N · g of soil−1 day−1) |
|---|---|---|
| Soil from rice cultivar | ||
| KDML 105 | 0.25 A | 1.24 A |
| Mahsuri | NSDb | 1.28 A |
| IR63087-1-17 | 1.22 B | 1.35 A |
| Unplanted soil | NSD | 0.93 |
Means (n = 3) followed by the same letter are not significantly different based on least significant difference comparison of means (α = 0.05).
NSD, no significant dilution of the label was detected.
We did not attempt to estimate the abundance of AOB in the rhizosphere soil. PCR for amoA, however, did not yield significantly more product from the rhizosphere soil of IR63087-1-17 (data not shown). Since the PCR conducted in this study was not meant to be quantitative, we cannot make reliable generalizations based on these results. We also failed to detect significant differences in N mineralization rates obtained by the different treatments (Table 2). Collectively, therefore, our data indicate that root surface populations of AOB play a major role in determining the nitrification rates in the root environment.
It is recognized that AOB tend to be surface attached rather than free living (1, 3), and the degree of surface attachment is stronger than that of heterotrophic bacteria (1). In general, bacteria attached within biofilms are more resistant to a variety of environmental stresses (19). Such an advantage would be necessary for slow-growing bacteria such as AOB. Notably, it has been shown that biofilm populations of N. europaea recover faster from starvation than do their free-living counterparts (12). Another response to starvation is the production of exopolysaccharides (48), which would promote stronger surface attachment. This characteristic would be relevant for AOB attached to rice roots, since we would expect the bacteria to be in a starved state between periods of fertilization. Between the two genera of AOB, enhanced aggregate formation has been observed for Nitrosomonas in the marine environment (37) and on membrane filters (26). Since cell aggregates within biofilms may be more resistant to extraction, the failure of DGGE to accurately reflect the levels of Nitrosomonas associated with the rice roots may have arisen from difficulties in extracting the Nitrosomonas DNA from the root samples.
Oxygen excretion from roots.
The existence of intervarietal differences in the flux of oxygen from rice roots has been recognized since 1969 (8), although such differences have never been convincingly linked to the ecology of any group of soil aerobes. Nitrosomonas strains have been described as r strategists, with low substrate affinities and high maximum activity compared to the K strategist Nitrosospira (6, 45, 46). In pure culture, N. europaea exhibits Km (O2) values between 6.9 and 17.4 μM. Oxygen concentration measurements on the rice root surface were generally higher than these values (Fig. 5), but in the case of Mahsuri, oxygen could become more limiting in the presence of heterotrophic bacteria, which are generally superior competitors for oxygen compared to AOB (38, 54). We have observed large, active, and presumably heterotrophic bacteria on root biofilms derived from Mahsuri (Fig. 4E). At present, no kinetic data with respect to oxygen are available for Nitrosospira spp. However, Nitrosospira spp. occurring in a nitrifying fluidized bed reactor exhibited Km (NH4+) values that were 1 to 2 orders of magnitude lower than values for N. europaea (45). This might also imply that Nitrosospira spp. have a higher affinity for oxygen, which would explain their predominance in Mahsuri. On the other hand, aside from the kinetic data cited for N. europaea, experiments conducted on a membrane-bound biofilm system also demonstrated the ability of N. europaea to outcompete Nitrosospira spp. at high substrate and O2 concentrations (44). Therefore, the greater abundance of Nitrosomonas spp. on the roots of IR63087-1-17 can be partly explained by the higher O2 concentration on the root surface. However, since Nitrosomonas spp. also require relatively high NH4+ concentrations, we also expect the turnover of NH4+ in the root environment of IR63087-1-17 to be different from those of other rice varieties.
FIG. 5.
Oxygen concentration profiles from the surface of roots of 3-week-old rice plants grown in culture solution.
In conclusion, we have demonstrated, using molecular methods, an enrichment of AOB on rice root surfaces. The levels of AOB root abundance detected in this study are 2 to 3 orders of magnitude higher on a volume basis than densities usually encountered in soils, including estimates in an aerobic soil obtained by a FISH technique (2, 25, 40). In soils, AOB typically comprise ≪1% of the total bacterial community, while on rice roots, they constituted >10%. Based on the known physiologies of Nitrosomonas and Nitrosospira, their respective abundances on the root surfaces of the different rice varieties could be partially explained by the differences in oxygen release from rice roots, which ultimately resulted in different rates of gross nitrification associated with each variety. We have not yet determined the temporal stability of the associations described. However, since biofilm populations are generally resistant to environmental stresses (19), these are likely to be stable through time. Studies to determine whether the nature of N uptake by the plant can affect the nature and activity of AOB on the root surface will be presented in a separate report. In an intensively cultivated crop such as rice, the microbial activities associated with roots are not likely to be trivial. Further characterization of this component of the rhizosphere communities should provide valuable insight into the nature of N cycling in the rice root environment.
REFERENCES
- 1.Aakra, A., M. Hesselsoe, and L. R. Bakken. 2000. Surface attachment of ammonia-oxidizing bacteria in soil. Microb. Ecol. 39:222-235. [DOI] [PubMed] [Google Scholar]
- 2.Adhya, T. K., P. Patnaik, V. R. Rao, and N. Sethunathan. 1996. Nitrification of ammonium in different components of a flooded rice soil system. Biol. Fertil. Soils 23:321-326. [Google Scholar]
- 3.Allison, S. M., and J. I. Prosser. 1993. Ammonia oxidation at low pH by attached populations of nitrifying bacteria. Soil Biol. Biochem. 25:935-941. [Google Scholar]
- 4.Amann, R., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amann, R. I. 1995. In situ identification of micro-organisms by whole-cell hybridization with rRNA-targeted nucleic acid probes, p. 1-15. In A. D. L. Akkermans, J. D. van Elsas, and F. J. de Bruin (ed.), Molecular microbial ecology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 6.Andrews, J. H., and R. F. Harris. 1986. r- and K- selection and microbial ecology. Adv. Microb. Ecol. 9:99-147. [Google Scholar]
- 7.Armstrong, W. 1979. Aeration in higher plants. Adv. Bot. Res. 7:225-232. [Google Scholar]
- 8.Armstrong, W. 1969. Rhizosphere oxidation in rice; an analysis of intervarietal differences in oxygen flux from roots. Physiol. Plant 22:296-303. [Google Scholar]
- 9.Armstrong, W., S. H. F. W. Justin, P. M. Beckett, and S. Lythe. 1990. Root adaptation to soil waterlogging. Aquat. Bot. 39:57-73. [Google Scholar]
- 10.Arth, I., P. Frenzel, and R. Conrad. 1998. Denitrification coupled to nitrification in the rhizosphere of rice. Soil Biol. Biochem. 30:509-515. [Google Scholar]
- 11.Barraclough, D. 1991. The use of mean pool abundances to interpret 15N tracer experiments. I. Theory. Plant Soil 131:89-96. [Google Scholar]
- 12.Batchelor, S. E., M. Cooper, S. R. Chhabra, L. A. Glover, G. S. A. B. Stewart, P. Williams, and J. I. Prosser. 1997. Cell density-regulated recovery of starved biofilm populations of ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 63:2281-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bedford, B. L., D. R. Bouldin, and B. D. Beliveau. 1991. Net oxygen and carbon dioxide balances in solutions bathing roots of wetland plants. J. Ecol. 79:943-959. [Google Scholar]
- 14.Bouldin, D. R. 1986. The chemistry and biology of flooded soils in relation to the nitrogen economy in rice fields, p. 1-14. In S. K. de Datta and W. H. Patrick (ed.), Nitrogen economy of flooded rice soils. Martinus Nijhoff, Dordrecht, The Netherlands.
- 15.Brooks, P. D., J. M. Stark, B. B. McInteer, and T. Preston. 1989. Diffusion method to prepare soil extracts for automated nitrogen-15 analysis. Soil Sci. Soc. Am. J. 53:1707-1711. [Google Scholar]
- 16.Bruns, M. A., J. R. Stephens, G. A. Kowalchuk, J. I. Prosser, and E. A. Paul. 1999. Comparative diversity of ammonia oxidizer 16S rRNA gene sequences in native, tilled and successional soils. Appl. Environ. Microbiol. 65:2994-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cassman, K. G., M. J. Kropf, J. Gaunt, and S. Peng. 1993. Nitrogen use efficiency of rice reconsidered: what are the key constraints? Plant Soil 155/156:359-362. [Google Scholar]
- 18.Cassman, K. G., S. Peng, D. C. Olk, J. K. Ladha, W. Reichardt, A. Dobermann, and U. Singh. 1998. Opportunities for increased nitrogen-use efficiency from improved resource management in irrigated rice systems. Field Crops Res. 56:7-39. [Google Scholar]
- 19.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 20.de Beer, D., and J. C. van den Heuvel. 1988. Gradients in immobilized biological systems. Anal. Chim. Acta 213:259-265. [Google Scholar]
- 21.George, T., J. K. Ladha, R. J. Buresh, and D. P. Garrity. 1993. Nitrate dynamics during the aerobic soil phase in lowland rice-based cropping systems. Soil Sci. Soc. Am. J. 57:1526-1532. [Google Scholar]
- 22.Gieseke, A., U. Purkhold, M. Wagner, R. Amann, and A. Schramm. 2001. Community structure and activity dynamics of nitrifying bacteria in a phosphate-removing biofilm. Appl. Environ. Microbiol. 67:1351-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hastings, R. C., M. T. Ceccherini, N. Miclaus, J. R. Saunders, M. Bazzicalupo, and A. J. McCarthy. 1997. Direct molecular biological analysis of ammonia oxidizing bacteria populations in cultivated soil plots treated with swine manure. FEMS Microbiol. Ecol. 23:45-54. [Google Scholar]
- 24.Head, I. M., W. D. Hiorns, T. M. Embley, A. J. McCarthy, and J. R. Saunders. 1993. The phylogeny of autotrophic ammonia-oxidizing bacteria as determined by analysis of 16S ribosomal RNA gene sequences. J. Gen. Microbiol. 139:1147-1153. [DOI] [PubMed] [Google Scholar]
- 25.Hesselsoe, M., K. K. Brandt, and J. Sorensen. 2001. Quantification of ammonia-oxidizing bacteria in soil using microcolony technique combined with fluorescence in situ hybridization (MCFU-FISH). FEMS Microbiol. Ecol. 38:87-95. [Google Scholar]
- 26.Hesselsoe, M., and J. Sorensen. 1999. Microcolony formation as a viability index for ammonia-oxidizing bacteria: Nitrosomonas europaea and Nitrosospira sp. FEMS Microbiol. Ecol. 28:383-391. [Google Scholar]
- 27.Hiorns, W. D., R. C. Hastings, I. M. Head, A. J. McCarthy, J. R. Saunders, R. W. Pickup, and G. H. Hall. 1995. Amplification of 16S ribosomal-RNA genes of autotrophic ammonia oxidizing bacteria demonstrates the ubiquity of Nitrosospiras in the environment. Microbiology (Reading) 141:2793-2800. [DOI] [PubMed] [Google Scholar]
- 28.Kowalchuk, G. A., Z. S. Naoumenko, P. J. L. Derikx, A. Felske, J. R. Stephen, and I. A. Arkhipchenko. 1999. Molecular analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in compost and composted materials. Appl. Environ. Microbiol. 65:396-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kowalchuk, G. A., J. R. Stephen, W. de Boer, J. I. Prosser, T. M. Embley, and J. W. Woldendorp. 1997. Analysis of ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in coastal sand dunes by denaturing gradient gel electrophoresis and sequencing of PCR-amplified 16S ribosomal DNA fragments. Appl. Environ. Microbiol. 63:1489-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCaig, A. E., C. J. Phillips, J. R. Stephen, G. A. Kowalchuk, S. M. Harvey, R. A. Herbert, T. M. Embley, and J. I. Prosser. 1999. Nitrogen cycling and community structure of proteobacterial β-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl. Environ. Microbiol. 65:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mendum, T. A., R. E. Sockett, and P. R. Hirsch. 1999. Use of molecular and isotopic techniques to monitor the response of autotrophic ammonia-oxidizing populations of the β subdivision of the class Proteobacteria in arable soils to nitrogen fertilizer. Appl. Environ. Microbiol. 65:4155-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mobarry, B. K., M. Wagner, V. Urbain, B. E. Rittmann, and D. A. Stahl. 1996. Phylogenetic probes for analyzing abundance and spatial organization of nitrifying bacteria. Appl. Environ. Microbiol. 62:2156-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mosier, A. R., S. K. Mohanty, A. Bhadrachalam, and S. P. Chakravorti. 1990. Evolution of dinitrogen and nitrous oxide from the soil to the atmosphere through rice plants. Biol. Fertil. Soils 9:61-67. [Google Scholar]
- 34.Muyzer, G., T. Brinkhoff, U. Nubel, C. Santegoeds, H. Schafer, and C. Wawer. 1997. Denaturing gradient gel electrophoresis (DGGE) in microbial ecology, p. 1-27. In A. D. L. Akkermans, J. D. Van Elsas, and F. J. De Bruijn (ed.), Molecular microbial ecology manual, vol. 3.4.4. Kluwer Academic Publishers, Dordrecht, The Netherlands. [Google Scholar]
- 35.Okabe, S., H. Satoh, and Y. Watanabe. 1999. In situ analysis of nitrifying biofilms as determined by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65:3182-3191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oved, T., A. Shaviv, T. Goldrath, R. T. Mandelbaum, and D. Minz. 2001. Influence of effluent irrigation on community composition and function of ammonia-oxidizing bacteria in soil. Appl. Environ. Microbiol. 67:3426-3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Phillips, C. J., Z. Smith, T. M. Embley, and J. I. Prosser. 1999. Ammonia-oxidizing bacteria of the β subdivision of the class Proteobacteria in the Northwestern Mediterranean Sea. Appl. Environ. Microbiol. 65:779-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prosser, J. I. 1989. Autotrophic nitrification in bacteria. Adv. Microbiol. Physiol. 30:125-181. [DOI] [PubMed] [Google Scholar]
- 39.Purkhold, U., A. Pommerening-Roser, S. Juretschko, M. C. Schmid, H.-P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reichardt, W., A. Briones, R. de Jesus, and B. Padre. 2001. Microbial population shifts in experimental rice systems. Appl. Soil Ecol. 17:151-163. [Google Scholar]
- 41.Revsbech, N. P., and B. B. Jorgensen. 1986. Microelectrodes: their use in microbial ecology. Adv. Microb. Ecol. 9:293-352. [Google Scholar]
- 42.Revsbech, N. P., O. Pedersen, W. Reichardt, and A. Briones. 1999. Microsensor analysis of oxygen and pH in the rice rhizosphere under field and laboratory conditions. Biol. Fertil. Soils 29:379-385. [Google Scholar]
- 43.Rotthauwe, J. H., K.-P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing populations. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schramm, A., D. de Beer, A. Gieseke, and R. Amann. 2000. Microenvironments and distribution of nitrifying bacteria in a membrane-bound biofilm. Environ. Microbiol. 2:680-686. [DOI] [PubMed] [Google Scholar]
- 45.Schramm, A., D. de Beer, J. C. van den Heuvel, S. Ottengraf, and R. Amann. 1999. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: quantification by in situ hybridization and the use of microelectrodes. Appl. Environ. Microbiol. 65:3690-3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schramm, A., D. de Beer, M. Wagner, and R. Amann. 1998. Identification and activities in situ of Nitrosospira and Nitrospira spp. as dominant populations in a nitrifying fluidized bed reactor. Appl. Environ. Microbiol. 64:3480-3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen, T. C. 1969. Induction of nitrate reductase and the preferential assimilation of ammonium in germinating rice seedlings. Plant Physiol. 44:1650-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stehr, G., S. Zorner, B. Bottcher, and H.-P. Koops. 1995. Exopolymers: an ecological characteristic of a floc-attached, ammonia-oxidizing bacterium. Microb. Ecol. 30:115-126. [DOI] [PubMed] [Google Scholar]
- 49.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rDNA sequences related to β-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stephen, J. R., Y.-J. Chang, S. J. MacNaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil β-subgroup Proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stephen, J. R., G. A. Kowalchuk, M.-A. V. Bruns, A. E. McCaig, C. J. Phillips, T. M. Embley, and J. I. Prosser. 1998. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl. Environ. Microbiol. 64:2958-2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teske, A., E. Alm, J. M. Regan, S. Toze, B. E. Rittmann, and D. A. Stahl. 1994. Evolutionary relationship among ammonia- and nitrite-oxidizing bacteria. J. Bacteriol. 176:6623-6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tripathi, B. P., J. K. Ladha, J. Timsina, and S. R. Pascua. 1997. Nitrogen dynamics and balance in intensified rainfed lowland rice-based cropping systems. Soil Sci. Soc. Am. J. 61:812-821. [Google Scholar]
- 54.van Niel, E. W. J., L. A. Robertson, and J. G. Kuenen. 1993. A mathematical description of the behavior of mixed chemostat cultures of an autotrophic nitrifier and a heterotrophic nitrifier/aerobic denitrifier: a comparison with experimental data. FEMS Microbiol. Ecol. 102:99-108. [Google Scholar]
- 55.Wagner, M., G. Rath, R. Amann, H.-P. Koops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 56.Wang, M. Y., M. Y. Siddiqi, T. J. Ruth, and A. D. M. Glass. 1993. Ammonium uptake by rice roots. I. Fluxes and subcellular distribution of 13NH4+. Plant Physiol. 103:1249-1258. [DOI] [PMC free article] [PubMed] [Google Scholar]