Abstract
The thyA gene, which encodes thymidylate synthase (TS), of Lactococcus lactis CHCC373 was sequenced, including the upstream and downstream regions. We then deleted part of thyA by gene replacement. The resulting strain, MBP71 ΔthyA, was devoid of TS activity, and in media without thymidine, such as milk, there was no detectable dTTP pool in the cells. Hence, DNA replication was abolished, and acidification by MBP71 was completely unaffected by the presence of nine different phages tested at a multiplicity of infection (MOI) of 0.1. Nonreplicating MBP71 must be inoculated at a higher level than CHCC373 to achieve a certain pH within a specified time. For a pH of 5.2 to be reached in 6 h, the inoculation level of MBP71 must be 17-fold higher than for CHCC373. However, by adding a limiting amount of thymidine this could be lowered to just 5-fold the normal amount, while acidification was unaffected with MBP71 up to an MOI of 0.01. It was found that nonreplicating MBP71 produced largely the same products as CHCC373, though the acetaldehyde production of the former was higher.
Lactic acid bacteria are employed in dairy fermentations for acidification and flavor formation. The lactic acid produced is important for the texture and taste of the product, and without sufficient lactic acid formation the result may be products of poor quality. Since the milk and the conditions in dairies are generally not sterile, bacteriophage constitutes a serious threat in the production of dairy products. The phage infects the lactic acid bacteria and propagates in an explosive manner, leading to cell death and thereby defective acidification. Lactococcal phage has been classified into 12 species according to morphology and DNA homology (26). Three of the phage species, 936, c2, and P335, are encountered in dairy plants worldwide (32). Bacterial strains are generally only susceptible to some of the 12 phage species.
Traditionally the number of infected batches has been kept down by employing naturally resistant bacterial strains. For Lactococcus lactis these naturally occurring resistance mechanisms can be divided in four groups: interference with phage adsorption, inhibition of injection of DNA, restriction and/or modification (R/M), and abortive infection (Abi) systems. Abi systems interfere with the phage cycle and result in a reduction of efficiency of plaquing (EOP) to 0.5 to 10−9 (2). However, most Abi systems do not target all the phage species the host is susceptible to, or they have at least a reduced efficiency for one of the main phage species. R/M systems in general function against a broad range of bacteriophages but with an EOP substantially higher than those obtained with the Abi systems, i.e., 10−1 to 10−6 (2). In addition, phage escaping the restriction is protected against the same R/M system.
Alternative methods for phage resistance have been engineered. Receptor mutants lacking the phage infection protein (Pip) are resistant to c2-type phage (3), and phage mutants overcoming the resistance phenotype could not be isolated. However, the resistance mechanism was limited to c2 phage. Another engineered system promotes resistance against P335-type phage by combining a bacteriophage-inducible promoter with a restriction cassette (16). In this system the phage infection triggers the expression of the LlaR restriction gene, resulting in cell death and an EOP of 10−4 for phage φ31. Another means of disturbing the phage life cycle is by providing false targets of origin of replication of the phage genome, which then compete for the replication functions (38). When the origin of replication from phage φ50, ori50, was cloned on a high-copy-number plasmid, the EOP for φ50 was reduced to 2.5 × 10−4. Antisense mRNA has also been employed as a means of phage resistance (31). It was shown that the expression of the antisense transcript of a complete gene reduced the total PFU for a range of isometric-headed phage by more than 99%. However, recent efforts employing the antisense strategy have met little or no success (2, 48), though a combination of the false-target and antisense strategies, i.e., a low copy-number plasmid with the φ31 origin of replication transcribing antisense mRNA, resulted in an EOP of 0.11 (49).
All the described mechanisms of phage resistance have a relatively limited scope, since a vast number of types of phage variants exist in nature. Furthermore, the phage may evolve and circumvent the defense mechanisms. Hence, there is a demand for a more robust method of avoiding phage infections, so dairy fermentations may be run with consistent results.
When a phage infects a cell it uses the cell system of DNA replication to replicate its own DNA. If DNA replication is abolished the phage will not be able to proliferate and the culture is completely protected (37). A mutant host unable to carry out DNA replication should thus be resistant to all phage species. One method of abolishing DNA replication is by limiting one or more of the four deoxynucleoside triphosphates (dNTPs). Thymidylate synthase (TS), which converts dUMP to dTMP, is an essential enzyme in the de novo synthesis of dTTP (Fig. 1). TS is encoded by the thyA gene, and a strain defective in this gene will not be able to synthesize dTTP unless thymidine is present in the medium (1). Since milk is devoid of thymidine, DNA replication is abolished for a thyA mutant culture when it is inoculated into milk. Here we show that the acidification by a thyA mutant is completely unaffected by phage.
FIG. 1.
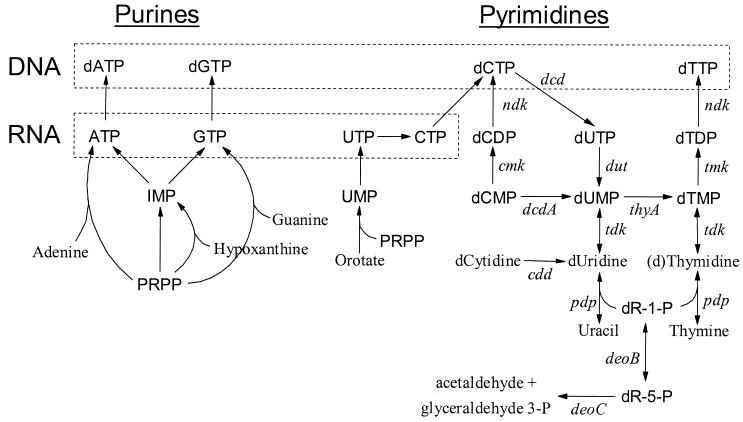
Overview of the nucleotide metabolism of L. lactis. Purine and pyrimidine metabolism is shown to the left and right of the figure, respectively. Only the interconversions of the deoxyribonucleotide pyrimidines and precursors are shown in detail. Other reaction arrows may comprise more than one enzymatic reaction. All genes indicated have been localized in the genome of L. lactis IL1403, except ndk and dcd (7). In the IL1403 genome sequence tdk is referred to as yfiG and tmk is referred to yeaB. Genes: deoB, phosphopentomutase; deoC, deoxyriboaldolase; pdp, thymidine phosphorylase; cdd, cytidine deaminase; tdk, thymidine kinase; dcdA, dCMP deaminase; thyA, TS; cmk, CMP kinase; dut, dUTPase; tmk, thymidylate kinase; ndk, NDP kinase; dcd, dCMP deaminase. PRPP, 5-phosphoribosyl-1-pyrophosphate.
MATERIALS AND METHODS
Bacterial strains, bacteriophages, plasmids, media, and growth conditions.
The bacterial strains, phages, and plasmids used in the present study are listed in Table 1. Cultures of L. lactis were routinely grown at 30°C in LM17, i.e., M17 supplemented with 0.5% of lactose (46). SA is a defined medium with 19 amino acids (28). LSA indicates SA with 1% lactose. For acidification experiments Milex 240 skim milk powder (Arla Food Ingredients, Viby, Denmark) was used after it had been reconstituted at 9.5% (wt/vol) and boiled for 30 min. This is referred to herein as reconstituted skim milk (RSM). During the manipulations to construct MBP71, erythromycin (5 mg/liter) was added. Thymidine was added at the nonlimiting concentration of 20 mg/liter when required for propagation. For cloning of pGhost9 derivatives Escherichia coli DH5α (Life Technologies Inc., Carlsbad, Calif.) was grown at 30°C in Luria-Bertani medium (5) supplemented with 250 mg of erythromycin per liter. Phage was purified as described by Terzaghi and Sandine (46).
TABLE 1.
Bacteria, phages, and plasmids used in this research
| Bacterium, phage, or plasmid | Comment | Reference or source |
|---|---|---|
| Bacteria | ||
| Lactococcus lactis subsp. lactis | ||
| NCDO712 | Wild type | 20 |
| CHCC373 | Wild type | Chr. Hansen Culture Collectiona |
| MBP65-3 | CHCC373 with pMBP63 integrated | This study |
| MBP71 | CHCC373 with ΔthyA | This study |
| Escherichia coli DH5α | Life Technologies Inc. | |
| Phages | ||
| CHPC12 | P335 species | Chr. Hansen Phage Collection |
| CHPC412 | P335 species | Chr. Hansen Phage Collection |
| CHPC708 | 936 species | Chr. Hansen Phage Collection |
| CHPC710 | 936 species | Chr. Hansen Phage Collection |
| CHPC733 | P335 species | Chr. Hansen Phage Collection |
| CHPC783 | 936 species | Chr. Hansen Phage Collection |
| CHPC795 | 936 species | Chr. Hansen Phage Collection |
| CHPC814 | 936 species | Chr. Hansen Phage Collection |
| CHPC836 | 936 species | Chr. Hansen Phage Collection |
| Plasmids | ||
| pGhost9 | Gene inactivation vector | 36 |
| pMBP57 | pGhost9 derivative with 400 bp of thyA | This study |
| pMBP63 | pGhost9 derivative with ΔthyA (two 800-bp fragments) | This study |
Chr. Hansen A/S, Hørsholm, Denmark.
Oligonucleotide primers.
The oligonucleotide primers used in the present study are listed in Table 2. The primers were obtained from TAG Copenhagen A/S, Copenhagen, Denmark, or MWG-Biotech, Ebersberg, Germany.
TABLE 2.
Oligonucleotide primers used in this studya
| Primer | Sequence | Positionb | Details | Restriction site |
|---|---|---|---|---|
| Pmb12 | TAATAAGAATTCCGCAGATCAAGTTTTTAAAC | 1148→ | Initial thyA fragment | EcoRI |
| Pmb15 | TAATAATCTAGAGGCTCATAGTCCACAAGTTC | 1904← | Initial thyA fragment | XbaI |
| Pmb20 | NNNNNNNNNNGAATTC | Random primer for EGWc | ||
| Pmb21 | NNNNNNNNNNAAGCTT | Random primer for EGW | ||
| Pmb22 | NNNNNNNNNNTCTAGA | Random primer for EGW | ||
| Pmb39 | GGAATTGGACGCAAAGTGG | 1290← | EGW upstream (step 1) | |
| Pmb38 | CATAAGTAACGAAAGAGCC | 1244← | EGW upstream (step 2) | |
| Pmb37 | ACTATCTGCGGTTTGACC | 1214← | EGW upstream (step 3) | |
| Pmb24 | CCTTATACTTGGGACGTGC | 1193← | EGW upstream (initial sequence) | |
| Pmb66 | CTTGGATGCGACTCTTATTC | 1649→ | EGW downstream (step 1) | |
| Pmb67 | CAAATGATATGCTTGTTGCGC | 1676→ | EGW downstream (step 2) | |
| Pmb27 | GATAATCAGTTTGAGCAGGC | 1803→ | EGW downstream (step 3) | |
| Pmb40 | GAGTTAGTTAAGCGAACAGC | 1827→ | EGW downstream (step 4) | |
| Pmb41 | GCTTAATGTTCCTGACGG | 1869→ | EGW downstream (initial sequence) | |
| Pmb55 | TATAATCTGCAGGGTCACACTATCAGTAATTG | 336→ | Left fragment in pMBP63 | PstI |
| Pmb63 | TATTTTAAGCTTCACAGTCTGCTATTTTGATTC | 1066← | Left fragment in pMBP63 | HindIII |
| Pmb64 | TAAATTAAGCTTCGCAGACAAGATTTTTAAAC | 1148→ | Right fragment in pMBP63 | HindIII |
| Pmb65 | ATTTAAGTCGACGGCTCATAGTCCACAAGTTC | 1904← | Right fragment in pMBP63 | SalI |
| Pmb43 | GCTTCGATTTTAGTATATGG | 817→ | Sequence across thyA deletion | |
| Pmb36 | GACTGTTGCCCCATAGCG | 1439← | Sequence across thyA deletion |
For primers that contain a restriction site, this site is underlined and the name of the respective enzyme is shown in the rightmost column. The primers are not written in a strictly numerical order.
Data in this column indicate the binding position of the 3′-end nucleotide of the primer according to Fig. 2. The arrow indicates the orientation.
EGW, easy gene walking.
DNA sequencing.
Sequencing was carried out using an ABI Prism 310 Genetic Analyzer according to the manufacturer's recommendations (PE Applied Biosystems, Foster City, Calif.).
Sequencing of thyA and the upstream and downstream regions.
Chromosomal DNA was purified as described earlier (15). About 800 bp of the 840-bp thyA gene of L. lactis subsp. lactis CHCC373 was amplified by PCR on chromosomal DNA using the primers Pmb12 and Pmb15 designed from the thyA sequence of L. lactis subsp. lactis NCDO712 (43). This fragment was then purified and sequenced. To obtain the sequence upstream of thyA in CHCC373, easy gene walking was employed as recommended previously (21). The nested primers Pmb39, Pmb38, and Pmb37 (Table 2) were used consecutively. Three parallel series were run with each of the random primers Pmb20, Pmb21, and Pmb22. In the first step the template was chromosomal DNA. The resulting mixture of bands was purified with the QIAquick PCR purification kit (QIAGEN GmbH, Hilden, Germany), and initially sequenced with Pmb24. Easy gene walking was also employed in sequencing the region downstream of thyA, but with four nested steps instead of three, i.e., the primers Pmb66, Pmb67, Pmb27, and Pmb40 followed by sequencing with Pmb41 (Table 2). The 2,417-bp sequence that was obtained is available from GenBank (accession no. AF336368).
Construction of MBP71 ΔthyA.
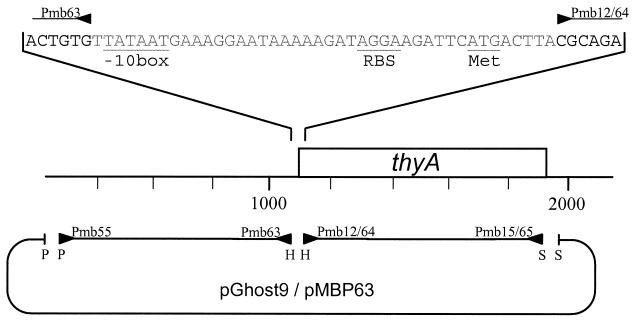
An approximately 800-bp fragment upstream of thyA from CHCC373 was obtained by PCR on chromosomal DNA with the primers Pmb55 and Pmb63 (Table 2), which furthermore introduced a PstI and a HindIII site upstream and downstream, respectively. The resulting fragment includes the putative −35 box of the thyA promoter but not the −10 box (Fig. 2). Another approximately 800-bp fragment comprising an internal part of the thyA coding region was obtained by PCR on chromosomal DNA with the primers Pmb64 and Pmb65 (Table 1), which introduced a HindIII and a SalI site upstream and downstream, respectively (Fig. 2). The resulting fragment includes the whole coding region of thyA except the first 8 bp and the last 34 bp. The PCR was carried out using the Pwo polymerase with proofreading (Boehringer Mannheim, Basel, Switzerland). The two PCR fragments were purified with the QIAquick PCR purification kit (QIAGEN GmbH), cut with the indicated restriction enzymes (Boehringer Mannheim), and purified again. The pGhost9 vector (36) was cut with the restriction enzymes PstI and SalI and purified. Thereafter, approximately 150 ng of the vector and 50 ng of each of the two fragments were ligated overnight at 16°C in a total volume of 20 μl using T4 DNA ligase (Boehringer Mannheim). From this mixture 10 μl was used to transform E. coli DH5α as described earlier (44). Plasmid DNA was isolated from 16 clones using the QIAprep spin miniprep kit protocol (QIAGEN GmbH). The purified plasmid DNAs were cut with PstI and SalI, and one of them, pMBP63, yielded a band of approximately 1,600 bp. This construct was further verified by PCR with the primers that were used to produce the two fragments and also with the two outer primers. Finally, the construct was verified by sequencing both strands over the region of the deletion with the two primers Pmb36 and Pmb43. Approximately 200 ng of pMBP63 was used to transform CHCC373 as described earlier (24). By employing the homologous integration and excision gene replacement feature of the pGhost9 vector (6) the chromosomal thyA gene of CHCC373 was thereafter successfully inactivated and the strain was cured of the plasmid. This resulted in MBP71 ΔthyA, in which the deletion on the chromosome was verified by sequencing.
FIG. 2.
Construction of pMBP63 for inactivation of thyA in CHCC373. Shown is a schematic of how pMBP63 was constructed from pGhost9 and PCR fragments from chromosomal DNA of CHCC373. The 5′ end of primers is indicated by a solid triangle. Bases in gray have been deleted in MBP71 ΔthyA. The base numbering is as in the sequence submitted to GenBank (accession no. AF336368). Abbreviations for restriction enzymes: P, PstI; H, HindIII; S, SalI.
Southern blot hybridization.
Chromosomal DNA was cut with EcoRI and run on a 1% agarose gel overnight, after which the fragments were blotted on GeneScreen Plus charged nylon membrane (NEN976; NEN Life Science Products Inc., Boston, Mass.). The plasmid probes were labeled and hybridized to the blotted fragments according to the instructions of the ECL direct nucleic acid labeling and detection system (Amersham Pharmacia Biotech, Hørsholm, Denmark). This was followed by 1.5 h of exposure to a Cronex 4 medical X-ray film (DuPont, Wilmington, Del.) after which the film was developed.
TS assay.
Cells from stationary cultures were inoculated at an optical density at 600 nm (OD600) of about 0.030 absorbing unit (AU) into fresh LM17 with or without thymidine. At an OD600 of 1.0 AU the cells were harvested, washed in cold buffer of an equal volume (50 mM Tris, 10 mM mercaptoethanol, and 1 mM Na2EDTA-pH 7.4 HCl), and resuspended in 1/100 of the original volume. Of this cell suspension, aliquots of 0.6 ml were added to 1.2 g of 0.25- to 0.50-mm-diameter glass beads in a 1.5-ml tube (25). The cells were disrupted by shaking the tubes in precooled canisters at 30 s−1 for 6 min in an MM200 mixer-mill (Retsch, Haan, Germany). Cell debris was removed by centrifugation at 12,000 × g for 10 min. From each tube about 0.2 ml of crude cell extract was obtained. The TS assay was adapted from previous work (12, 33) and was carried out as follows. A 0.1-ml aliquot of the extract was preheated to 30°C and mixed with 0.1 ml of preheated assay mix to yield the following final concentrations (stock solution concentrations are indicated in parentheses): 50 mM Tris (1 M pH 7.4 HCl), 25 mM MgCl2 (1 M), 100 mM mercaptoethanol (1 M pH 7.4 HCl), 1 mM Na2EDTA (0.1 M pH 7.4 HCl), 0.25 mM tetrahydrofolic acid (50 mM in 1 M Tris with 1 M mercaptoethanol-pH 7.4 HCl) (Sigma-Aldrich Denmark A/S, Vallensbæk Strand, Denmark), 15 mM HCHO (1 M), 0.1 mM dUMP (1 mM; Sigma-Aldrich Denmark), and 1-μCi/ml dUMP-5-3H (1.0 mCi/ml; Amersham Pharmacia Biotech). At suitable time points 5-μl samples were spotted on no. 25 25-mm-diameter glass filter disks (Schleicher & Schuell Inc., Keene, N.H.). To immediately stop the enzymatic reaction, 5 μl of 20% sodium dodecyl sulfate had been applied to filter disks prior to spotting. The filter disks were transferred to plastic vials, and the tritiated H2O released from the dUMP-5-3H during the reaction was evaporated in a vacuum for 15 min. Finally, 5 ml of Ultima Gold scintillation cocktail (Packard Instrument Company, Meriden, Conn.) was added to each vial, after which the activity was counted in a Beckman LS 1801 scintillation counter (Beckman Coulter Inc., Fullerton, Calif.). Since the counts per minute for each vial increased over the first several hours, they were not read until at least 24 h after sampling, when the values had stabilized. Each vial was then read three times, with a standard deviation of less than 5% for all samples and less than 2.5% for 92% of the samples. The protein concentration in the crude extracts was determined by the method of Lowry et al. (34).
Determination of triphosphate nucleotide pools.
MBP71 was grown at 30°C in LSA medium with thymidine to an OD450 of 0.7 AU. The cells were harvested, washed, and inoculated at 0.3 AU into 100 ml of preheated LSA medium with or without thymidine. Immediately after inoculation 220 μl from each culture was added to 2 μl of 33PO43− (10 mCi/ml; about 1 week old at the time of use; Amersham Pharmacia Biotech) for incorporation of labeled phosphate. The main cultures were used to monitor OD450. Growth for the culture with thymidine was exponential, whereas it was linear for the one without thymidine. At 0.6 AU 200-μl samples were withdrawn from the two labeled cultures for determination of dTTP pools and processed by a newly developed procedure kindly provided prior to publication (J. Martinussen, S. L. L. Wadskov-Hansen, and K. Hammer, submitted for publication). Thin-layer chromatography was carried out on polyethyleneimine plates in two dimensions as described earlier (27).
Acidification of RSM by CHCC373 and MBP71.
LM17 stationary cultures with thymidine were washed with an isotonic solution to remove residual thymidine. The cells were then resuspended in fresh solution to be about five times concentrated. A stationary culture has an OD600 of about 3.0 AU. Inoculation levels in milk are shown as the percentage of how much of an overnight culture of the same volume has been used for inoculation. Per definition, 100% inoculation is set to be that which equals an inoculation level of 3.0 AU. The cells were inoculated into 200 ml of cold RSM, to which thymidine and/or phage had already been added when present. The cultures were then transferred to a water bath of the relevant temperature, and pH was monitored automatically over time.
RESULTS
Sequencing of thyA and the upstream and downstream regions in CHCC373.
The DNA sequence of thyA from L. lactis NCDO712 was published earlier (43). Primers from this sequence were employed to obtain an internal part of thyA from CHCC373. By employing easy gene walking (21) approximately 1,150 bp upstream and 500 bp downstream of thyA were obtained. An overview of the complete 2,417-bp sequence obtained is shown in Fig. 2. The nucleotide sequence identity between thyA of CHCC373 and thyA of NCDO712 was 88%. In either case the gene encodes a 279-amino-acid TS with 97% identity between the two strains. The identity between thyA of CHCC373 and L. lactis subsp. lactis IL1403 was 100% (7).
Construction of MBP71 ΔthyA from CHCC373.
The pGhost9 gene inactivation vector (6, 35, 36) was used to inactivate the thyA gene of CHCC373 by homologous integration and excision controlled through the temperature-dependent replication of the vector. Since the whole sequence mentioned above was not available at the time, just 42 bp of thyA was deleted, including the putative −10 box, the ribosome binding site, and the first 8 bases of the coding region (Fig. 2). An 800-bp PCR fragment upstream and a similar one downstream of the intended deletion were cloned into pGhost9 (Fig. 2). The resulting plasmid, pMBP63, was transformed into CHCC373, and the gene inactivation feature of the pGhost9 vector was employed. This work resulted in MBP71 ΔthyA (Fig. 2), which requires thymidine for DNA replication and thereby for propagation (Fig. 1; see also below). Since pMBP63 was excised from the chromosome and cured from MBP71, the strain was sensitive to erythromycin and the 42-bp deletion in ΔthyA was stable.
MBP71 contained no residual fragments from the pGhost9 vector.
Since the pGhost9 vector contains DNA foreign to L. lactis (6, 35, 36), there could be food safety concerns that residual fragments had remained in the chromosome of MBP71.
DNA from CHCC373, MBP71, and MBP65-3 was blotted. The latter strain was an intermediate in the construction of MBP71, in which pMBP63 was integrated into the chromosome of CHCC373. The plasmids pGhost9, pMBP63 (1,600-bp insert), and pMBP57 (400-bp insert) were used as probes (Table 1). pMBP63 and pMBP57 hybridized strongly to the chromosomal DNA from all three strains. pGhost9 hybridized to the DNA from MBP65-3, but not CHCC373 and MBP71, strongly indicating that MBP71 does not contain pGhost9 DNA.
There is no TS activity in MBP71.
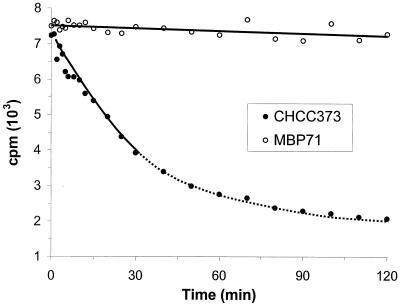
The TS assay, based on the release of tritiated H2O from dUMP-5-3H in the conversion to dTMP, was carried out to confirm that there was no TS activity in MBP71 ΔthyA. The H2O was removed by evaporation, and remaining radioactivity, in counts per minute, was a measure of remaining dUMP-5-3H. In Fig. 3 the result is shown for the TS assay of CHCC373 and MBP71 both grown in LM17 in the presence of thymidine, which must be added for exponential growth of MBP71. For CHCC373 the radioactivity decreased over time, confirming the presence of TS activity. On the other hand the radioactivity was constant for MBP71, showing that there was no TS activity in this mutant.
FIG. 3.
TS assay for CHCC373 and MBP71. Remaining radioactivity is indicated on the secondary axis in counts per minute (cpm). Thymidine was added to the growth medium for both CHCC373 and MBP71. The solid lines indicate exponential fits of the data. The protein concentration in the extract was 6.9 and 6.5 mg/ml for CHCC373 and MBP71, respectively.
The reaction for the crude extract of CHCC373 followed first-order kinetics over 50% of its course with respect to dUMP removal, and an exponential fit is shown in Fig. 3. The first-order rate constant was 2.0 × 10−2 min−1, which is comparable to the 3.1 × 10−2 min−1 found for purified TS from Lactobacillus casei (12).
MBP71 grown without thymidine has no detectable dTTP pool.
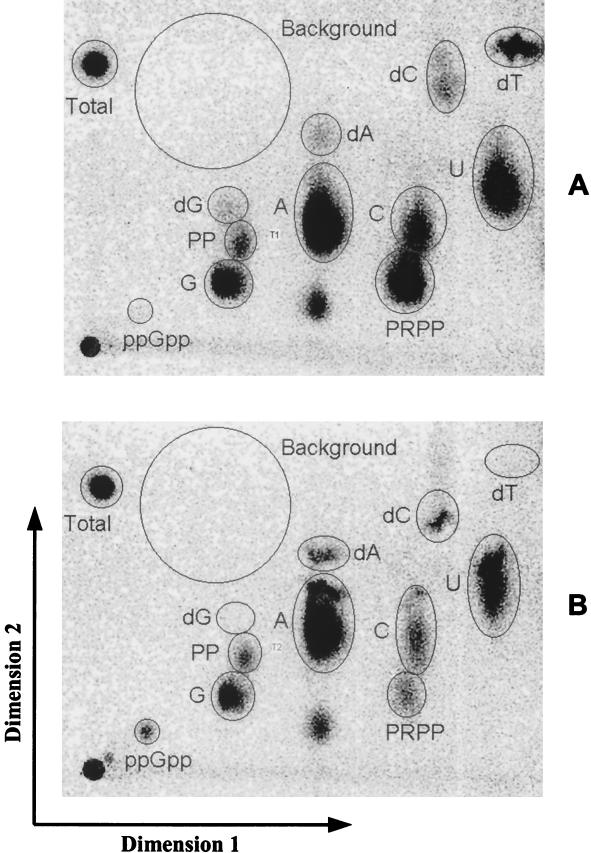
As described in the previous section, MBP71 has no TS activity, and without thymidine in the medium, synthesis of dTTP should not be possible (Fig. 1). To verify this, the intracellular triphosphate nucleotide pools in MBP71, grown in LSA with or without thymidine, were analyzed using a new protocol kindly provided prior to publication (Martinussen et al., submitted for publication). As can be seen in Fig. 4a there was a substantial dTTP pool in the cells grown with thymidine. For the cells grown without thymidine, there was no detectable dTTP pool, showing that it cannot be synthesized (Fig. 4B).
FIG. 4.
Nucleotide pools in MBP71 grown with or without thymidine. For each of the triphosphate nucleotide spot designations the “TP” part of the name has been omitted, so that, e.g., dTTP is referred to as dT. (A) Triphosphate nucleotide pool for MBP71 grown in LSA medium with thymidine. (B) Same as panel A except that thymidine was omitted. The various spots were identified from earlier work (27). Spot labels: Total, 1/10 of the total amount of activity run on each plate; PP, pyrophosphate; PRPP, 5-phosphoribosyl-1-pyrophosphate; ppGpp, guanosine 5′-diphosphate 3′-diphosphate.
The spots were generally weaker for the culture without thymidine, including dGTP and 5-phosphoribosyl-1-pyrophosphate. However, some spots showed the opposite tendency, including the spot for dATP. Also, ppGpp, which indicates a stress response (41), was absent in the culture with thymidine but was clearly identifiable in the culture without thymidine.
The number of CFU did not increase for MBP71 grown without thymidine.
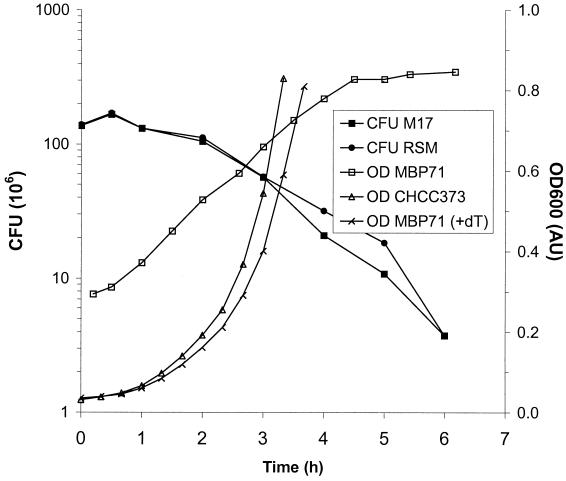
When dTTP cannot be synthesized, DNA replication and cell division are abolished. To verify this, the number of CFU was determined for a stationary culture of MBP71 inoculated at 10% into LM17 and RSM (Fig. 5). For both media the initial cell count of 2 × 108 CFU/ml increased by about 20% from 0 to 0.5 h, which has also been observed in similar experiments. After 0.5 to 1.0 h the number of CFU started decreasing, and after about 2 h the decrease appeared to be exponential (Fig. 5). These results show that MBP71 is nondividing and hence nonreplicating in LM17 and RSM.
FIG. 5.
CFU and OD600 for CHCC373 and MBP71 in M17 and RSM. A stationary culture of MBP71 was inoculated at 10% into LM17 and RSM. The OD600 was monitored for the LM17 culture. The number of CFU was determined at the time points shown by diluting the sample 50 times in three consecutive steps and plating 0.1-ml aliquots in triplicate. This resulted in an initial value of around 150 CFU. The standard deviation was <12 CFU. In a separate experiment CHCC373 and MBP71 were inoculated at 1% into LM17 and LM17 plus 20 mg of thymidine (dT) per liter, respectively.
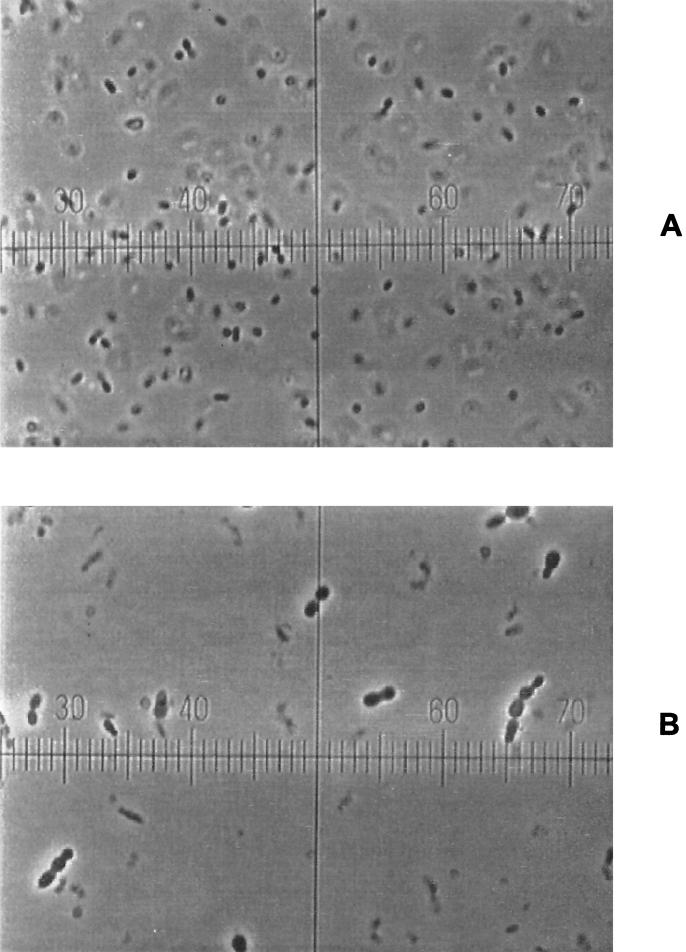
For the LM17 culture of MBP71, the OD600 was also monitored (Fig. 5). The OD increased linearly as opposed to exponentially, and the OD stopped increasing after 4.5 h when it had increased about threefold. In another experiment the OD was monitored over 24 h (data not shown). The OD started decreasing after around 6 h, and after 24 h the OD had decreased to around 30% of the maximum, showing that MBP71 lyses in the nonreplicating state. In Fig. 6 it is shown how nonreplicating cells of MBP71 greatly increase in size compared to exponentially grown MBP71, and this is presumably what causes the lysis.
FIG. 6.
MBP71 grown overnight in LM17 with or without thymidine. (A) MBP71 inoculated at 10% and grown overnight in LM17 with thymidine. (B) Cells grown without thymidine. Magnification for both panels, ×900.
In a separate experiment also shown in Fig. 5, thymidine was added to the LM17. When 20 mg of thymidine per liter was added, MBP71 grew exponentially with a specific growth rate, μ, of 0.96 h−1, compared to 1.05 h−1 for CHCC373 (Fig. 5). After 24 h the OD was about 90% of the maximum, showing that exponentially grown MBP71 does not lyse substantially. Increasing the thymidine concentration to 100 mg/liter did not increase μ for MBP71, but lowering the concentration to 5 mg/liter decreased μ to 0.86 h−1. That MBP71 grows more slowly than CHCC373 indicates that the uptake of exogenous thymidine is insufficient to sustain the maximum growth rate of MBP71.
MBP71 acidifies almost twice as fast at 37°C as it does at 30°C.
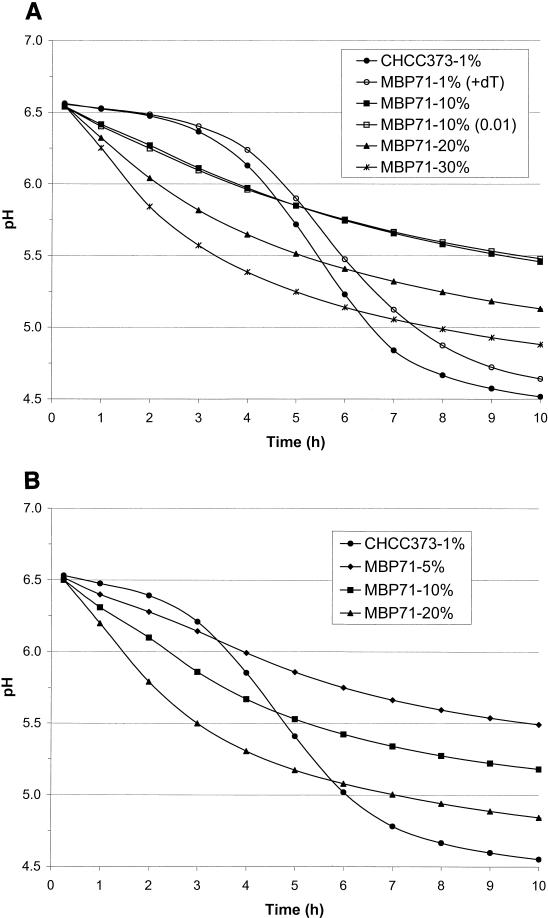
It was of interest to determine how much of nonreplicating MBP71 should be added to RSM to yield the same pH as for CHCC373 at a specified time. Hence, stationary cultures were washed and inoculated into RSM at various levels, and pH was monitored over time. The results are shown in Fig. 7A and B for 30 and 37°C, respectively. One checkpoint could be that pH 5.2 must be reached after 6 h (39). As can be seen in Fig. 7A, this pH was reached in 6 h for wild-type CHCC373 inoculated at 1%, which is also the approximate cell density often used in dairies (data not shown). Since MBP71 is not able to divide in the RSM, the cell number will not increase over time, and the inoculation level must be higher than that for CHC373. For inoculation levels of 10, 20, and 30%, pH 5.2 was reached in 17, 8.5, and 5.5 h, respectively (Fig. 7A).
FIG. 7.
Acidification curves for CHCC373 and MBP71 at 30°C (A) and 37°C (B). RSM (200 ml) with or without 20 mg of thymidine (dT) per liter was inoculated with CHCC373 or MBP71 at the concentrations indicated. The levels of inoculation are as designated in Materials and Methods.
As shown in the previous section MBP71 grew exponentially when 20 mg of thymidine per liter was added to LM17 but grew 8% more slowly than CHCC373. As can be seen from Fig. 7A, MBP71 inoculated at 1% into RSM with added thymidine reached pH 5.2 about 0.5 h later than CHCC373.
One could have chosen to harvest exponentially growing cultures and used them for the above acidification experiments. However, exponentially growing cultures are not employed in dairies. To investigate if the age of the stationary culture influenced acidification, the M17 culture inoculated with MBP71 had also been diluted to 0.01 (vol/vol) in another flask at the time of inoculation, and it would thus become stationary about 5 h later than the mother culture. As can be seen from Fig. 7A, this culture acidifies like the mother culture.
In a dairy fermentation, such as the production of Cheddar cheese, the temperature is only around 30°C at the onset of the fermentation. Normally, after about an hour the temperature is raised gradually to 37 to 38°C (39). Hence, it was important that nonreplicating MBP71 be able to acidify at, e.g., 37°C, and acidification curves at this temperature are shown in Fig. 7B. For CHCC373 the acidification curve at 37°C looks similar to the one at 30°C, and pH 5.2 was reached in 5.5 h (Fig. 7B). Acidification by nonreplicating MBP71 was almost twice as fast at 37°C compared to that at 30°C. For the inoculation levels of 10 and 20%, pH 5.2 was reached in 9.5 and 5 h, respectively. For pH 5.2 to be reached in 6 h the inoculation level should be around 17%.
Acidification by nonreplicating MBP71 was completely unaffected by all the phage it was challenged with.
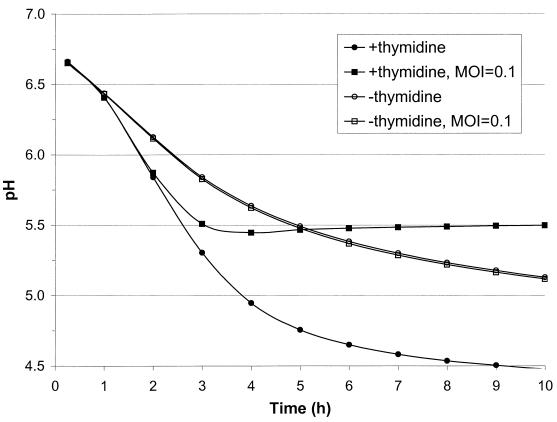
Through the abolishment of DNA replication, acidification by MBP71 should be unaffected by the presence of phage. To test this, MBP71 was inoculated at 10%, corresponding to 2 × 108 CFU/ml, into RSM with or without thymidine and with or without phage CHPC733 from species P335 (Table 1), which lyses CHCC373. Phage CHPC733 was added at 3 × 107 PFU/ml, resulting in an MOI of about 0.1. When no phage was present but thymidine was, pH 5.2 was reached in 3.5 h (Fig. 8). However, under these conditions the cells were susceptible to phage attacks, and in the culture with phage, the pH only went slightly below 5.5 before acidification stopped (Fig. 8). For the nonreplicating cultures of MBP71 it took 8.5 h to reach pH 5.2, both with and without phage added (Fig. 8), showing that acidification is completely unaffected.
FIG. 8.
Effect of phage on acidification by MBP71 with or without thymidine. Cells of MBP71 were inoculated at 10% into 200 ml of RSM with or without phage CHPC733 at an MOI of 0.1. These cultures were set up at 37°C both with and without 20 mg of thymidine per liter.
In another similar experiment, MBP71 was challenged with eight phage infecting CHCC373. These phage belong to the P335 or 936 species and were CHPC12, -412, -708, -710, -783, -795, -814, and -836 (Table 1). In these experiments acidification by nonreplicating MBP71 was also found to be completely unaffected by the phage, whereas the addition of thymidine made the cells susceptible to phage attacks (data not shown). All nine phage listed in Table 1 were routinely propagated to a titer of 1010 to 1011 PFU/ml, and all had a high EOP.
Although it was shown in the previous section that the age of the applied overnight cultures did not affect acidification, it should be noted that there is some variation in acidification time from experiment to experiment. For RSM inoculated with MBP71 at 10% and 37°C, pH 5.2 was reached in 9.5 h for the experiment shown in Fig. 7B, but for the experiment shown in Fig. 8 it took 8.5 h.
The phage injects its DNA into nonreplicating MBP71.
One could argue that the nonreplicating cells of MBP71 were resistant due to an altered cell wall structure caused by the increase in the cell size shown in Fig. 6B. However, it was verified, using phage CHPC733 with fluorescently labeled DNA (19), that the phage adsorbed to the cell wall of MBP71, which had been in the nonreplicating state for 1 h, and phage DNA was injected (data not shown).
Using the above visualization one may view MBP71 as a phage vacuum cleaner wherein phage DNA is trapped upon infection. Thus, MBP71 may be used to remove phage from milk, after which CHCC373 can safely be added for acidification. To test this, MBP71 was inoculated at 1%, i.e., 2 × 107 CFU/ml, into separate RSM cultures with the eight phage mentioned in the previous section at a titer of 6 × 106 PFU/ml, resulting in an MOI of 0.3. The cultures were incubated for 2 h at 30°C for MBP71 to inactivate the phage. During this time the pH dropped 0.1 unit. After this, CHCC373 was added at 1%, and the pH dropped to 5.7 to 6.0 over the following 4 to 5 h before acidification stopped. In control cultures to which MBP71 had not been added initially, pH stopped at around 6.5, showing that MBP71 indeed inactivates a substantial fraction of the phage, but that the phage population quickly recovers.
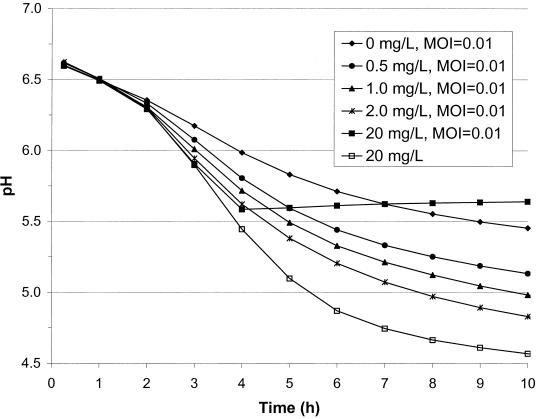
Increasing acidification of MBP71 by addition of thymidine.
MBP71 grows exponentially when 20 mg of thymidine per liter is added but is nonreplicating when thymidine is not added. However, by adding a limiting amount of thymidine to the RSM one may temporarily restore DNA replication. Until the thymidine is used up the cells may carry out DNA replication and acidification should be relatively faster, but naturally the phage will also be able to propagate during this time.
To test this approach, MBP71 was inoculated at 5%, corresponding to 108 CFU/ml, into RSM with various concentrations of thymidine and at various MOIs, and the pH was monitored over time at 37°C. All combinations of 0, 0.5, 1, 2, and 20 mg of thymidine per liter and phage at 0, 1.5 × 105, 1.5 × 106, and 1.5 × 107 PFU/ml, corresponding to MOIs of about 0, 0.001, 0.01, and 0.1, were set up. Selected acidification curves are presented in Fig. 9. As can be seen the addition of thymidine greatly increased acidification. For the culture without thymidine it took 16 h to reach pH 5.3. When 2 mg of thymidine per liter was added, it took only 6 h to reach pH 5.2, even with phage at an of MOI 0.01, the same as the time it took when no phage had been added (Fig. 9). When the MOI was increased to 0.1, acidification in the cultures with limiting thymidine was affected to a greater or lesser extent (data not shown). If nonlimiting thymidine, i.e., 20 mg/liter, had been added to the milk, acidification stopped at around pH 5.6, but without phage pH 5.2 was reached in 4.5 h.
FIG. 9.
Acidification by MBP71 with phage and limiting thymidine. Various concentrations of thymidine were added to 200 ml of RSM. At the same time phage was added at various MOIs. MBP71 was inoculated at 5%, and pH was monitored over time at 37°C.
Nonreplicating MBP71 was homolactic like CHCC373.
In a dairy fermentation it is important that nonreplicating MBP71, which has a physiology different from that of CHCC373, produces the same end products. The most important variable is that the strain remain homolactic, i.e., the cells produce primarily lactic acid, as opposed to, e.g., formate, acetate, and ethanol, indicating mixed-acid fermentation (18, 45). To investigate the end product pattern, cultures similar to the ones in Fig. 7 were set up, i.e., 1% CHCC373 at both 30 and 37°C and 20 and 10% MBP71 at 30°C and 37°C, respectively. Various inoculation levels of MBP71 were used to yield similar acidification curves. At 10 h end product concentrations were determined by high-performance liquid chromatography and gas chromatography (13, 42) (Table 3).
TABLE 3.
Product patterns of CHCC373 and MBP71a
| Strain | Temp (°C) | Inoculum (%) | Concn of added thymidine (mg/liter) | pH | Acid (%) | Mean lactic acid concn ± SD (g/liter) | Mean ethanol concn ± SD (mg/liter) | Mean acetaldehyde concn ± SD (mg/liter) |
|---|---|---|---|---|---|---|---|---|
| CHCC373 | 30 | 1 | 4.5 | 204 | 6.1 ± 0.1 | 32 ± 2 | 2.6 ± 0.3 | |
| 37 | 1 | 4.5 | 204 | 6.0 ± 0.0 | 32 ± 3 | 2.3 ± 0.4 | ||
| MBP71 | 30 | 20 | 5.2 | 112 | 3.2 ± 0.0 | 28 ± 1 | 4.0 ± 0.2 | |
| 37 | 10 | 5.3 | 100 | 2.8 ± 0.3 | 32 ± 1 | 3.4 ± 0.2 | ||
| 30 | 1 | 20 | 4.8 | 162 | 5.1 ± 0.1 | 45 ± 2 | 4.3 ± 0.5 | |
| 37 | 1 | 20 | 4.7 | 177 | 5.0 ± 0.1 | 65 ± 4 | 5.0 ± 0.5 | |
| 37 | 5 | 0.5 | 5.3 | 100 | 2.5 | 76 ± 3 | 4.7 ± 0.4 | |
| 37 | 5 | 1.0 | 5.0 | 138 | 3.6 ± 0.1 | 102 ± 1 | 5.4 ± 0.1 |
MBP71 and CHCC373 were inoculated at various levels into 200 ml of RSM with various amounts of thymidine as indicated. The cultures were set up at the indicated temperature, as were two flasks with no cells added. At 10 h two samples were taken from each culture, and the end product concentrations were determined. The standard deviation is shown for each value, except for one value, for which a sample was lost. The pH at 10 h is also shown, and so is the amount of acid produced, based on a titration curve of RSM with HCl, relative to the highest observed pH, i.e., 5.3.
First, it is important to note that the pH at the sampling time was not identical for all cultures. The amount of acid produced varied twofold (Table 3). Most importantly, the two MBP71 cultures produced as much lactic acid, relative to total acid, as did CHCC373. No acetic acid production was detected in any of the cultures, and the MBP71 cultures produced only ethanol in the low milligrams-per-liter range, as did the CHCC373 cultures (Table 3). Hence, nonreplicating MBP71 was homolactic. However, the acetaldehyde production was relatively higher for the MBP71 cultures than for the CHCC373 cultures. These elevated acetaldehyde concentrations were also found when MBP71 was inoculated at 1% into RSM with 20 mg of thymidine per liter, i.e., when grown similarly to CHCC373 (Table 3). And they were also found when MBP71 was inoculated at 5% into RSM with limiting thymidine. No significant diacetyl or acetoin production was detected in the eight cultures.
DISCUSSION
In this work we constructed the mutant L. lactis MBP71 ΔthyA, which has no TS activity. Thus, thymidine is required for synthesis of dTTP and thereby DNA replication. Milk did not support propagation of MBP71, indicating that milk is devoid of thymidine, and in this medium the acidification by MBP71 was completely unaffected by the presence of phage at an MOI of 0.1. When thymidine was added, acidification stopped. Due to the abolishment of DNA replication, cells of MBP71 are nondividing and they must be inoculated at a higher level than CHCC373 to achieve a certain pH within a specific time. At 37°C MBP71 must be inoculated at levels around 17-fold higher than those of CHCC373. However, by adding a limiting amount of thymidine to the milk, and thereby temporarily restoring DNA replication, the inoculation level could be lowered to 5-fold that of CHCC373, while acidification was still unaffected up to an MOI of 0.01. Nonreplicating MBP71 produced the same products as CHCC373, except that the acetaldehyde production was higher.
Nonreplicating cells of MBP71 are susceptible to TLD.
In Fig. 5 it was shown how the number of CFU for nonreplicating MBP71 starts decreasing exponentially after the initial 1 to 2 h. This is in agreement with earlier results from thyA mutants of various species and is referred to as thymineless death (TLD), which occurs due to a breakdown of the chromosomal DNA (1). However, the OD keeps increasing linearly for more than 2 h after TLD commences (Fig. 5). Hence, it is possible that TLD has little influence on the growth and acidification of nonreplicating MBP71, and it is furthermore possible to select for mutants which are insensitive to TLD (14). Instead of being caused by TLD, the stop in the OD increase after 4.5 h may be caused by cell lysis or a regulatory mechanism.
Applying nonreplicating cultures in practice.
By examining, e.g., Fig. 7B, it is clear that MBP71 has an altered acidification profile compared to that of CHCC373 and that the production parameters must be changed for MBP71. However, a direct consequence of the acidification profile is that it makes it easy to control the end pH of a fermentation by adjusting the inoculation level. This could be an advantage when it is desirable to control the acidity of the product.
Although the cells do not increase in number, the high level of inoculum for MBP71 could affect the organoleptic properties of the product. However, when, e.g., L. lactis lyses intracellular enzymes, e.g., peptidases, are released into the cheese matrix, and this has a positive effect on maturation (4). The OD of nonreplicating MBP71 decreased to 30% of the maximum OD after 24 h, whereas the value was 90% for exponentially grown MBP71. Hence, maturation may be improved compared to that observed for traditional cultures, if the total amount of peptidases and proteinases is similar for the two types of cultures.
It was found that nonreplicating MBP71 produced more acetaldehyde than did CHCC373. Earlier it was found that the dUMP pool of thyA mutants of E. coli was increased, and the dUMP was broken down to uracil and deoxyribose-1-phosphate (dR1P) (1). The dR1P is further catabolized to glyceraldehyde-3-phosphate and acetaldehyde (Fig. 1), and this may indeed be where the observed acetaldehyde was derived from. Alternatively, the acetaldehyde could be a product of the mixed-acid branch, where it may be produced in elevated amounts under certain conditions (9). However, then the acetaldehyde would not be expected to occur at elevated concentrations when thymidine was supplied at 20 mg/liter, unless the thymidine was catabolized (Fig. 1). Although acetaldehyde may affect the taste of the product, it should not affect growth at concentrations up to at least 18 mg/liter (9). In either of the above-mentioned cases the genes encoding the enzymes responsible for its production may be inactivated.
It should be noted that MBP71 has been constructed using genetic engineering and is considered a genetically modified organism (GMO) in the European Union, thus requiring approval for its use in food fermentations. Instead one could construct a thyA mutant of CHCC373 using classical mutagenesis. Since MBP71 does not contain any foreign DNA, it is not considered a genetically modified organism in, e.g., the United States, and it may be applied in this instance.
Potential ways by which phage could overcome the resistance mechanism.
The phage-resistant thyA mutant described in this work has two distinct advantages over other phage resistance mechanisms. First, with our current knowledge it should be functional against all phage species infecting CHCC373, or other strains of L. lactis, either as presented or slightly enhanced, i.e., as a conditional tmk mutant (see below). This is unlike, e.g., the Abi systems or pip mutants. Secondly, it does not allow some attacking phage to slip through the defense mechanism, which is a problem with the R/M systems. However, it is still possible to envisage ways whereby phage may overcome this mechanism of protection.
First, it has been reported that the Bacillus subtilis phage φ3T has a thy gene, thyP3, in its chromosome (30). If such a phage infected a thyA mutant, synthesis of dTTP and thus DNA replication would be restored, possibly leading to lysis of the culture. However, none of the nine phage MBP71 was challenged with affected acidification. Furthermore, of the phage infecting L. lactis which have currently been sequenced, none were found to contain a thy gene (data not shown). Also, a possible solution to this potential problem is to inactivate more of the genes necessary for the de novo synthesis of dTTP, i.e., dut, dcdA, and tmk, though in the third instance a conditional mutant would be necessary (Fig. 1). Even with these genes inactivated certain phage could have one or more of them in their chromosome. Specifically, several phage infecting L. lactis have a gene homologous to the dut gene (11). Also, the genome of the T4 phage infecting E. coli encodes most of the enzymes for dNTP synthesis (22). As a solution to this one could, instead of limiting the availability of dNTPs, target the DNA replication machinery itself so that it would not be active, or the required enzymes would not be synthesized, when the cells were inoculated into the milk.
Secondly, even if no established phage infecting L. lactis has a thy gene the phage may acquire genes from the host it infects (23). With the strain MBP71 this poses a particular risk, since only 42 bp of thyA has been deleted. If the remaining part of the thyA gene was integrated in a phage chromosome in front of a promoter, ribosome binding site, and start codon, the result could be an active thyA gene. With thyA mutants being used on a large scale there would be a selection pressure for such phage, though incorporating other phage resistance mechanisms could lower the potential risk. As acquirement of host DNA appears to occur through homologous recombination between the infecting phage and prophage in the host chromosome (8, 17), specific regions may not be readily acquirable for the infecting phage.
Finally, phage may overcome the lack of dTTP for DNA replication inside the cell by hydrolyzing the host chromosome and reutilizing the nucleotides, and indeed phage c6A, which infects L. lactis, does utilize the host nucleotides (40). However, none of the nine phages tested in this research were able to slow acidification (Fig. 8), though it is possible that none of them break down the host chromosome. Another explanation could be that the infecting phage cannot initiate its cycle upon infection due to the cell's physiological state. Again this potential problem can be overcome by making a conditional tmk mutant or targeting the DNA replication machinery itself.
Increasing acidification of nonreplicating MBP71.
It is clear that the culture cost greatly increases if a dairy has to use, e.g., 17-fold as much inoculum. However, a dairy may not necessarily use MBP71 all the time. It may be used in dairies with extensive phage problems or when acidification times get longer, indicating the presence of phage. And its use should be seen in contrast to failure of acidification, resulting in products of reduced value or complete loss of the batch.
Also, we have described how the inoculation level could be lowered to 5%, i.e., fivefold the normal amount, by adding a limiting amount of thymidine to the milk, while acidification remained unaffected by the presence of phage up to 106 PFU/ml (Fig. 9). In the whey sample from a milk fermentation in which phage has lysed the culture the phage titer normally does not exceed 5 × 109 PFU/ml (data not shown). Considering that the volumetric carryover from one fermentation to another is in the area of 10−8 (29), less than 5 × 101 PFU/ml of the following batch would originate from an infected batch. The phage titer in raw milk is normally less than 103 PFU/ml (47), which is not necessarily specific for the strain employed. Thus, the initial phage titer in a fresh batch should be less than 10−3 of the titer tested here, and the inoculation level of MBP71 might be lowered even further without risking failure of acidification. However, thymidine is currently not approved as an additive in cheese production. An alternative to adding thymidine would be to construct a temperature-sensitive mutant in which the TS is not active above, e.g., 34°C. For the first 1 to 2 h of, e.g., Cheddar cheese fermentation, the temperature is below this mark and the TS would be active. However, after this point the temperature is above 34°C for several hours and DNA replication would be abolished.
There are alternatives to increasing acidification by temporarily allowing DNA replication. First, it was observed in Fig. 5 that the number of CFU for nonreplicating MBP71 increased about 20% over the first 0.5 h. This could be due to stationary cells, with two complete chromosomes, which then divide in the fresh medium and thus increase the total number of CFU. This in turn could be exploited to increase acidification. When E. coli is grown under certain conditions there are several chromosomes per cell in the stationary phase (10). If such cells of L. lactis were inoculated into milk, they should be able to divide one or more times, even if DNA replication were not possible, thus increasing the total number of CFU and thereby presumably acidification.
Secondly, in E. coli growing under aerobic conditions the specific rate of glycolysis, or flux, was increased by 71% by incorporating ATPase activity (B. J. Koebmann, H. V. Westerhoff, J. L. Snoep, D. Nilsson, and P. R. Jensen, submitted for publication). The ATPase nonspecifically hydrolyzes ATP and drives up the glycolytic flux to keep up with the demand. In L. lactis an increased glycolytic flux would lead to a higher lactic acid flux, and we are currently working on the incorporation of ATPase activity in MBP71.
Acknowledgments
This work was supported by a grant from The Danish Academy of Technical Sciences (ATV).
We thank Ida Jørring for her excellent and expeditious work in the laboratory. We furthermore thank the following people for their help with various tasks: Marie Koefoed, Pia Rasmussen, Lone Riisberg, Per Strøman, Abdallah Albayasli, Marianne Richelieu, Jan Neuhard, Jan Martinussen, and Casper Jørgensen.
REFERENCES
- 1.Ahmad, S. I., S. H. Kirk, and A. Eisenstark. 1998. Thymine metabolism and thymineless death in prokaryotes and eukaryotes. Annu. Rev. Microbiol. 52:591-625. [DOI] [PubMed] [Google Scholar]
- 2.Allison, G. E., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226. [Google Scholar]
- 3.Babu, K. S., W. S. Spence, M. R. Montville, and B. L. Geller. 1995. Characterization of a cloned gene (pip) from Lactococcus lactis required for phage infection, p. 569-575. In J. J. Ferretti, M. S. Gilmore, T. R. Klaenhammer, and F. Brown (ed.), Genetics of streptococci, enterococci and lactococci. Karger, Basel, Switzerland. [PubMed]
- 4.Bartels, H. J., M. E. Johnson, and N. F. Olson. 1987. Accelerated ripening of Gouda cheese. 1. Effect of heat-shocked thermophilic lactobacilli and streptococci on proteolysis and flavor development. Milchwissenschaft 42:83-88. [Google Scholar]
- 5.Bertani, G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 62:293-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Biswas, I., A. Gruss, S. D. Ehrlich, and E. Maguin. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J. Bacteriol. 175:3628-3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolotin, A., P. Wincker, S. Mauger, O. Jaillon, K. Malarme, J. Weissenbach, S. D. Ehrlich, and A. Sorokin. 2001. The complete genome sequence of the lactic acid bacterium Lactococcus lactis. Genome Res. 11:731-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bouchard, J. D., and S. Moineau. 2000. Homologous recombination between a lactococcal bacteriophage and the chromosome of its host strain. Virology 270:65-75. [DOI] [PubMed] [Google Scholar]
- 9.Boumerdassi, H., C. Monnet, M. Desmazeaud, and G. Corrieu. 1997. Isolation and properties of Lactococcus lactis subsp. lactis biovar diacetylactis CNRZ 483 mutants producing diacetyl and acetoin from glucose. Appl. Environ. Microbiol. 63:2293-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boye, E., and A. Lobner-Olesen. 1991. Bacterial growth control studied by flow cytometry. Res. Microbiol. 142:131-135. [DOI] [PubMed] [Google Scholar]
- 11.Brøndsted, L., S. Østergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 12.Crusberg, T. C., R. Leary, and R. L. Kisliuk. 1970. Properties of thymidylate synthase from dichloromethotrexate-resistant Lactobacillus casei. J. Biol. Chem. 245:5292-5296. [PubMed] [Google Scholar]
- 13.Curic, M., B. Stuer-Lauridsen, P. Renault, and D. Nilsson. 1999. A general method for selection of α-acetolactate decarboxylase-deficient Lactococcus lactis mutants to improve diacetyl production. Appl. Environ. Microbiol. 65:1202-1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Crecy-Lagard, V. A., J. Bellalou, R. Mutzel, and P. Marliere. 2001. Long term adaptation of a microbial population to a permanent metabolic constraint: overcoming thymineless death by experimental evolution of Escherichia coli. BMC Biotechnol. 1:10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dickely, F., D. Nilsson, E. B. Hansen, and E. Johansen. 1995. Isolation of Lactococcus lactis nonsense suppressors and construction of a food-grade cloning vector. Mol. Microbiol. 15:839-847. [DOI] [PubMed] [Google Scholar]
- 16.Djordjevic, G. M., D. J. O'Sullivan, S. A. Walker, M. A. Conkling, and T. R. Klaenhammer. 1997. A triggered-suicide system designed as a defense against bacteriophages. J. Bacteriol. 179:6741-6748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durmaz, E., and T. R. Klaenhammer. 2000. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl. Environ. Microbiol. 66:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fordyce, A. M., V. L. Crow, and T. D. Thomas. 1984. Regulation of product formation during glucose or lactose limitation in nongrowing cells of Streptococcus lactis. Appl. Environ. Microbiol. 48:332-337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furukawa, H., T. Kuroiwa, and S. Mizushima. 1983. DNA injection during bacteriophage T4 infection of Escherichia coli. J. Bacteriol. 154:938-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gasson, M. J. 1983. Plasmid complements of Streptococcus lactis NCDO712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrison, R. W., J. C. Miller, M. J. D'Souza, and G. Kampo. 1997. Easy gene walking. BioTechniques 22:650-653. [DOI] [PubMed] [Google Scholar]
- 22.Hazebrouck, S., F. Maley, V. Machtelinckx, P. Sonigo, and J. J. Kupiec. 1999. Structural and functional analysis of surface domains unique to bacteriophage T4 thymidylate synthase. Biochemistry 16:2094-2101. [DOI] [PubMed] [Google Scholar]
- 23.Hill, H., L. A. Miller, and T. R. Klaenhammer. 1991. In vivo genetic exchange of a functional domain from a type II A methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 173:4363-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holo, H., and I. F. Nes. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl. Environ. Microbiol. 55:3119-3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hummel, W., and M. Kula. 1989. Simple method for small-scale disruption of bacteria and yeast. J. Microbiol. Methods 9:201-209. [Google Scholar]
- 26.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 27.Jensen, K. F., U. Houlberg, and P. Nygaard. 1979. Thin-layer chromatographic methods to isolate 32P-labeled 5-phosphoribosyl-α-1-pyrophosphate (PRPP): determination of cellular PRPP pools and assay of PRPP synthetase activity. Anal. Biochem. 98:254-263. [DOI] [PubMed] [Google Scholar]
- 28.Jensen, P. R., and K. Hammer. 1993. Minimal requirements for exponential growth of Lactococcus lactis. Appl. Environ. Microbiol. 59:4363-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Josephsen, J., N. Andersen, H. Behrndt, E. Brandsborg, G. Christiansen, M. B. Hansen, S. Hansen, E. W. Nielsen, and F. K. Vogensen. 1994. An ecological study of lytic bacteriophages of Lactococcus lactis subsp. cremoris isolated in a cheese plant over a five year period. Int. Dairy J. 4:123-140. [Google Scholar]
- 30.Kenny, E., T. Atkinson, and B. S. Hartley. 1985. Nucleotide sequence of the thymidylate synthase gene (thyP3) from the Bacillus subtilis phage φ3T. Gene 34:335-342. [DOI] [PubMed] [Google Scholar]
- 31.Kim, S. G., and C. A. Batt. 1991. Antisense mRNA-mediated bacteriophage resistance in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 57:1109-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labrie, S., and S. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lomax, M. I., and G. R. Greenberg. 1967. A new assay of thymidylate synthetase activity based on the release of tritium from deoxyuridylate-5-3-H. J. Biol. Chem. 10:109-113. [PubMed] [Google Scholar]
- 34.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 35.Maguin, E., T. Duwat, S. G. Hege, S. D. Ehrlich, and A. Gruss. 1992. New thermosensitive plasmid for gram-positive bacteria. J. Bacteriol. 174:5633-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maguin, E., H. Prevost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nilsson, D., and T. Janzen. July 1998. Method of preventing bacteriophage infection of bacterial cultures. International patent application no. PCT/DK99/00382.
- 38.O'Sullivan, D. J., C. Hill, and T. R. Klaenhammer. 1993. Effect of increasing the copy number of bacteriophage origins of replication, in trans, on incoming-phage proliferation. Appl. Environ. Microbiol. 59:2449-2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pearce, L. E. 1980. Description of the activity test for cheese starters. Bull. Int. Dairy Found. 129:11. [Google Scholar]
- 40.Powell, I. B., D. L. Tulloch, A. J. Hillier, and B. E. Davidson. 1992. Phage DNA synthesis and host DNA degradation in the life cycle of Lactococcus lactis bacteriophage c6A. J. Gen. Microbiol. 138:945-950. [DOI] [PubMed] [Google Scholar]
- 41.Rallu, F., A. Gruss, S. D. Ehrlich, and E. Maguin. 2000. Acid- and multistress-resistant mutants of Lactococcus lactis: identification of intracellular stress signals. Mol. Microbiol. 35:517-528. [DOI] [PubMed] [Google Scholar]
- 42.Richelieu, M., U. Houlberg, and J. C. Nielsen. 1997. Determination of α-acetolactic acid and volatile compounds by headspace gas chromatography. J. Dairy Sci. 80:1918-1925. [Google Scholar]
- 43.Ross, P., F. O'Gara, and S. Condon. 1990. Cloning and characterization of the thymidylate synthase gene from Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 56:2156-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 45.Sjöberg, A., I. Persson, M. Quednau, and B. Hahn-Hägerdal. 1995. The influence of limiting and nonlimiting growth conditions on glucose and maltose metabolism in Lactococcus lactis ssp. lactis strains. Appl. Microbiol. Biotechnol. 42:931-938. [Google Scholar]
- 46.Terzaghi, B. E., and W. E. Sandine. 1975. Improved medium for lactic streptococci and their bacteriophages. Appl. Environ. Microbiol. 29:807-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tsaneva, K. 1973. Occurrence of Str. lactis bacteriophages in dairy factories. Khranitelna Promishlenost 22:27-29. [Google Scholar]
- 48.Walker, S. A., and T. R. Klaenhammer. 1998. Molecular characterization of a phage-inducible middle promoter and its transcriptional activator from the lactococcal bacteriophage φ31. J. Bacteriol. 180:921-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Walker, S. A., and T. R. Klaenhammer. 2000. An explosive antisense RNA strategy for inhibition of a lactococcal bacteriophage. Appl. Environ. Microbiol. 66:310-319. [DOI] [PMC free article] [PubMed] [Google Scholar]