Abstract
Codon sequence design is crucial for generating mRNA sequences with desired functional properties for tasks such as creating novel mRNA vaccines or gene editing therapies. Yet existing methods lack flexibility and controllability to adapt to various design objectives. We propose a novel framework, ARCADE, that enables flexible control over generated codon sequences. ARCADE is based on activation engineering and leverages inherent knowledge from pretrained genomic foundation models. Our approach extends activation engineering techniques beyond discrete feature manipulation to continuous biological metrics. Specifically, we define biologically meaningful semantic steering vectors in the model’s activation space, which directly modulate continuous-valued properties such as the codon adaptation index, minimum free energy, and GC content without retraining. Experimental results demonstrate the superior performance and far greater flexibility of ARCADE compared to existing codon optimization approaches, underscoring its potential for advancing programmable biological sequence design.
Keywords: mRNA design, controllable sequence generation, foundation model post training, foundation model for genomics, activation engineering
1. Introduction
Codon design aims to select synonymous codons to construct mRNA sequences that encode the same protein, yet with custom functional attributes. This requires precise control over multiple properties such as codon usage bias, secondary structure stability, and desired protein expression levels. These properties are crucial for controlling mRNA translation dynamics and have direct implications in developing mRNA-based vaccines and medicines [1, 2, 3, 4].
Previous research has tackled the codon design problem on multiple fronts but has largely focused on optimizing a narrow set of properties: codon usage bias (codon adaptiveness index, CAI) and RNA structural stability (minimal free energy, MFE). A lack of explicit, flexible, and fine-grained control over diverse sequence properties is prevalent in both algorithmic approaches [5, 6] and deep learning-based methods [7, 8].
This gap is also present in the more general field of biological sequence generation. Prior works often lack explicit control mechanisms [9, 10, 11] or rely on training auxiliary prediction models for gradient-based guided generation [12, 13, 14, 15], where the performance of the predictors and the need for large-scale labeled data severely limit their utility in controlling diverse properties.
Among the techniques developed for controllable generation in language models (LMs), activation engineering fully utilizes the LM’s inherent knowledge, modifying the LM’s internal activations during inference to steer model outputs towards desired properties [16, 17]. Requiring no LM-specific designs or extensive training as earlier methods do [18, 19, 20, 21, 22], activation engineering promises to be a viable solution to controllability in codon design and biological sequence generation.
Still, there are two major hurdles in the development of controllable codon design methods. First, in natural language processing, controllable generation typically steers discrete and human-interpretable properties, such as sentiment, topic, or styles [20, 23, 21, 19, 24]. However, properties in genomics, such as biological functionality (e.g., protein expression), structural integrity, and translation rate, are often continuous and require computational models or experimental assays for quantification. This highlights a compelling need to extend controllable generation methods, in particular activation engineering, to fine-grained control over continuous properties for genomics sequence design tasks.
Second, current biological foundation models [1, 25, 26, 27, 28] largely adopt encoder-only architectures, which provide powerful pretrained contextual embeddings useful for various downstream tasks, but do not support direct generation as current decoder-only LMs do. This necessitates modifications to the biological foundation models to grant them generation-like capabilities that will facilitate the integration of controllable generation methods developed for LMs.
To bridge these gaps, we propose ARCADE (Activation engineeRing for ControllAble coDon dEsign), a framework that enables attribute-specific control over codon sequences through activation engineering on pretrained biological foundation models. Instead of optimizing for individual attributes, ARCADE aims to adapt to various desired biological properties such as codon adaptive index (CAI), minimized free energy (MFE), protein expression, and their combinations.
ARCADE leverages activation engineering to enable controllable codon sequence generation under synonymy constraints, which guarantee the codons translate to the same protein sequence. We prepare and inject biologically meaningful semantic steering vectors into a foundation model’s latent activation space, allowing direct and flexible control over diverse continuous attributes during generation. To enable generation-like control for codon design, ARCADE augments encoder-only biological foundation models with a token classification objective, reframing codon sequence generation as a position-wise prediction problem.
ARCADE offers flexibility across multiple objectives, fine-grained, continuous control over each attribute, and requires no training or large-scale labeled data. Our implementation is available at https://github.com/Kingsford-Group/arcade.
Our contributions are as follows:
We develop the first deep learning method for controllable codon sequence design, enabling flexible multi-attribute steering without model training.
We enable generation-like control of the outputs of encoder-only biological foundation models through token classification, which leverages their reconstruction capabilities.
We establish a generalizable activation engineering framework capable of handling continuous-valued biological properties, creating biologically meaningful steering vectors that enable explicit control over key biological attributes such as protein expression, mRNA stability, and codon usage diversity.
We extensively validate our method on various codon design objectives and their combinations, showing that it outperforms previous methods in a wide range of settings, allowing far more flexibility to adapt to new design goals.
2. Related work
Controllable text generation
Controllable text generation enables the steering of language model outputs in terms of style, content, or factuality. CTRL [21] introduces control codes in conditional transformers. Li et al. propose prefix tuning [29], which enables lightweight adaptation by optimizing small continuous prompts. PPLM [19] offers inference-time control using external attribute classifiers. Subramani et al. [16] and Hernandez et al. [30] develop learnable activation-based methods that modify internal representations to reflect specific knowledge or behavior.
Meanwhile, training-free activation engineering techniques have gained popularity for their efficiency and flexibility. ActAdd [17] steers generation using the activation difference between prompt pairs. CAA [31] refines this using the average of the activation differences between contrast prompt sets to control behaviors like hallucination or sentiment. Konen et al. [24] allow nuanced control of text style by averaging activations from examples with desired stylistic attributes. These approaches mark significant progress toward efficient, flexible, and precise text generation control, though none have been applied to the codon design task and biological sequence generation in general.
Genomic language models
Advancements in large language models (LLMs) have spurred the development of Genomic Language Models (gLMs), which are large-scale pretrained foundation models capable of performing multiple tasks in understanding and interpreting biological sequences, particularly DNA and RNA sequences. For example, DNABERT [25], Nucleotide Transformer [26], HyenaDNA [32], and EVO [33] are all large-scale gLMs for DNA sequence modeling.
For RNA sequence modeling, gLMs like RNABERT [34] and BigRNA [35] are trained on general RNA sequences, while Li et al. [1] developed a gLM that is more suitable for mRNA and protein expression-related tasks by using the codons of RNA sequences as input. We leverage the representation power of gLMs to extract embeddings of codon sequences, which lie at the basis of ARCADE.
Codon design
Early computational methods for codon design focus on matching the codon usage frequency of the target gene to that of the host organism. For instance, JCAT [5] greedily selects the most frequent synonymous codons in the identified highly expressed genes of the host. More recent algorithms address the complexity and multi-objective nature of codon design. LinearDesign [6] uses lattice parsing to navigate the massive search space for possible codon sequences and jointly optimizes the codon adaptation index (CAI) and minimum free energy (MFE), but its decoding complexity scales quadratically with input length, making it computationally expensive to design long sequences. These non-machine learning methods lack flexibility and generalizability, and can be time-consuming when exploring many variations.
Machine learning approaches learn latent features directly from sequences to guide codon design. They have also achieved linear-time inference and benefited from GPU acceleration. Fu et al. [7] used a BiLSTM-CRF network and treated codon design as a sequence annotation problem, where they mapped amino acid sequences to codon sequences based on the codon usage patterns in the host genome. CodonBERT [8], a BERT-based architecture, applies cross-attention between amino acid and codon sequences to learn contextual codon preferences. RNADiffusion [14], a latent diffusion model that maps RNA sequences to a low-dimensional latent space, enables guided generation by leveraging gradients from reward models operating on the same latent space.
While these machine learning methods can capture generalizable latent sequence features, they lack flexibility in codon design objectives. Our work bridges this gap by exploring activation engineering to flexibly steer codon sequences, imbuing them with a broad spectrum of desired properties.
3. Method
3.1. The codon design problem
The codon design problem is defined as rewriting a given input codon sequence to promote specific biological properties while preserving the encoded protein sequence. Formally, let be an input codon sequence, where each codon translates to amino acid via the translation function . The objective is to construct an output sequence , such that for each and exhibits more favorable biological properties. This latter goal is a variable and multi-objective function that depends on the use case and users’ desires, which are manifested by the user selecting a set of properties for which they want to optimize. The varied properties in Section 4.2 provide extensive examples of these objective functions.
3.2. Activation engineering for codon design
To optimize a set of user-specified biological properties , we apply activation engineering (Figure 1). For an input sequence , we modify the activations of the frozen foundation model to steer its output toward a desired direction, such as increasing or decreasing the value of a specified property .
Figure 1:
Overview of ARCADE. Given an input codon sequence, (a) when there is no steering, the foundation model reconstructs the sequence using a token classification head. (b, c) To guide generation toward desired properties, activation engineering is performed by injecting the learned steering vectors into the layer activations of a frozen foundation model. (d) Steering vectors are constructed from the activation differences between high- and low-value examples, mutated from a set of seed sequences, and scored with a property-specific function.
To steer towards properties , a steering vector is created for each layer and each . A linear combination of these vectors is added to the original activations at layer (Figure 1(c)):
| (1) |
where denotes the modified activation at layer , denotes the original activation, and the values control the steering strengths for each goal.
To construct the steering vector for a property , we label training sequences with high values of the property by and low values as . We denote by and the mean activation vectors at layer for sequences with high and low values of , respectively. The steering vector toward a higher value of the property is defined as
| (2) |
To obtain and samples for each property , we propose a data-driven construction method tailored for continuous-valued biological attributes, as illustrated in Figure 1(d). Specifically, given a set of seed codon sequences, we introduce property-specific directed mutations to generate variants of high and low values of that property. The mutation method depends on the property of interest (see Appendix A.2 for details). The high- and low-value variants are treated as positive and negative examples, respectively, and the steering vector is computed as the difference in average layer-wise activations between these two groups.
In summary, our activation engineering method enables gradient-free, property-aware control over biological sequences, fully eliciting the inherent knowledge in foundation models to fulfill fine-grained, continuous biological design objectives.
3.3. Token classification for sequence reconstruction
While activation engineering is typically applied to decoder-only models with generative capabilities, genomics foundation models are typically encoder-only models [36, 32, 25, 1]. To adapt an encoder-only model for codon sequence generation, we introduce a token classification head that reconstructs the mRNA codon sequences, effectively approximating a decoder-like function (Figure 1(a)).
Let denote a codon sequence of length , where each belongs to the vocabulary of the 64 possible codons. The model encodes the input into contextual representations , where denotes the hidden representation at position . A token classification head maps each to a categorical distribution over the possible codons:
| (3) |
where and are learnable parameters. The model is trained to minimize the cross-entropy loss between the predicted distribution and the ground-truth codon label :
| (4) |
To ensure synonymy, we apply a codon-level mask during training that restricts predictions to codons encoding the same amino acid as the original input (see Algorithm 1 in Appendix A.3). This formulation casts codon reconstruction as a constrained token-level classification task, enabling biologically valid and controllable sequence generation through subsequent activation steering.
4. Experiments
For all our experiments, we build ARCADE on the codon-level foundation model introduced by Li et al. [1], which we then fine-tuned on the codon token classification task (see Appendix A.1 for more details of the foundation model). We demonstrate that ARCADE enables effective and flexible control over a wide range of attributes for codon design.
4.1. Datasets
Training and evaluation data
We extract protein-coding transcripts from the GENCODE Human Release 47 (GRCh38.p14) annotation [37] (abbreviated “GENCODE”). We isolate the coding regions (CDSs) from 85,099 transcripts. To conform to the input length limit of 1,024 codons of the foundation model [1], we filter out sequences longer than 3,072 base pairs. The dataset is split into training and testing sets at a 9:1 ratio, while ensuring all transcript variants of the same gene are assigned to the same split to prevent information leakage between transcripts of the same gene. The training set is used both to fine-tune the token classification head and to compute activation statistics for steering vector construction. The test set contains 8,508 sequences and is held out for evaluation.
mRFP evaluation dataset
For evaluation on experimentally measured monomeric Red Fluorescent Protein (mRFP) expression, we use the data from Nieuwkoop et al. [38], which contains 1,459 variants of the mRFP gene annotated with their measured expression levels. This benchmark enables us to assess the effectiveness of steering methods on a real-world, functionally grounded biological property.
4.2. Diverse codon properties
To evaluate whether ARCADE generalizes across diverse biological and functional properties of codon sequences, we applied it to generate sequences that optimize a wide range of metrics. Details of the metric calculations are provided in Appendix A.8.
Codon usage metrics
The Codon Adaptation Index (CAI) is widely used to evaluate how closely a codon sequence aligns with the preferred codon usage in a given organism [39]. CAI values range from 0 to 1 (unitless), with higher values implying greater compatibility with the host’s translational machinery. Optimizing CAI has been shown to enhance the efficiency in protein translation [40, 41].
Structure-related metrics
Minimum Free Energy (MFE) [42], measured in kcal/mol, reflects the thermodynamic stability of mRNA folding. We report Negated Minimum Free Energy (NMFE) in the results, the additive inverse of MFE. Thus, higher NMFE values correspond to more stable mRNA structures. GC content measures the fraction of guanine and cytosine nucleotides in the sequence (range 0-1). Higher GC content is typically correlated with increased structural stability [43].
Immunity-related metrics
CpG and UpA dinucleotide densities are known to affect the innate immune response. The CpG density and UpA density metrics are computed as the number of CG or UA dinucleotides per 100 base pairs(bp) of sequence [44]. Modulating these densities can attenuate viral replication and enhance vaccine safety [45].
Functional outputs
The expression level of mRFP (metric mRFP) serves as a proxy for evaluating the functional impact of codon optimization strategies. Increasing the expression of a gene (such as mRFP) is a typical goal for improving the efficacy of gene therapies [46, 38]. The distribution of mRFP expression values in the training data is presented in Appendix Figure 5.
4.3. Steering single properties
For each codon property, we apply ARCADE to steer toward either higher (“Positive”) or lower (“Negative”) values. Positive steering results in consistently elevated values, while negative steering leads to decreased values relative to the wild-type (“WT”) sequence (see mean values in Table 1 and distributions in Figure 4, Appendix A.4). This demonstrates the controllability of ARCADE and its broad applicability across multiple aspects of codon design objectives.
Table 1:
Effective control of the metrics in Section 4.2 with ARCADE. “WT” denotes the unmodified wild-type input sequences. mRFP expression is evaluated on the dataset from Nieuwkoop et al. [38], and all other metrics are averages over GENCODE test set (see Section 4.1). .
| Translation (CAI) | Stability (NMFE) | GC content | CpG density | UpA density | mRFP expression | |
|---|---|---|---|---|---|---|
| Positive | 0.964 | 350.42 | 0.671 | 8.168 | 7.024 | 11.110 |
| WT | 0.778 | 331.09 | 0.522 | 3.376 | 3.159 | 10.162 |
| Negative | 0.568 | 305.22 | 0.325 | 1.330 | 1.915 | 8.129 |
We examine the effect of steering strength on CAI and NMFE. Increasing consistently alters the generated sequences in the intended direction (Figure 2). For CAI, values of below approximately 0.25 have a negligible effect, followed by a rapid increase as rises from 0.25 to 2, and saturates around . NMFE shows a similar monotonic trend, with saturation also occurring at a threshold around . This indicates that serves as an effective tuning knob for balancing native sequence properties and desired objectives. For both metrics, we observe that the range is the most appropriate regime: strong enough to meaningfully influence sequence generation yet without exceeding the model’s representation capacity. This provides some practical guidance on how to choose (see more discussion in Section 4.5).
Figure 2:
Effect of steering strength on sequence-level properties. Left: CAI improves rapidly for and plateaus for . Right: NMFE increases with larger and also saturates around . The exact values for each point are provided in the Tables 7 and 8 in Appendix A.5.
4.4. Steering pairs of properties
Flexible steering of pairs of desired properties is important for practical applications where codon sequences must satisfy multiple biological constraints, such as increasing expression while maintaining structural stability. We demonstrate ARCADE’s effectiveness in simultaneously steering towards the property pairs CAI + NMFE, CAI + GC content, and mRFP + NMFE.
As shown in Table 2, joint steering improves both target metrics relative to the wild-type (WT) baseline. For example, in CAI + NMFE, CAI increases from 0.778 to 0.978, while NMFE improves from 331.09 to 424.05. Similar trends hold for joint steering of CAI + GC content, and mRFP expression + NMFE, where both a structural and a functional property are jointly improved over the WT baseline, and matching or exceeding results by single-property steering. These results demonstrate that ARCADE can support robust, multi-objective control across translational and structural features and even real-valued functional outputs.
Table 2:
Joint steering across multiple objective pairs. Each row block corresponds to a pair of target properties. “WT” denotes the unmodified wild-type input sequences. “Single” steers toward a single objective . “Joint Steering” jointly steers the two directions, both using (Eq. 1).
| Objective Pair | Metric | WT | Single | Joint Steering |
|---|---|---|---|---|
| CAI + NMFE | CAI | 0.778 | 0.964 | 0.978 |
| NMFE | 331.09 | 350.42 | 424.05 | |
| CAI + GC | CAI | 0.778 | 0.964 | 0.978 |
| GC | 0.522 | 0.671 | 0.672 | |
| mRFP + NMFE | mRFP | 10.162 | 11.110 | 11.144 |
| NMFE | 195.97 | 223.37 | 225.93 |
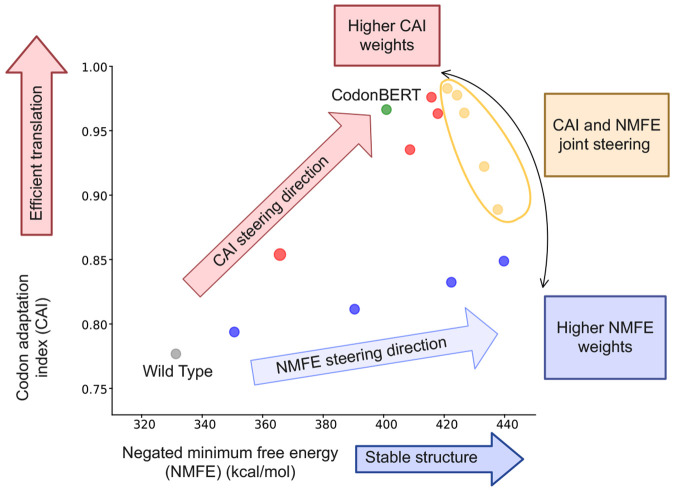
To better understand the effectiveness of multi-objective steering, we analyze the geometry of steering directions in the metrics space. Figure 3 visualizes the average steering effect induced by ARCADE for CAI, NMFE, and their combination with varied steering strength.
Figure 3:
Visualization of steering directions in the metrics space. The vectors represent average value shifts steered by ARCADE for CAI, NMFE, and their combination (CAI+NMFE). This structure suggests the existence of a controllable region in the model’s latent space (top right), where multi-objective trade-offs can be navigated via vector-based interpolation.
Although CAI and NMFE are generally considered to be in trade-off, ARCADE is able to improve both CAI and NMFE at the same time by steering solely for CAI or NMFE (see also Section 4.5). This suggests that, within the model’s representation space, there exists a region where the two objectives can be jointly optimized without strong conflict. ARCADE also exposes a locally controllable subspace within the model’s internal representation, allowing navigation of the trade-offs between objectives. Within this subspace, the steering strength acts as a continuous control parameter, enabling flexible balancing between objectives without model retraining or architectural modification. Instead of collapsing toward one objective, the model responds to joint steering by exploring a shared region of the representation space where both objectives can improve.
4.5. Comparison with other methods
Other methods are typically limited to optimizing a narrow selection of metrics. JCAT [5] only optimizes for CAI. CodonBERT [8] and BiLSTM-CRF [7] only target CAI and MFE, yet they do not allow control of the trade-off between the two metrics.
Though ARCADE is designed for flexible control, it is competitive with these baseline methods on the metrics for which they aim to optimize (Tables 3 and 4). For each objective (CAI, NMFE), we select the best-performing single-property models via a grid search on the training data over steering strengths , denoted as CAIsingle and NMFEsingle. The joint steering model (CAI+NMFE) applies both directions simultaneously with and achieves competitive or superior results on both metrics compared to the other methods.
Table 3:
Comparison of ARCADE with CodonBERT and JCAT on CAI and NMFE metrics. Within ARCADE, CAIsingle and NMFEsingle denote the best-performing single-property models (see Table 9 in Appendix A.6), and “CAI+NMFE” applies both steering directions simultaneously. “unit NMFE” is NMFE normalized by sequence length. The best results among the last three columns are in bold.
| WT | ARCADE | CodonBERT | JCAT | |||
|---|---|---|---|---|---|---|
| CAIsingle | NMFEsingle | CAI+NMFE | ||||
| CAI | 0.773 | 0.978 | 0.849 | 0.978 | 0.967 | 1.0 |
| NMFE | 331.09 | 401.27 | 439.55 | 424.05 | 400.71 | 369.18 |
| unit NMFE | 0.324 | 0.384 | 0.435 | 0.413 | 0.399 | 0.363 |
Table 4:
Comparison of ARCADE with BiLSTM-CRF on a subset of sequences with coding sequence length < 1500, to match the input length constraints of the BiLSTM-CRF model. The best results of the last two columns are marked in bold.
| WT | ARCADE | BiLSTM-CRF | |||
|---|---|---|---|---|---|
| CAIsingle | NMFEsingle | CAI+NMFE | |||
| CAI | 0.778 | 0.981 | 0.851 | 0.979 | 0.750 |
| NMFE | 220.26 | 341.68 | 377.87 | 362.20 | 245.56 |
| unit NMFE | 0.322 | 0.381 | 0.436 | 0.411 | 0.362 |
We also compared ARCADE with LinearDesign [6], a recent non-machine learning method for jointly optimizing CAI and MFE. For fairness, we adopt their combination mode (with internal parameter , distinct from our method). Due to the high computational cost of LinearDesign, we evaluate the methods in Table 5 on a randomly selected subset of 100 coding sequences from the full test set (size 8,508). We also run the second-best method, CodonBERT, from Tables 3 and 4 on this data set.
Table 5:
Comparison with LinearDesign. For mRFP expression, ARCADE applies steering vectors that improve mRFP expression. For CAI and NMFE, ARCADE adopts joint steering combining CAI and NMFE vectors (both with ).
| ARCADE | CodonBERT | LinearDesign | |
|---|---|---|---|
| mRFP expression | 11.186 | 9.916 | 9.570 |
| CAI | 0.977 | 0.964 | 0.944 |
| NMFE | 416.30 | 395.46 | 568.172 |
As shown in Table 5, all methods achieve comparable CAI values, while LinearDesign produces the most stable sequences in terms of NMFE. However, for mRFP expression, ARCADE significantly outperforms both LinearDesign and CodonBERT. This result underscores the flexibility of ARCADE in steering properties beyond CAI and MFE, including experimentally measured functional outcomes.
Even for 100 sequences, LinearDesign requires over 8 hours using two Intel(R) Xeon(R) CPU E5-2690v2 3.00GHz (20 cores), highlighting its limitations for large-scale codon design tasks. In contrast, ARCADE enjoys superior scalability, optimizing these 100 sequences in 16 seconds, including model loading time, using a single NVIDIA A100 GPU with 40GB of memory.
4.6. Ablation studies
To verify that the observed effects are attributable to meaningful steering directions rather than mere perturbation, we compare steering on CAI and NMFE with random steering perturbations and also with steering strength . As shown in Table 1, positive steering raises CAI by 24.7%, increases GC content by 28.5%, and improves NMFE by 5.8% relative to the wild-type baseline, and negative steering reduces CAI by 26.5%, decreases GC content by 37.7%, and lowers NMFE by 7.8%.
In contrast, applying a random steering vector, which is constructed to have the same shape and magnitude as the learned steering vector but filled with random values, or setting the steering strength to zero, results in values nearly identical to those of the wild-type sequence, within 0.01% change relative to the value of wild-type (Table 6). These results confirm that the observed effects do not arise from arbitrary perturbations or noise, corroborating that our steering directions are both informative and closely linked to the biological properties of interest.
Table 6:
Ablation study. “Random” applies random steering vectors with the same norm as the original vectors. “” applies no steering at all.
| CAI | NMFE | GC content | |
|---|---|---|---|
| WT | 0.778 | 331.09 | 0.522 |
| Random | 0.778 | 331.12 | 0.522 |
| 0.778 | 331.12 | 0.522 |
5. Discussion
In this paper, we introduce ARCADE, a flexible and generalizable framework for controllable codon sequence generation. Unlike prior codon design methods, which focus on optimizing a fixed, narrow set of metrics, typically CAI and MFE, ARCADE enables fine-grained, multi-objective control over diverse biological properties, including codon usage, secondary structure stability, nucleotide composition, immune-related motifs, and even experimentally measured expression levels. This process only depends on steering vectors extracted from pretrained foundation models. Other methods that optimize CAI and MFE do not incidentally optimize other metrics such as mRFP expression.
One key insight is that the latent space of pretrained codon language models captures biologically meaningful directions. ARCADE exploits this structure to enable flexible control, supporting both single- and multi-objective generation. Even for properties often considered in trade-off (e.g., CAI vs. MFE), ARCADE identifies directions that jointly improve both metrics towards a Pareto-like trade-off space, offering a new perspective on optimization in genomics design tasks.
Despite its flexibility, ARCADE has some constraints. Its effectiveness depends on the inductive biases of the underlying foundation model and the natural sequences from which the steering vectors were constructed. As a result, steering is confined to biologically plausible regions of the sequence space, which may be good for some applications and limiting for others. We have not evaluated the performance of the model on more than two objectives simultaneously, and it is likely that as the number of objectives increases, the approach may degrade. Moreover, ARCADE applies only global, whole-sequence control and does not provide sequence-specific or position-aware editing.
Future work could address these gaps by integrating local feedback mechanisms or gradient-based refinement to support instance-level control. We also plan to extend this approach to other molecular metrics, such as DNA regulatory elements, and to explore nonlinear or learned steering vectors that adapt to different sequence contexts. Finally, integrating ARCADE into iterative design loops with experimental validation would further validate its utility for synthetic biology.
Acknowledgment
This work was supported in part by the US National Science Foundation [III-2232121] and the US National Institutes of Health [R01HG012470]. Conflict of Interest: C.K. is a co-founder of Ellumigen, Inc.
A. Appendix
A.1. Foundation model backbone
We build ARCADE upon the codon-level foundation model introduced by Li et al. [1], a BERT [47] encoder pretrained on over 10 million mRNA codon sequences spanning a broad range of species, including mammals, bacteria, and human viruses. We use its publicly released checkpoint and tokenizer (https://github.com/Sanofi-Public/CodonBERT). The authors imposed a custom artifact license and software license, which grants permission free of charge, for academic research purposes only and for non-commercial uses only, to any person from academic research or non-profit organizations to copy, use, or modify the software, models, software, datasets and/or algorithms, and associated documentation files. We fully adhere to these two licenses.
The model uses a 12-layer transformer encoder with 12 self-attention heads per layer and a hidden size of 768. Input sequences are tokenized at the codon level using a 64-token vocabulary representing the standard genetic codons. The model is pretrained using a masked codon modeling objective to capture contextual patterns of codon usage across diverse organisms.
We fine-tune the model using Low-Rank Adaptation (LoRA) [48] on the codon token classification task to enable sequence reconstruction. The model parameters are then frozen and used as the fixed backbone for all activation steering experiments. We use learning rate 5 × 10−5, lora rank 32, lora scaling alpha 32, dropout 0.1. The accuracy of the fine-tuned model on the token classification test set is as high as 99.9%.
A.2. Deriving and samples via sequence mutations
To construct steering vectors for each metric, we adopt a metric-specific strategy. We randomly sample a subset of 77 coding sequences from the training set and apply controlled mutations to generate sequence variants.
For CAI, we create by mutating codons with high usage frequencies, and by replacing codons with less frequently used synonyms. A similar strategy is applied for GC content, CpG density, and UpA density.
For NMFE, which is a structural property that is not directly interpretable in terms of codon substitutions, we use LinearDesign [6] to generate high-stability sequences for (high NMFE), while (low NMFE) is generated via random synonymous perturbations.
For the mRFP dataset, which contains mutated coding sequences of the mRFP gene along with experimentally measured expression values, we directly use the top and bottom 3% of sequences ranked by expression as the (high-expression) and (low-expression) sets, respectively.
A.3. Synonymous codon mask
To ensure that only synonymous codon variations are considered during reconstruction, we define a mask of shape (, seqlen, 4) on output logits of token classification head. The mask is constructed as Algorithm 1.
| Algorithm 1 Generating synonymous codon mask | |
|---|---|
|
|
A.4. Performance across single-property steering tasks
To complement the mean values reported in Table 1, Figure 4 presents the full distribution of property values under each steering direction: Positive, Negative, and WT (wild-type baseline). By showing the distributional effects across all sequences, this figure highlights ARCADE’s ability to steer individual sequences, not just shift the mean.
A.5. Effect of steering strength
Tabel 7 and Table 8 show the exact values in Figure 2.
Table 7:
Performance of CAI only steering with different steering strength .
| Metric | WT | CodonBERT | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMFE | 331.09 | 331.12 | 331.29 | 365.49 | 408.50 | 417.69 | 415.59 | 405.43 | 401.27 | 399.46 | 396.58 | 394.47 | 400.71 |
| CAI | 0.777 | 0.778 | 0.7790 | 0.8544 | 0.9358 | 0.9638 | 0.9765 | 0.9765 | 0.9777 | 0.9776 | 0.9760 | 0.974 | 0.9669 |
Figure 4:
Distribution of property values across the test set. Boxplots for each steering direction (Positive, Negative) and the wild-type (WT) baseline are presented. Variability in the distributions reflects differences in sequence length across the dataset.
Table 8:
Performance of NMFE only steering with different steering strength .
| WT | CodonBERT | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NMFE | 331.09 | 331.12 | 331.42 | 350.42 | 390.18 | 422.16 | 438.94 | 439.55 | 440.19 | 400.71 |
| CAI | 0.777 | 0.778 | 0.779 | 0.794 | 0.812 | 0.833 | 0.860 | 0.849 | 0.849 | 0.967 |
A.6. Steering strength in comparison
In Table 9, we present the steering strengths used for the ARCADE models in Tables 3 and 4.
Table 9:
| CAIsingle | NMFEsingle | CAI+NMFE | |
|---|---|---|---|
| 4 | 10 | (1,1) |
A.7. Details in Figure 3
We report the steering strengths represented by the dots in Figure 3.
For the red dots (CAI steering), from bottom-left to top-right, the steering strengths are 0.5, 0.75, 1, 2, respectively.
For the red dots (NMFE steering), from bottom-left to top-right, the steering strengths are 1, 1, 5, 2, 10, respectively.
The combination of the s for the yellow dots, as well as their performance in terms of CAI and NMFE, is presented in Table 10.
Table 10:
The performance of joint steering in Figure 3.
| (1,4) | (1,2) | (1,1) | (4,1) | (10,1) | |
|---|---|---|---|---|---|
| MFE | 433.03 | 426.45 | 424.05 | 405.11 | 397.80 |
| CAI | 0.923 | 0.964 | 0.978 | 0.981 | 0.977 |
A.8. Calculation of metrics
Codon Adaptation Index (CAI)
The CAI quantifies synonymous codon usage bias across species [39]. It is commonly used as a proxy for translational efficiency. Given an mRNA coding sequence of length nucleotides, the number of codons is . Let denoe the -th codon in the sequence, for . Each codon is assigned a relative adaptiveness weight , which reflects its frequency relative to the most used synonymous codon for the same amino acid, based on a reference table (e.g., from the Codon Statistics Database [49]).
Then, the CAI of a sequence is defined as the geometric mean of the adaptiveness values over all codons:
| (5) |
which ranges from 0 to 1, with higher values indicating better alignment with the host’s codon usage preferences [39].
Minimum Free Energy (MFE)
The MFE of an RNA sequence reflects its predicted thermodynamic stability. A lower MFE indicates a more stable RNA secondary structure. In our work, we compute MFE for each mRNA using the RNAfold algorithm from the ViennaRNA package (v2.7.0) with default settings [50]. Results are reported in kcal/mol.
To facilitate interpretation, we use the Negated minimum free energy (NMFE) as defined in Section 4.2, such that higher NMFE values correspond to greater structural stability.
GC content
The GC content of an mRNA sequence is defined as the percentage of guanine and cytosine nucleotides it contains. Let denote the count of nucleotide in .
| (6) |
CpG density and UpA density
The CpG density is defined as the number of CG dinucleotides per 100 base pairs(bp) [44]. It is calculated as follows:
| (7) |
The UpA density is calculated analogously.
mRFP expression prediction
Since mRFP expression is an experimentally measured value without a known analytical formula, we train a predictive model by fine-tuning the pretrained model from Li et al. [1]. We follow the default hyperparameters and training settings described in their paper. The distribution of mRFP expression values in the training set is shown in Figure 5.
B. Safety
B.1. Broader impacts
Our method, ARCADE, introduces controllable codon design by enabling fine-grained manipulation of biological properties such as expression level and structural stability. This can support applications in synthetic biology, gene therapy, and mRNA vaccine development, offering potential societal benefits by accelerating therapeutic design and improving biological sequence performance.
Figure 5:
Distribution of mRFP expression
However, the same controllability could raise risks if used to design sequences with unintended or malicious properties, such as avoiding immunogenicity detection or bypassing biosafety checks. Furthermore, because our approach builds on pretrained models, its behavior is ultimately constrained by the biases and limitations of the underlying training data, which may lead to unreliable results in poorly represented sequence regimes.
While this work remains foundational and is not intended for direct deployment, future applications should consider integrating biosafety filters, rigorous interpretability tools, and transparent sequence audit trails to ensure responsible use.
B.2. Responsible code release
We plan to release the source code for ARCADE under a research-only open-source license (CC BY 4.0) to encourage reproducibility and further academic research. We will include documentation specifying that the code is intended solely for non-commercial, academic use.
To prevent unintended use, we claim that the code is not intended for clinical or therapeutic deployment. Additionally, users are encouraged to conduct proper validation before applying the code to any real-world biological or medical contexts.
References
- [1].Li Sizhen, Moayedpour Saeed, Li Ruijiang, Bailey Michael, Riahi Saleh, Kogler-Anele Lorenzo, Miladi Milad, Miner Jacob, Pertuy Fabien, Zheng Dinghai, et al. CodonBERT large language model for mRNA vaccines. Genome Research, 34(7):1027–1035, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].L Frelin, Ahlen G, Alheim M, Weiland O, Barnfield C, Liljeström P, and Sällberg M. Codon optimization and mrna amplification effectively enhances the immunogenicity of the hepatitis c virus nonstructural 3/4a gene. Gene therapy, 11(6):522–533, 2004. [DOI] [PubMed] [Google Scholar]
- [3].Wang Zijun, Schmidt Fabian, Weisblum Yiska, Muecksch Frauke, Barnes Christopher O, Finkin Shlomo, Schaefer-Babajew Dennis, Cipolla Melissa, Gaebler Christian, Lieberman Jenna A, et al. mrna vaccine-elicited antibodies to sars-cov-2 and circulating variants. Nature, 592(7855):616–622, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Miao Lei, Zhang Yu, and Huang Leaf. mrna vaccine for cancer immunotherapy. Molecular cancer, 20(1):41, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Grote Andreas, Hiller Karsten, Scheer Maurice, Münch Richard, Nörtemann Bernd, Hempel Dietmar C, and Jahn Dieter. Jcat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic acids research, 33(suppl_2):W526–W531, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhang He, Zhang Liang, Lin Ang, Xu Congcong, Li Ziyu, Liu Kaibo, Liu Boxiang, Ma Xiaopin, Zhao Fanfan, Jiang Huiling, et al. Algorithm for optimized mrna design improves stability and immunogenicity. Nature, 621(7978):396–403, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Fu Hongguang, Liang Yanbing, Zhong Xiuqin, Pan ZhiLing, Huang Lei, Zhang HaiLin, Xu Yang, Zhou Wei, and Liu Zhong. Codon optimization with deep learning to enhance protein expression. Scientific reports, 10(1):17617, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ren Zilin, Jiang Lili, Di Yaxin, Zhang Dufei, Gong Jianli, Gong Jianting, Jiang Qiwei, Fu Zhiguo, Sun Pingping, Zhou Bo, et al. Codonbert: a bert-based architecture tailored for codon optimization using the cross-attention mechanism. Bioinformatics, 40(7):btae330, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Killoran Nathan, Lee Leo J, Delong Andrew, Duvenaud David, and Frey Brendan J. Generating and designing dna with deep generative models. arXiv preprint arXiv:1712.06148, 2017. [Google Scholar]
- [10].Repecka Donatas, Jauniskis Vykintas, Karpus Laurynas, Rembeza Elzbieta, Rokaitis Irmantas, Zrimec Jan, Poviloniene Simona, Laurynenas Audrius, Viknander Sandra, Abuajwa Wissam, et al. Expanding functional protein sequence spaces using generative adversarial networks. Nature Machine Intelligence, 3(4):324–333, 2021. [Google Scholar]
- [11].Madani Ali, McCann Bryan, Naik Nikhil, Keskar Nitish Shirish, Anand Namrata, Eguchi Raphael R, Huang Po-Ssu, and Socher Richard. Progen: Language modeling for protein generation. arXiv preprint arXiv:2004.03497, 2020. [Google Scholar]
- [12].Senan Simon, Reddy Aniketh Janardhan, Nussbaum Zach, Wenteler Aaron, Bejan Matei, Love Michael I, Meuleman Wouter, and Pinello Luca. Dna-diffusion: Leveraging generative models for controlling chromatin accessibility and gene expression via synthetic regulatory elements. In ICLR 2024 Workshop on Machine Learning for Genomics Explorations, 2024. [Google Scholar]
- [13].Zongying Lin, Hao Li, Liuzhenghao Lv, Bin Lin, Junwu Zhang, Yu-Chian Chen Calvin, Li Yuan, and Yonghong Tian. Taxdiff: taxonomic-guided diffusion model for protein sequence generation. arXiv preprint arXiv:2402.17156, 2024. [Google Scholar]
- [14].Huang Kaixuan, Yang Yukang, Fu Kaidi, Chu Yanyi, Cong Le, and Wang Mengdi. Latent diffusion models for controllable rna sequence generation. arXiv preprint arXiv:2409.09828, 2024. [Google Scholar]
- [15].Tang Sophia, Zhang Yinuo, Tong Alexander, and Chatterjee Pranam. Gumbel-softmax flow matching with straight-through guidance for controllable biological sequence generation. arXiv preprint arXiv:2503.17361, 2025. [Google Scholar]
- [16].Peters Matthew E. Subramani Nishant, Suresh Nivedita. Extracting latent steering vectors from pretrained language models. arXiv preprint arXiv:2205.05124, 2022. [Google Scholar]
- [17].Turner Alexander Matt, Thiergart Lisa, Leech Gavin, Udell David, Vazquez Juan J., Mini Ulisse, and MacDiarmid Monte. Steering language models with activation engineering. arXiv preprint arXiv:2308.10248, 2023. [Google Scholar]
- [18].Brown Tom, Mann Benjamin, Ryder Nick, Subbiah Melanie, Jared D Kaplan Prafulla Dhariwal, Neelakantan Arvind, Shyam Pranav, Sastry Girish, Askell Amanda, et al. Language models are few-shot learners. Advances in neural information processing systems, 33:1877–1901, 2020. [Google Scholar]
- [19].Dathathri Sumanth, Madotto Andrea, Lan Janice, Hung Jane, Frank Eric, Molino Piero, Yosinski Jason, and Liu Rosanne. Plug and play language models: A simple approach to controlled text generation. arXiv preprint arXiv:1912.02164, 2019. [Google Scholar]
- [20].Hu Zhiting, Yang Zichao, Liang Xiaodan, Salakhutdinov Ruslan, and Xing Eric P. Toward controlled generation of text. In International conference on machine learning, pages 1587–1596. PMLR, 2017. [Google Scholar]
- [21].Keskar Nitish Shirish, McCann Bryan, Varshney Lav R., Xiong Caiming, and Socher Richard. Ctrl: A conditional transformer language model for controllable generation. arXiv preprint arXiv:1909.05858, 2019. [Google Scholar]
- [22].Panigrahi Abhishek, Saunshi Nikunj, Zhao Haoyu, and Arora Sanjeev. Task-specific skill localization in fine-tuned language models. In International Conference on Machine Learning, pages 27011–27033. PMLR, 2023. [Google Scholar]
- [23].Liang Xun, Wang Hanyu, Wang Yezhaohui, Song Shichao, Yang Jiawei, Niu Simin, Hu Jie, Liu Dan, Yao Shunyu, Xiong Feiyu, et al. Controllable text generation for large language models: A survey. arXiv preprint arXiv:2408.12599, 2024. [Google Scholar]
- [24].Konen Kai, Jentzsch Sophie, Diallo Diaoulé, Schütt Peer, Bensch Oliver, Baff Roxanne El, Opitz Dominik, and Hecking Tobias. Style vectors for steering generative large language model. arXiv preprint arXiv:2402.01618, 2024. [Google Scholar]
- [25].Ji Yanrong, Zhou Zhihan, Liu Han, and Davuluri Ramana V. DNABERT: pre-trained bidirectional encoder representations from transformers model for DNA-language in genome. Bioinformatics, 37(15):2112–2120, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Dalla-Torre Hugo, Gonzalez Liam, Mendoza-Revilla Javier, Carranza Nicolas Lopez, Grzywaczewski Adam Henryk, Oteri Francesco, Dallago Christian, Trop Evan, de Almeida Bernardo P, Sirelkhatim Hassan, et al. The nucleotide transformer: Building and evaluating robust foundation models for human genomics. BioRxiv, pages 2023–01, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Hsu Chloe, Verkuil Robert, Liu Jason, Lin Zeming, Hie Brian, Sercu Tom, Lerer Adam, and Rives Alexander. Learning inverse folding from millions of predicted structures. ICML, 2022. [Google Scholar]
- [28].Lin Zeming, Akin Halil, Rao Roshan, Hie Brian, Zhu Zhongkai, Lu Wenting, Smetanin Nikita, Costa Allan dos Santos, Fazel-Zarandi Maryam, Sercu Tom, Candido Sal, et al. Language models of protein sequences at the scale of evolution enable accurate structure prediction. bioRxiv, 2022. [Google Scholar]
- [29].Liang Percy Li Xiang Lisa. Prefix-tuning: Optimizing continuous prompts for generation. arXiv preprint arXiv:2101.00190, 2021. [Google Scholar]
- [30].Andreas Jacob Hernandez Evan, Li Belinda Z.. Inspecting and editing knowledge representations in language models. arXiv preprint arXiv:2304.00740, 2023. [Google Scholar]
- [31].Rimsky Nina, Gabrieli Nick, Schulz Julian, Tong Meg, Hubinger Evan, and Turner Alexander. Steering llama 2 via contrastive activation addition. In Proceedings of the 62nd Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 15504–15522, Bangkok, Thailand, 2024. Association for Computational Linguistics. [Google Scholar]
- [32].Nguyen Eric, Poli Michael, Faizi Marjan, Thomas Armin, Wornow Michael, Birch-Sykes Callum, Massaroli Stefano, Patel Aman, Rabideau Clayton, Bengio Yoshua, et al. HyenaDNA: Long-range genomic sequence modeling at single nucleotide resolution. Advances in Neural Information Processing Systems, 36, 2024. [Google Scholar]
- [33].Nguyen Eric, Poli Michael, Durrant Matthew G, Thomas Armin W, Kang Brian, Sullivan Jeremy, Ng Madelena Y, Lewis Ashley, Patel Aman, Lou Aaron, et al. Sequence modeling and design from molecular to genome scale with Evo. BioRxiv, pages 2024–02, 2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Akiyama Manato and Sakakibara Yasubumi. Informative RNA base embedding for RNA structural alignment and clustering by deep representation learning. NAR Genomics and Bioinformatics, 4(1):lqac012, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Celaj Albi, Gao Alice Jiexin, Lau Tammy TY, Holgersen Erle M, Lo Alston, Lodaya Varun, Cole Christopher B, Denroche Robert E, Spickett Carl, Wagih Omar, et al. An RNA foundation model enables discovery of disease mechanisms and candidate therapeutics. bioRxiv, pages 2023–09, 2023. [Google Scholar]
- [36].Benegas Gonzalo, Ye Chengzhong, Albors Carlos, Li Jianan Canal, andSong Yun S. Genomic language models: opportunities and challenges. Trends in Genetics, 2025. [DOI] [PubMed] [Google Scholar]
- [37].Frankish Adam, Diekhans Mark, Ferreira Anne-Maud, Johnson Rory, Jungreis Irwin, Loveland Jane, Mudge Jonathan M, Sisu Cristina, Wright James, Armstrong Joel, et al. Gencode reference annotation for the human and mouse genomes. Nucleic acids research, 47(D1):D766–D773, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Nieuwkoop Thijs, Terlouw Barbara R, Stevens Katherine G, Scheltema Richard A, De Ridder Dick, Van der Oost John, and Claassens Nico J. Revealing determinants of translation efficiency via whole-gene codon randomization and machine learning. Nucleic acids research, 51(5):2363–2376, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sharp Paul M and Wen-Hsiung Li. The codon adaptation index-a measure of directional synonymous codon usage bias, and its potential applications. Nucleic acids research, 15(3):1281–1295, 1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhou Zhiyong, Schnake Paul, Xiao Lihua, and Lal Altaf A. Enhanced expression of a recombinant malaria candidate vaccine in escherichia coli by codon optimization. Protein expression and purification, 34(1):87–94, 2004. [DOI] [PubMed] [Google Scholar]
- [41].Al-Hawash Adnan B, Zhang Xiaoyu, and Ma Fuying. Strategies of codon optimization for high-level heterologous protein expression in microbial expression systems. Gene Reports, 9:46–53, 2017. [Google Scholar]
- [42].Dirks Robert M, Bois Justin S, Schaeffer Joseph M, Winfree Erik, and Pierce Niles A. Thermodynamic analysis of interacting nucleic acid strands. SIAM review, 49(1):65–88, 2007. [Google Scholar]
- [43].Jing Zhang CC Kuo Jay, and Chen Liang. Gc content around splice sites affects splicing through pre-mrna secondary structures. BMC genomics, 12:1–11, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Beck Daniel, Maamar Millissia Ben, and Skinner Michael K. Genome-wide cpg density and dna methylation analysis method (medip, rrbs, and wgbs) comparisons. Epigenetics, 17(5):518–530, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sharp Colin P, Thompson Beth H, Nash Tessa J, Diebold Ola, Pinto Rute M, Thorley Luke, Lin Yao-Tang, Sives Samantha, Wise Helen, Hendry Sara Clohisey, et al. Cpg dinucleotide enrichment in the influenza a virus genome as a live attenuated vaccine development strategy. PLoS Pathogens, 19(5):e1011357, 2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Alexaki Aikaterini, Hettiarachchi Gaya K, Athey John C, Katneni Upendra K, Simhadri Vijaya, Hamasaki-Katagiri Nobuko, Nanavaty Puja, Lin Brian, Takeda Kazuyo, Freedberg Darón, et al. Effects of codon optimization on coagulation factor ix translation and structure: Implications for protein and gene therapies. Scientific reports, 9(1):15449, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Devlin Jacob, Chang Ming-Wei, Lee Kenton, and Toutanova Kristina. Bert: Pretraining of deep bidirectional transformers for language understanding. In Proceedings of the 2019 conference of the North American chapter of the association for computational linguistics: human language technologies, volume 1 (long and short papers), pages 4171–4186, 2019. [Google Scholar]
- [48].Edward J Hu Yelong Shen, Wallis Phillip, Allen-Zhu Zeyuan, Li Yuanzhi, Wang Shean, Wang Lu, Chen Weizhu, et al. Lora: Low-rank adaptation of large language models. ICLR, 1(2):3, 2022. [Google Scholar]
- [49].Subramanian Krishnamurthy, Payne Bryan, Feyertag Felix, and Alvarez-Ponce David. The codon statistics database: A database of codon usage bias. Molecular Biology and Evolution, 39(8):msac157, 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lorenz Ronny, Stephan H Bernhart, Höner Zu Siederdissen Christian, Tafer Hakim, Flamm Christoph, Stadler Peter F, and Hofacker Ivo L. ViennaRNA Package 2.0. Algorithms for Molecular Biology, 6(1):26, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]