Abstract
Parachlamydiaceae are endosymbionts of free-living amoeba first identified in 1997. Two developmental stages, elementary and reticulate bodies, were observed; however, their localization and proportions according to culture condition and duration remain unknown. The life cycle of Parachlamydia acanthamoeba within Acanthamoeba polyphaga was studied by transmission electron microscopy of 8-, 36-, and 144-h coculture. Morphometry and quantification were performed using SAMBA software. The elementary body, the predominant stage within the amoebae, was located mainly within their vacuoles. The multiplication of Parachlamydia bacteria by binary fission of reticulate bodies was independently associated with culture in PYG broth (odds ratio [OR] = 4.4; 95% confidence interval [CI], 1.55 to 12.46) and with the presence of reticulate bodies within the amoebae (OR = 2.10; 95% CI, 1.53 to 2.89). A third developmental stage was observed, the crescent body. Its presence outside and inside the amoebae was associated mainly with prolonged incubation time (OR = 3.98; 95% CI, 1.49 to 10.68, and OR = 5.98; 95% CI, 1.75 to 20.4, respectively). Elementary and crescent bodies were released into the extracellular medium within vesicles or after amoebal lysis. For both, phagocytosis was their mode of entry. This electron micrograph study revealed another infective developmental stage, the crescent body, and provided quantitative analysis of the life cycle of P. acanthamoeba within A. polyphaga.
Legionella was the first pathogen demonstrated to multiply and persist in free-living amoebae (21), which are common inhabitants of natural aquatic environments and water systems (11, 19). Since then, several other intracellular bacterial pathogens were shown to infect free-living amoebae, including Chlamydia pneumoniae (6), Mycobacterium avium (24), Listeria monocytogenes (13), an Ehrlichia-like organism (15), the Legionella-like amoebal pathogens (3, 9), Afipia species (12), and Chlamydia-like endosymbionts (1, 4). The latter, Parachlamydia acanthamoeba, could be pathogenic for humans (7a). However, there are only eight strains of Parachlamydiaceae described to date, including the BN9 endosymbiont (1) and Hall's coccus (4). Amann et al. demonstrated the presence of two developmental stages, a thin-walled gram-negative stage and a gram-positive infective stage (1). These two stages, i.e., reticulate and elementary bodies (well known for Chlamydia spp. [16, 17]), were also subsequently reported by Horn et al. and by Fritsche et al. for additional Parachlamydiaceae strains (7, 10). However, the localization of these bodies within free-living amoebae according to culture condition and duration was not yet studied. It is unknown how they exit the protozoan host. Fritsche et al. showed the presence of Chlamydia-like inclusions in a cyst of Acanthamoeba (7), suggesting that amoebae could also act as Trojan horses for the Parachlamydiaceae, since amoebae are resistant to various disinfecting solutions (5, 20, 26) and to extreme temperature, pH, and osmolarity conditions while encysted (reviewed in reference 18).
In the present paper, we report on a third and as yet unknown developmental stage, the crescent body, and on the life cycle of P. acanthamoeba within Acanthamoeba polyphaga.
MATERIALS AND METHODS
Broth and media.
Broths used for amoebal coculture were obtained by the method of T. J. Rowbotham (22), slightly modified: peptone yeast-extract glucose (PYG), 100 g of proteose peptone (Difco, Sparks, Md.), 10 g of yeast extract (Merck, Darmstadt, Germany), 4.9 g of MgSO4 · 7H2O, 5 g of sodium citrate · 2H2O, 0.1 g of Fe(NH4)2(SO4)2 · 6 H2O, 1.7 g of KH2PO4, 1.97 g of Na2HPO4 · 7H20, 45 g of glucose, and 0.295 g of CaCl2 in 5 liters of distilled water; Page's modified Neff's amoeba saline (PAS), 120 mg of NaCl, 4 mg of MgSO4 · 7H2O, 4 mg of CaCl2 · 2H2O, 142 mg of Na2HPO4, and 136 mg of KH2PO4 in 1 liter of distilled water; encystment buffer was prepared as described by Steinert et al. (24); nonnutritive agar, 1.5 g of agar (Research Organics, Cleveland, Ohio) in 100 ml of PAS. All media were autoclaved for 15 min at 121°C, and broths were also filtered through a 0.22-μm-pore-size membrane (Corning, Corning N.Y.).
Strains, culture, and fixation.
P. acanthamoeba strain BN9 (ATCC VR 1471) was kindly provided by R. Amann. Parachlamydia sp. “Hall's coccus” and A. polyphaga Linc-AP1 were kindly provided by T. J. Rowbotham. Three days' coculture of bacterial strains with A. polyphaga in PYG, corresponding to about 106 bacteria/ml, was used for inoculation of 5 × 105 A. polyphaga organisms/ml. Cocultures in PAS were incubated at 32°C for 8 and 36 h, while PYG has been used for longer incubation (144 h), as amoebal lysis occurred after 3 to 5 days in PAS in the presence of Parachlamydia strains. Amoebae were washed twice in PAS (two centrifugation steps at 179 × g for 10 min; Heraeus Megafuge 1.0 R; Sepatech, Hanau, Germany) before fixation in 4% glutaraldehyde in monophosphate buffer (pH 7.2). A negative control (uninfected A. polyphaga) was treated similarly after 36 h of incubation.
Encystment procedures.
Ten milliters of a 1-day coculture of Parachlamydia with 5 × 105 A. polyphaga organisms/ml was centrifuged at 179 × g for 10 min, and the supernatant was discarded. The pellets were suspended in (i) 0.3 ml of PAS and inoculated on nonnutrient agar or (ii) suspended in 10 ml of encystment buffer. After 18 h of incubation at room temperature in a laminar flow hood (open petri plates), 4 to 5 ml of distilled water were dispersed on nonnutrient agar to resuspend cysts while swabbing with a sterile loop; this procedure was repeated twice to increase its efficiency. This suspension and the 18-h encystment buffer preparations were washed twice and fixed in 4% glutaraldehyde, as described above. Encystment of uninfected A. polyphaga on dried agar plates was performed similarly (negative control).
Electron microscopy.
Fixed samples were washed overnight in monophosphate buffer (pH 7.2) and then fixed for 1 h at 4°C at room temperature with 1% osmium tetroxide. Dehydration was performed by successive washes in increasing acetone concentrations (25 to 100%). After the preparations were incubated for 1 h in a vol/vol suspension of acetone-epon and overnight in epon, they were embedded in Araldite (Fluka, St Quentin Fallavier, France). Thin sections were cut from embedded blocks by an Ultracut microtome (Reichert-Leica, Marseille, France), deposited on copper grids coated with formvar (Sigma-Aldrich, Taufkirchen, Germany) and stained for 10 min with a solution of methanol-uranyl acetate and lead nitrate with sodium citrate in water. Grids were examined from magnifications of ×2,800 to ×180,000 with Jeol 1220 (Jeol, Croissy sur Seine, France) and Morgagni 268D (Philips, Eindhoven, The Netherlands) electron microscopes. Both microscopes were calibrated using shadowcast carbon replicas of diffraction parallel line gratings with a spacing of 463 nm (Euromedex, Souffleleweyersheim, France).
Morphometry and quantification.
The SAMBA IPS version 4.06 software (Alcatel TITN Answare, Toulouse, France) was used for morphometry of amoebae (surface of cytoplasm, vacuoles, and nucleus) and for quantification of each developmental stage of Parachlamydia according to their localization (intracytoplasmic, intravacuolar, and in the immediate proximity, but outside of A. polyphaga). For this purpose, 50 pictures (magnification, ×3,500; TIFF format) were acquired for each broth culture, including negative controls. Amoebae were selected randomly from the amoebae present at the upper left corner of each square of the grids. Fragments of amoebae without visible nuclei were not eligible for quantification. All round bacterial forms were classified as reticulate or elementary bodies according to their structure; crescent bodies were defined as any crescent form. The proportions of cysts and trophozoites were assessed for the 36- and 144-h cultures as well as for cysts preparation.
Statistical analysis.
Localization of the developmental stages of Parachlamydia and encystment efficiency were compared using the χ2 test. Using the Student t test, the mean numbers of developmental bodies were compared according to incubation time and their localization within amoebae. Comparison of median surfaces was performed with the Mann-Whitney test. Logistic regression was used to study the factors associated with the presence of crescent bodies inside and outside amoeba and those associated with multiplication of P. acanthamoeba. These multivariate analyses were adjusted for incubation time, culture broth, strain, and localization of other bacterial developmental stages within amoeba. STATA software (version 7.0; Stata Corporation, College Station, Tex.) was used for analysis.
RESULTS
Samples.
After 144 h of coculture of P. acanthamoebae strain BN9 (BN9) in PYG, there were not enough trophozoites (due to amoeba lysis by BN9) for electron microscopy. The poor quality of the sample of Parachlamydia sp. Hall's coccus in encystment buffer prevented its accurate analysis. Thus, a total of nine preparations were available for electron microscopy, including five samples of infected A. polyphaga trophozoites. Of the 250 trophozoites (50 per sample) for which complete quantification and morphometry was performed, 209 (83.6%) were infected by Parachlamydia and at least one bacterium was found outside, but in the immediate proximity of, 88 (35%) of them.
Developmental stages of P. acanthamoeba organisms and their localization.
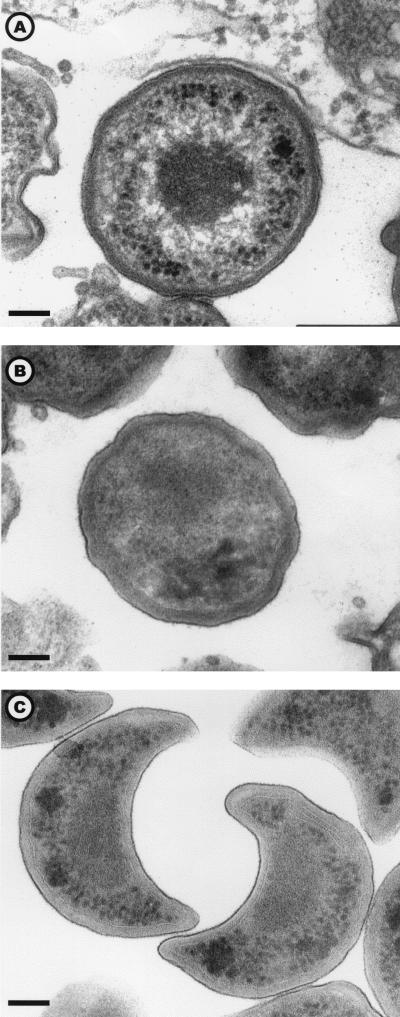
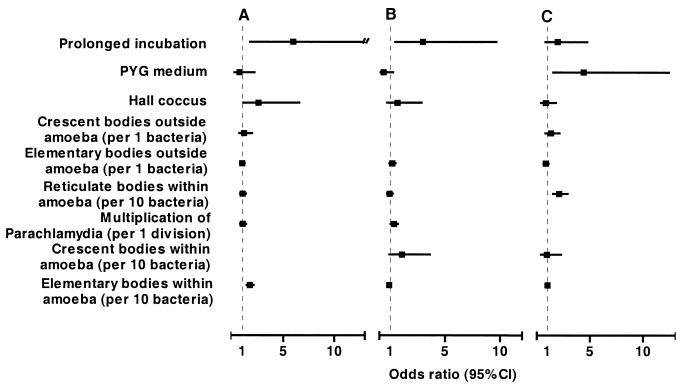
Three morphological stages were observed: the reticulate body (Fig. 1A), the elementary body (Fig. 1B), and the crescent body (Fig. 1C). The elementary body was the predominant stage within the amoeba. Per amoeba, the mean numbers of elementary, reticulate, and crescent bodies and of reticulate bodies undergoing binary fission were 19.4, 12.7, 1.4, and 0.8, respectively. Elementary bodies were located mainly within vacuoles, and their number increased with incubation time. For instance, for Hall's coccus we found a mean number of 39.72 bodies per amoeba after 144 h of incubation, compared to 0.86 (P < 0.001) and 23.26 (P = 0.005) after 8 and 36 h incubation, respectively (Table 1). The reticulate body was the predominant stage within the cytoplasm, but it was also numerous within vacuoles (Table 1). The crescent body was never found within the cytoplasm of the amoeba, but it was found repeatedly within their vacuoles. Even after adjustment, the presence of intravacuolar crescent bodies was associated with prolonged incubation time (odds ratio [OR] = 5.98; 95% confidence interval [CI], 1.75 to 20.4), the presence of intra-amoebal elementary bodies (OR = 1.74; 95% CI, 1.42 to 2.12 [per 10 bacteria]) and, marginally, of Hall's coccus strain (OR = 2.57; 95% CI, 1.004 to 6.59 [compared to the BN9 endosymbiont]) (Fig. 2A). In a similar model, the presence of a crescent body outside but in immediate proximity to A. polyphaga was associated with a prolonged incubation time (OR = 3.98; 95% CI, 1.49 to 10.68), and the presence of reticulate bodies undergoing binary fission within amoebae (OR = 1.33; 95% CI, 1.04 to 1.72 [per bacterium]) (Fig. 2B). Except for the marginal association of Hall's coccus with the presence of intra-amoebal crescent bodies, these observations remained statistically significant when we further adjusted for amoebal surface. The reticulate bodies were rarely observed outside the amoeba: 4 (1.6%) of 250 amoeba. Conversely, elementary bodies were observed exceptionally within the amoebal cytoplasm, and crescent bodies did not seem to invade it at all. The size of the crescent bodies, about 500 nm, is similar to that of the elementary bodies, while reticulate bodies are slightly larger (600 nm). Intermediate states of the transformation of reticulate forms into elementary forms or into crescent bodies were also observed.
FIG. 1.
The three developmental stages of Parachlamydia sp. Hall's coccus: the reticulate body (A), the elementary body (B), and the crescent body (C). Electron micrograph; magnification, ×45,600 (bar = 100 nm).
TABLE 1.
Developmental stages of Parachlamydia in 250 infected trophozoites of Acanthamoeba polyphaga
| Culture group (n) and stagesa | Mean no. of developmental stages/amoeba
|
|||
|---|---|---|---|---|
| Inside Acanthamoeba
|
Outside Acanthamoeba | |||
| Intravacuolar | Intracytoplasmic | Bothb | ||
| All cocultures (250) | ||||
| Elementary bodies | 19.24 | 0.13 | 19.37 | 0.52 |
| Reticulate bodies | 10.21 | 2.54 | 12.75 | 0.02 |
| Crescent bodies | 1.45 | 0 | 1.45 | 0.23 |
| Divisionsc | 0.72 | 0.11 | 0.83 | 0 |
| Hall coccus, PAS 8 h (50) | ||||
| Elementary bodies | 0.86 | 0 | 0.86 | 0.14 |
| Reticulate bodies | 2.02 | 3.20 | 5.22 | 0.02 |
| Crescent bodies | 0.02 | 0 | 0.02 | 0.10 |
| Divisionsc | 0.04 | 0.08 | 0.12 | 0 |
| BN9, PAS 8 h (50) | ||||
| Elementary bodies | 2.30 | 0 | 2.30 | 0.02 |
| Reticulate bodies | 0.46 | 0.10 | 0.66 | 0.02 |
| Crescent bodies | 0.18 | 0 | 0.18 | 0.04 |
| Divisions | 0.16 | 0.04 | 0.2 | 0 |
| Hall coccus, PAS 36 h (50) | ||||
| Elementary bodies | 23.20 | 0.06 | 23.26 | 0.18 |
| Reticulate bodies | 10.66 | 2.52 | 13.18 | 0 |
| Crescent bodies | 2.56 | 0 | 2.56 | 0.44 |
| Divisions | 0.44 | 0.06 | 0.50 | 0 |
| BN9, PAS 36 h (50) | ||||
| Elementary bodies | 30.20 | 0.46 | 30.66 | 0.20 |
| Reticulate bodies | 20.18 | 5.76 | 25.94 | 0.04 |
| Crescent bodies | 1.74 | 0 | 1.74 | 0.34 |
| Divisions | 1.06 | 0.34 | 1.40 | 0 |
| Hall coccus, PYG 144 h (50) | ||||
| Elementary bodies | 39.62 | 0.10 | 39.72 | 2.04 |
| Reticulate bodies | 17.72 | 1.12 | 18.84 | 0.04 |
| Crescent bodies | 2.74 | 0 | 2.74 | 0.24 |
| Divisions | 1.92 | 0.04 | 1.96 | 0 |
n, number of cultures.
Both, intravacuolar and intracytoplasmic.
Divisions, reticulate bodies undergoing binary fission.
FIG. 2.
OR of having crescent bodies inside (A) and outside (B) A. polyphaga and of the presence of reticulate bodies undergoing binary fission (C). Data are adjusted for all variables listed.
Life cycle of P. acanthamoeba within A. polyphaga.
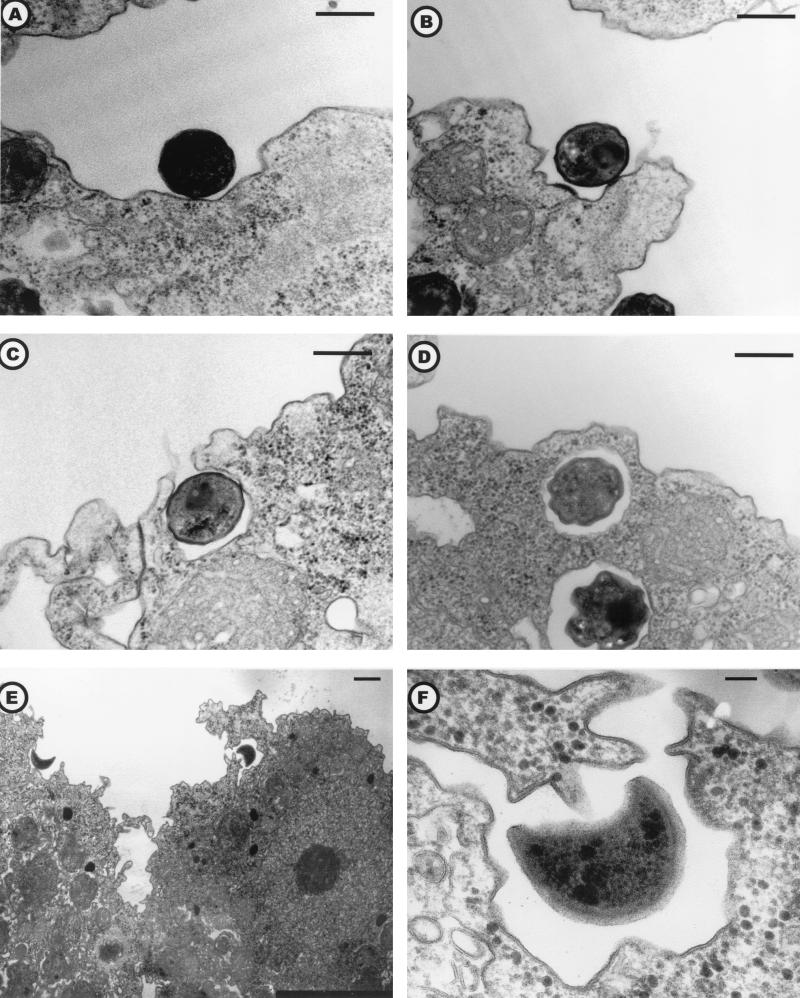
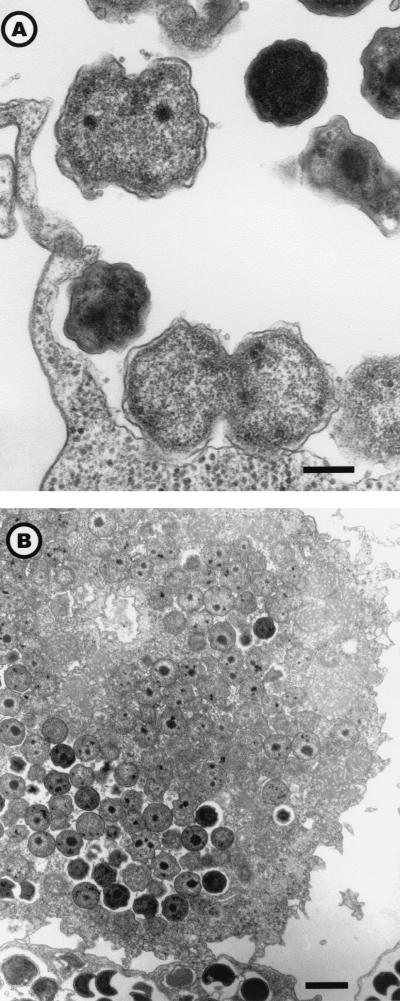
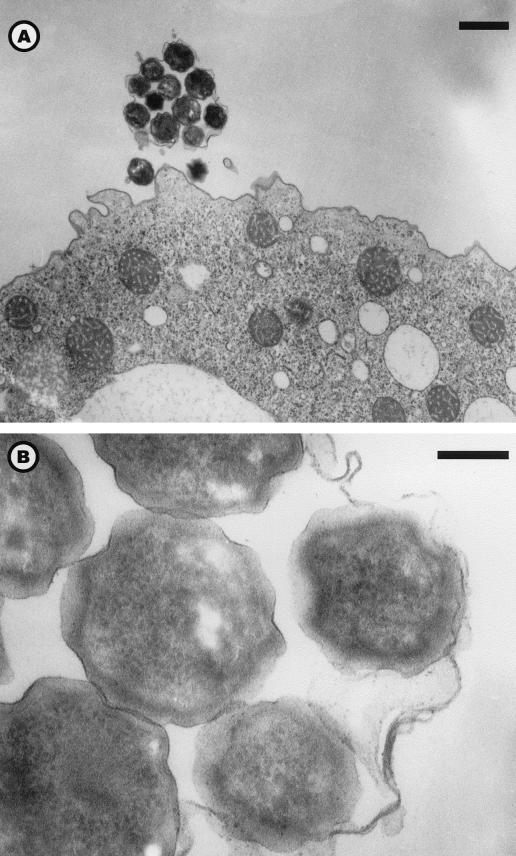
Entry by phagocytosis of elementary bodies (Fig. 3, A to D) or crescent bodies (Fig. 3, E to F) revealed their infective nature. Elementary and crescent bodies could be differentiated in less than 8 h in reticulate bodies (Table 1). The reticulate bodies could divide by binary fission (Fig. 4A) and were able to “invade” the amoebal cytoplasm (Fig. 4B). Multiplication of Parachlamydia, assessed by the presence and number of reticulate bodies in division, occurred mainly in the vacuoles (mean of 0.72 per amoeba; n = 250) and less frequently within the amoebal cytoplasm (mean of 0.11 per amoeba; n = 250). Multiplication was greater in the 144-h PYG broth (mean of 1.92 division per amoeba; n = 50). That observation was confirmed by multivariate analysis: after adjustment, binary fission of reticulate bodies remained associated with culture in PYG broth (OR = 4.4; 95% CI; range, 1.55 to 12.46 [compared to PAS broth]) and with the presence of reticulate bodies within the amoeba (OR = 2.10; 95% CI; range, 1.53 to 2.89 per 10 bacteria) (Fig. 2C). In the vacuoles, the reticulate bodies could be condensed in elementary and/or crescent bodies, which are released after amoebal lysis or expelled within vesicles (Fig. 5A and B). The amoebal size increased with infection; thus, the median total surface for uninfected amoebae (n = 50) was 118 μm2, compared to 147.7 μm2 (P = 0.038) for Parachlamydia-infected samples (n = 250). This increased size was partially due to vacuole formation and/or dilatation. Indeed, the vacuole surface was 10.0 (n = 100) and 31.8 (n = 100) μm2 after 8 and 36 h of incubation in PAS broth (P < 0.001), respectively. However, the cytoplasm surface also was enlarged significantly with prolonged incubation (P < 0.001); it was 62.9 (n = 100) and 125.2 (n = 100) μm2 after 8 and 36 h, respectively. No differences in cytoplasm or vacuole surfaces were observed between A. polyphaga infected with Hall's coccus and the BN9 endosymbiont.
FIG. 3.
Electron micrographs illustrating the phagocytosis of Hall's coccus elementary bodies (A to D) or BN9 endosymbiont crescent bodies (E and F). Magnifications: panels A to D, ×18,700 (bar = 500 nm); panel E, ×4,250 (bar = 1 μm); and panel F, ×51,000 (bar = 100 nm).
FIG. 4.
Binary fission (A) and invasion of the amoebal cytoplasm (B) by reticulate bodies. Hall’s coccus (A) and the BN9 endosymbiont (B) are shown. Electron micrograph; magnifications, ×18,400 (A) (bar = 300 nm) and ×4,600 (B) (bar = 1 μm).
FIG. 5.
Expelled vesicles filled with Parachlamydia sp. Hall's coccus. Electron micrograph; magnifications, ×5,000 magnification (bar = 1 μm) (A) and ×60,000 (bar = 100 nm) (B).
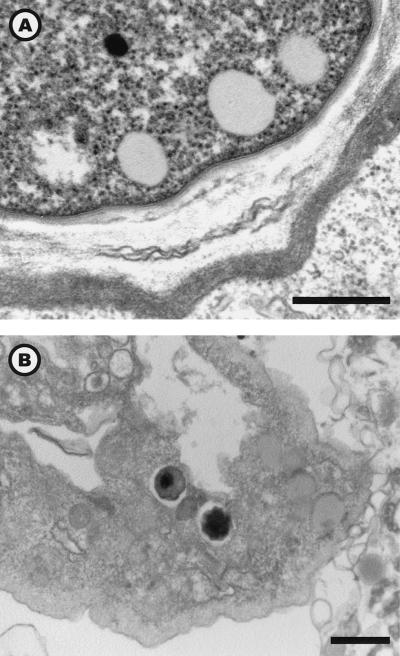
Encystment.
During encystment, some amoebae showed the presence of a few Parachlamydia organisms within vacuoles (Fig. 6B). However, no Parachlamydia organisms were observed in mature cysts (Fig. 6A). The number of cysts was inversely proportional to that of bacteria: 0 of 565 and 2 of 591 (0.34%) for the heavily infected BN9 36-h PAS coculture and Hall 144-h PYG coculture, respectively, compared to 5 of 620 (0.8%) for the Hall 36-h PAS coculture (P = 0.055). However, no differences in encystment efficiency were found between Hall and BN9 with the drying procedure: 11 of 388 (2.8%) and 14 of 452 (3.1%) for BN9 and Hall, respectively. The drying technique was more efficient than encystment buffer (P < 0.001), yet both procedures gave limited encystment: 25 of 840 (2.98%) and 8 of 563 (1.42%), respectively. This low encystment rate contrasted with the 99.6% cysts obtained with the dried agar plate procedure for uninfected sample (P < 0.001 compared to any infected sample). Coalescent cysts were observed in all preparations obtained with the encystment buffer or drying technique, but they were never found in cocultures.
FIG. 6.
Uninfected A. polyphaga cyst (A) and infected amoeba in the process of being encysted (B). Electron micrograph; magnifications, ×30,240 (bar = 500 nm) (A) and ×9,240 (B) (bar = 1 μm).
DISCUSSION
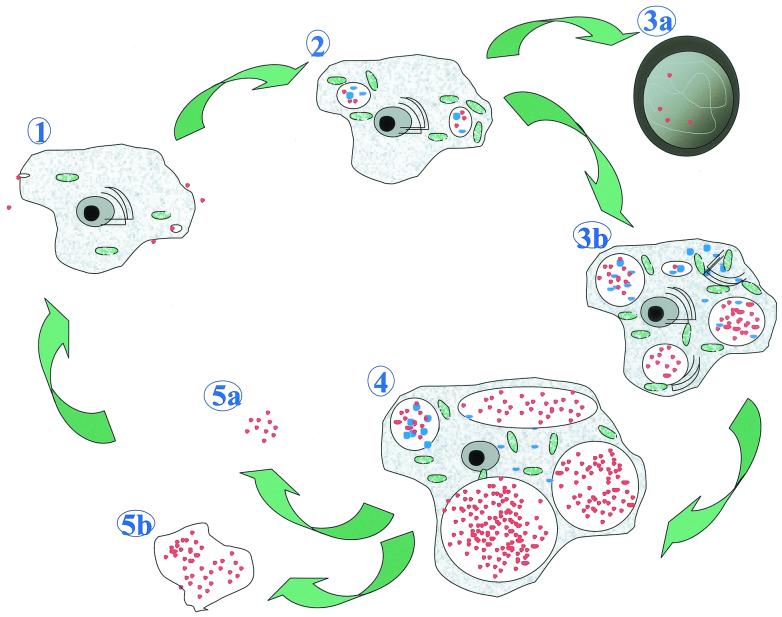
The present electron micrograph study described a third developmental stage, the crescent body, and provided a thorough description and quantitative analysis of the life cycle of P. acanthamoeba within A. polyphaga (Fig. 7).
FIG. 7.
Life cycle of P. acanthamoeba within A. polyphaga. (1) Entry of reticulate or crescent bodies (red dots) within the protozoa. (2) Differentiation in reticulate bodies (blue dots) and multiplication by binary fission within vacuoles. (3a) Encystment: trophozoites heavily infected by Parachlamydia lose their ability to encyst. (3b) Multiplication of the bacteria (many reticulate bodies, intravacuolar and intracytoplasmic), with increased vacuole and cytoplasm sizes. Dilatation of the endoplasmic reticulum (black semicircles) and metabolic activation (mitochondria, in green) are shown. (4) Amoeba with dilated vacuoles filled with elementary and crescent bodies. (5a) Free Parachlamydia bacteria. (5b) Expelled vesicles filled with Parachlamydia bacteria.
The crescent body, though never described earlier, could be seen on several previously published electron micrograph pictures (1, 7, 10). The presence of crescent bodies is thus not restricted to the Parachlamydia species; they can also be found on electron micrographs of the UWE25 endosymbiont (7) and of Neochlamydia hartmanellae (10). Its absence within Chlamydophila and Chlamydia species (16, 23) suggests that it may be restricted to Parachlamydiaceae, and that its presence could be used as a taxonomic tool for this phylum.
Moreover, our data confirmed the hypothesis of Amann (1) that elementary and reticulate parachlamydial bodies behaved similarly to their homologue chlamydial bodies (23). We could show that the condensed infective gram-positive elementary body could be phagocytosed by the amoeba and then could reorganize within the vacuole, in less than 8 h, into reticulate bodies, which are the metabolically active stage. After division by binary fission, the reticulate body could be condensed in an elementary body. The crescent body, found to be gram-positive (data not shown), thus seems to be another infective stage. It is, however, found mainly outside amoeba or within large vacuoles filled with Parachlamydia bacteria. This suggests that the crescent form might be a further stage, besides the elementary body of Parachlamydia, enabling the bacterium to overcome conditions under which the reticulate body would not survive. The crescent bodies were associated with prolonged incubation, independently of their localization within amoeba and of other cofactors. Though its presence within the amoeba correlated with that of elementary bodies, its presence outside the amoeba was independent of that of elementary bodies. Since these associations remained strong in various models, including one adjusted for amoebal surface, and since amoeba were selected randomly, the risk of selection bias can be appreciably reduced. The main limitation of such quantification is a systematic error in classifying the developmental stage in crescent, elementary, and reticulate bodies. However, the same person (G. Greub) acquired all pictures and doubtful quantifications were reassessed, without significantly modifying the initial results.
Our data demonstrated that the phagocytosis of elementary or crescent bodies takes place in less than 8 h. These bodies then differentiated into reticulate bodies, which divides by binary fission, similar to Chlamydiaceae (17), resulting in an exponential increase of intravacuolar bacteria. The observed increased vacuoles' size with prolonged incubation could explain the mechanism of amoebal lysis in heavily infected cultures, such as the lysis observed with the 144-h BN9 coculture. Though lysis released large numbers of infective Parachlamydia organisms, some amoeba could also expel them within vesicles (Fig. 5), decreasing the risk of amoebal lysis. Such a mechanism had been reported only for Legionella pneumophila (2) and was viewed as a potential additional mode of Legionella transmission to humans (8).
Fritsche demonstrated the presence of Parachlamydia within cysts (7). However, encystment of infected amoebae seems to be exceptional, since the encystment rate decreased from 99.6 to 3% with infection with Parachlamydia and since this 3% of cysts were revealed to be uninfected despite systematic observation of all cysts found on a total of 16 electron microscopy grids. It was already reported for another endosymbiont that infected trophozoites lose their ability to encyst (14). Our data strongly suggest that encystment is prevented, at least partially, by Parachlamydia infection. The survival of Parachlamydia within cysts should be further studied, including after exposure to biocides.
The potential pathogenicity of Parachlamydia is not established (7a). However, the multiplication of Parachlamydia within amoebal vacuoles suggests that this bacterium can survive amoebal lytic enzymes and could possibly also survive those of macrophage phagosomes. As C. pneumoniae, whose pathogenicity is well established, was shown to infect A. castellanii, with vacuoles filled with elementary and reticulate bodies (6), one could hypothesize that Parachlamydia organisms will multiply within macrophages, as already has been shown with Chlamydia (25). Additional studies on macrophage permissivity to Parachlamydia should test this hypothesis.
In conclusion, the present report describes for the first time an additional developmental stage, the crescent body. It gives a comprehensive quantitative description of the life cycle of Parachlamydia within A. polyphaga, demonstrating that expelled vesicles are not restricted to the genus Legionella, and thus opening new views on the vector role of free-living amoeba.
Acknowledgments
We thank Lina Barrassi, Claude Cataldo, Mireille Vasserot, Claude Alazia, Bernard La Scola, Gary Burkhart, and Nicolas Aldrovandi for their help and technical assistance.
REFERENCES
- 1.Amann, R., N. Springer, W. Schönhuber, W. Ludwig, E. N. Schmid, K. D. Müller, and R. Michel. 1997. Obligate intracellular bacterial parasites of Acanthamoebae related to Chlamydia spp. Appl. Environ. Microbiol. 63:115-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berk, S. G., R. S. Ting, G. W. Turner, and R. J. Ashburn. 1998. Production of respirable vesicles containing live Legionella pneumophila cells by two Acanthamoeba spp. Appl. Environ. Microbiol. 64:279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birtles, R. J., T. J. Rowbotham, D. Raoult, and T. G. Harrison. 1996. Phylogenetic diversity of intra-amoebal legionellae as revealed by 16S rRNA gene sequence comparison. Microbiology 142:3525-3530. [DOI] [PubMed] [Google Scholar]
- 4.Birtles, R. J., T. J. Rowbotham, C. Storey, T. J. Marrie, and D. Raoult. 1997. Chlamydia-like obligate parasite of free-living amoebae. Lancet 349:925-926. [DOI] [PubMed] [Google Scholar]
- 5.Borazjani, R. N., L. L. May, J. A. Noble, S. V. Avery, and D. G. Ahearn. 2000. Flow cytometry for determination of the efficacy of contact lens disinfecting solutions against Acanthamoeba spp. Appl. Environ. Microbiol 66:1057-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essig, A., M. Heinemann, U. Simnacher, and R. Marre. 1997. Infection of Acanthamoeba castellanii by Chlamydia pneumoniae. Appl. Environ. Microbiol. 63:1396-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritsche, T. R., M. Horn, M. Wagner, R. P. Herwig, K. H. Schleifer, and R. K. Gautom. 2000. Phylogenetic diversity among geographically dispersed Chlamydiales endosymbionts recovered from clinical and environmental isolates of Acanthamoeba spp. Appl. Environ. Microbiol. 66:2613-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Greub, G., and D. Raoult. Parachlamydiaceae: potential emerging pathogens. Emerging Infect. Dis., in press. [DOI] [PMC free article] [PubMed]
- 8.Harb, O. S., and J. A. Kwaik. 2000. Interaction of Legionella pneumophila with protozoa provides lessons. ASM News 66:609-616. [Google Scholar]
- 9.Hay, J., D. V. Seal, B. Billcliffe, and J. H. Freer. 1995. Non-culturable Legionella pneumophila associated with Acanthamoeba castellanii: detection of the bacterium using DNA amplification and hybridization. J. Appl. Bacteriol. 78:61-65. [DOI] [PubMed] [Google Scholar]
- 10.Horn, M., M. Wagner, K. D. Müller, E. N. Schmid, T. R. Fritsche, K. H. Schleifer, and R. Michel. 2000. Neochlamydia hartmannellae gen. nov., sp. nov. (Parachlamydiaceae), an endoparasite of the amoeba Hartmannella vermiformis. Microbiology 146:1231-1239. [DOI] [PubMed] [Google Scholar]
- 11.Kurtz, J. B., C. L. Bartlett, U. A. Newton, R. A. White, and N. L. Jones. 1982. Legionella pneumophila in cooling water systems. J. Hyg. Camb. 88:369-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.La Scola, B., L. Barrassi, and D. Raoult. 2000. Isolation of new fastidious α Proteobacteria and Afipia felis from hospital water supplies by direct plating and amoebal co-culture procedures. FEMS Microbiol. Ecol. 34:129-137. [DOI] [PubMed] [Google Scholar]
- 13.Ly, T. M. C., and H. E. Müller. 1990. Ingested Listeria monocytogenes survive and multiply in protozoa. J. Med. Microbiol. 33:51-54. [DOI] [PubMed] [Google Scholar]
- 14.Michel, R., B. Hauröder-Philippczyk, K. D. Muller, and I. Weishaar. 1994. Acanthamoeba from human nasal mucosa infected with an obligate intracellular parasite. Eur. J. Protistol. 30:104-110. [Google Scholar]
- 15.Michel, R., K. D. Muller, and E. N. Schmid. 1995. Ehrlichia-like organisms (KSL1) observed as obligate intracellular parasites of Saccamoeba species. Endocytobiosis Cell Res. 11:69-80. [Google Scholar]
- 16.Moulder, J. W. 1984. Order II. Chlamydiales, p. 729-739. In N. R. Krieg and J. G. Holt (ed.), Bergey's manual of systematic bacteriology. Baltimore, Md.
- 17.Robinson, E. N. Jr., and Z. McGee. 1989. Chlamydiae and a common sexually transmitted disease, p. 330-337. In M. Schaechter, G. Medoff, and D. Schlessinger. (ed.), Mechanisms of microbial disease. Williams & Wilkins, Baltimore, Md.
- 18.Rodriguez-Zaragoza, S. 1994. Ecology of free-living amoebae. Crit. Rev. Microbiol. 20:225-241. [DOI] [PubMed] [Google Scholar]
- 19.Rohr, U., S. Weber, R. Michel, F. Selenka, and M. Wilhelm. 1998. Comparison of free-living amoebae in hot water systems of hospitals with isolates from moist sanitary areas by identifying genera and determining temperature tolerance. Appl. Environ. Microbiol. 64:1822-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohr, U., S. Weber, F. Selenka, and M. Wilhelm. 2000. Impact of silver and copper on the survival of amoebae and ciliated protozoa in vitro. Int. J. Hyg. Environ. Health 203:87-89. [DOI] [PubMed] [Google Scholar]
- 21.Rowbotham, T. J. 1980. Preliminary report on the pathogenicity of Legionella pneumophila for freshwater and soil amoebae. J. Clin. Pathol. 33:1179-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowbotham, T. J. 1983. Isolation of Legionella pneumophila from clinical specimens via amoebae, and the interaction of those and other isolates with amoebae. J. Clin. Pathol. 36:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rubin, S. J. 1987. Chlamydiae, p. 835-842. In B. J. Howard, J. Klass, A. S. Weissfeld, S. J. Rubin, and R. C. Tilton (ed.), Clinical and pathogenic microbiology. The C.V. Mosby Company, Washington, D.C.
- 24.Steinert, M., K. Birkness, E. White, B. Fields, and F. Quinn. 1998. Mycobacterium avium bacilli grow saprozoically in coculture with Acanthamoeba polyphaga and survive within cyst walls. Appl. Environ. Microbiol. 64:2256-2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyrick, P. B., and E. A. Brownridge. 1978. Growth of Chlamydia psittacci in macrophages. Infect. Immun. 19:1054-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zanetti, S., P. L. Fiori, A. Pinna, S. Usai, F. Carta, and G. Fadda. 1995. Susceptibility of Acanthamoeba castellanii to contact lens disinfecting solutions. Antimicrob. Agents Chemother. 39:1596-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]