Abstract
Historically, Vibrio parahaemolyticus infections have been characterized by sporadic cases caused by multiple, diverse serotypes. However, since 1996, V. parahaemolyticus serotype O3:K6 strains have been associated with several large-scale outbreaks of illness, suggesting the emergence of a “new” group of organisms with enhanced virulence. We have applied three different molecular subtyping techniques to identify an appropriate method for differentiating O3:K6 isolates from other serotypes. Pulsed-field gel electrophoresis (PFGE) following NotI digestion differentiated seven closely related subtypes among O3:K6 and related strains, which were distinct from PFGE patterns for non-O3:K6 isolates. Ribotyping and tdh sequencing were less discriminatory than PFGE, but further confirmed close genetic relationships among recent O3:K6 isolates. In vitro adherence and cytotoxicity studies with human epithelial cells showed that O3:K6 isolates exhibited statistically higher levels of adherence and cytotoxicity to host cells than non-O3:K6 isolates. Epithelial cell cytotoxicity patterns were determined with a lactate dehydrogenase release assay. At 3 h postinfection, high relative cytotoxicities (>50% maximum lactate dehydrogenase activity) were found among a greater proportion of recently isolated O3:K6 and closely related strains (75%) than among the non-O3:K6 isolates (23%). A statistically significant relationship between adherence and cytotoxicity suggests that the pathogenic potential of some isolates may be associated with increased adherence to epithelial cells. Our findings suggest that enhanced adherence and cytotoxicity may contribute to the apparent unique pathogenic potential of V. parahaemolyticus O3:K6 strains.
Vibrio parahaemolyticus is reported as an agent of food-borne illness in the United States (13) and around the globe. Human infection with this pathogen is associated most frequently with the consumption of seafood, primarily raw or improperly cooked shellfish (4). Consumption of sufficiently high numbers of organisms of virulent V. parahaemolyticus strains can cause gastroenteritis, septicemia, and even death. Since 1996, an increased incidence of gastroenteritis in many parts of the world (7, 19) has been associated with V. parahaemolyticus serotype O3:K6. The recent predominance of the O3:K6 serotype as a causative agent of multiple gastroenteritis outbreaks is noteworthy, because, in general, V. parahaemolyticus infections have been characterized by sporadic cases caused by multiple, diverse serotypes. The association of this “new” O3:K6 serotype with large-scale food-borne disease outbreaks suggests this organism may have an unusual capacity to be transmitted by foods and/or to cause human infection.
The current standard method for definitive identification of V. parahaemolyticus O3:K6 requires serotyping (9). This method is expensive, tedious, and time-consuming. Because O3:K6 isolates recovered since 1996 have been shown to be genetically distinct from previously isolated O3:K6 strains (3, 17, 19), we reasoned that molecular methods might provide more rapid and sensitive alternatives for differentiating among V. parahaemolyticus strains. Thus, we evaluated the abilities of pulsed-field gel electrophoresis (PFGE), ribotyping, and tdh sequence analysis to discriminate among V. parahaemolyticus isolates, with a specific focus on differentiating the “new” O3:K6 serotype. Our findings confirm close genetic relationships among recently isolated V. parahaemolyticus O3:K6 strains.
To identify possible mechanisms associated with the apparent enhanced virulence of V. parahaemolyticus O3:K6 strains, we measured swarming, adherence, and cytotoxicity phenotypes of various isolates. We hypothesized that V. parahaemolyticus O3:K6 motility may be associated with virulence, because the motility phenotype has been correlated with virulence factor expression in other Vibrio species (10). The acquisition of an open reading frame (ORF), referred to as ORF8 (17), in recent O3:K6 isolates also may confer additional virulence capabilities to this group of V. parahaemolyticus strains. Our results suggest that enhanced adherence and cytotoxicity may contribute to the apparent unique pathogenic potential of the recent O3:K6 isolates.
MATERIALS AND METHODS
Bacterial isolates and media.
All isolates used in this study were provided by the Food and Drug Administration (FDA). Table 1 lists the isolates, including serotype, source, and origin of isolation. Serotypes were determined previously by the Centers for Disease Control and Prevention (CDC). Upon receipt, all isolates were grown in tryptic soy agar (TSA; Difco Laboratories, Detroit, Mich.) supplemented with 2% NaCl (TSAS) and streaked for isolation of single colonies. Isolates were stored in tryptic soy broth supplemented with 2% NaCl (TSBS) and 20% glycerol at −80°C.
TABLE 1.
V. parahaemolyticus isolates used in this study
| Laboratory isolate identification | FDA identification (reference) | Serotype | Source | Place or circumstance of isolation | Yr of isolation |
|---|---|---|---|---|---|
| O3:K6 group | |||||
| FSL-Y1-016 | TX-2103 | O3:K6 | Clinical | Texas | 1998 |
| FSL-Y1-017 | NY-3064 | O3:K6 | Clinical | New York | 1998 |
| FSL-Y1-018 | CT-6636 | O3:K6 | Clinical | Connecticut | |
| FSL-Y1-019 | VP47 | O3:K6 | Clinical | India | |
| FSL-Y1-020 | KX-V225 | O3:K6 | Clinical | Thailand | |
| FSL-Y1-023 | VP86 | O3:K6 | Clinical | Calcutta | 1996 |
| FSL-Y1-024 | VP199 | O3:K6 | Clinical | Calcutta | 1997 |
| FSL-Y1-025 | VP208 | O3:K6 | Clinical | Calcutta | 1997 |
| FSL-Y1-026 | VP155 | O3:K6 | Clinical | Calcutta | 1996 |
| FSL-Y1-027 | VP96 | O3:K6 | Clinical | Calcutta | 1996 |
| FSL-Y1-014 | AN-5034 | O4:K68a | Clinical | Bangladesh | 1996 |
| FSL-Y1-015 | AN-16000 | O1:KUTab | Clinical | Bangladesh | 1998 |
| Non-O3:K6 group | |||||
| FSL-Y1-021 | U-5474 | Old O3:K6c | Clinical | Bangladesh | 1980 |
| FSL-Y1-022 | AQ4037 | Old O3:K6c | Clinical | Traveler arriving at Osaka, Japan, from Maldives | 1985 |
| FSL-Y1-005 | 5C-1C | O1 | Food (oyster) | Washington | 1988 |
| FSL-Y1-011 | 8332924 | O1:K56 | Food (oyster) | Gulf of Mexico | 1983 |
| FSL-Y1-012 | 48432 | O4:K12 | Clinical | Washington | 1991 |
| FSL-Y1-003 | T3980 | O4:K13 | Clinical | Japan | |
| FSL-Y1-002 | NY477 | O4:K8 | Food (oyster) | New York | 1977 |
| FSL-Y1-006 | M350A | O5 | Food (oyster) | Washington | 1994 |
| FSL-Y1-010 | T-3979 | O5:K15 | Clinical | Japan | |
| FSL-Y1-013 | 47978 | O6:K18 | Clinical | Washington | 1991 |
| FSL-Y1-001 | 92000713 (1) | O8 | Food (clam) | 1992 | |
| FSL-Y1-004 | CRAB | Unknown | Food | Washington | 1972 |
| FSL-Y1-008 | 92000713 (2) | Unknown | Food (clam) | 1992 |
Swarming.
To determine the relative swarming capabilities of isolates, bacteria were streaked initially for single-colony isolation from frozen stock onto T1N3 (1% tryptone, 3% NaCl) agar. Inoculum from single colonies was stabbed into plates of TSA supplemented with either 0.5% NaCl or 0.5% NaCl plus 0.15% MgSO4. Colony diameters were recorded after 24 h of incubation at 37°C. A strong swarming phenotype was defined as a colony diameter >200% that of FSL-Y1-002 (a nonswarmer, as determined previously by our group). A colony diameter of 175 to 200% that of FSL-Y1-002 was considered a weak swarming phenotype. Measurements were repeated in three independent trials. The presence of a strong swarming phenotype in all trials was considered a positive reaction (+) in Table 2. The absence of a swarming phenotype in all trials was considered a negative reaction (−). The presence of both strong and weak swarming phenotypes for the same isolate among different trials is indicated by +/−. The absence of a swarming phenotype in either one or two trials is represented as variable (v). The likelihood of isolates in the O3:K6 and non-O3:K6 groups to exhibit a specific swarming phenotype was assessed by chi-square analysis. The +, +/−, and v reactions were grouped into a single category prior to analysis.
TABLE 2.
Phenotypic, molecular, and virulence characterization of V. parahaemolyticus isolates
| Laboratory isolate identification | PCR resulta
|
Swarming phenotypeb
|
Ribotype (DuPoint identification) | PFGE type | No. of adhered V. parahaemolyticus cells per:
|
% Cytotoxicitye
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| tdh | ORF8 | NaCl | NaCl + MgSO4 | Coverslip (105)c | HeLa celld | 3 h post- infection | 6 h post-infection | |||
| O3:K6 group | ||||||||||
| FSL-Y1-016 | + | + | − | +/−f | C (DUP-6622) | A1 | 140 | 10.4 | 54 | 98 |
| FSL-Y1-017 | + | + | v | vf | A1 (DUP-6626) | A7 | 360 | 11.5 | 56 | 97 |
| FSL-Y1-018 | + | + | vf | +/−f | A1 (DUP-6626) | A7 | 307 | 10.5 | 65 | 100g |
| FSL-Y1-019 | + | + | vf | v | A2 (DUP-6626) | A2 | 400 | 8.1 | 79 | 100g |
| FSL-Y1-020 | + | + | vf | vf | A1 (DUP-6626) | A3 | 400 | 5.3 | 78 | 99 |
| FSL-Y1-023 | + | + | − | − | B | A4 | 467 | 14.5 | 26 | 100 |
| FSL-Y1-024 | + | + | vf | vf | A1 (DUP-6626) | A2 | 333 | 10.4 | 88 | 100g |
| FSL-Y1-025 | + | + | +/−f | + | A2 (DUP-6626) | NDh | 133 | 1.8 | 27 | 81 |
| FSL-Y1-026 | + | + | vf | +/−f | A1 (DUP-6626) | ND | ND | ND | 15 | 60 |
| FSL-Y1-027 | + | + | − | − | A1 (DUP-6626) | A3 | ND | ND | 69 | 100g |
| FSL-Y1-014 | + | + | vf | vf | A1 (DUP-6626) | A6 | 227 | 5.3 | 63 | 100g |
| FSL-Y1-015 | + | + | − | − | A1 (DUP-6626) | A5 | 191 | 6.9 | 67 | 96 |
| Non-O3:K6 group | ||||||||||
| FSL-Y1-021 | + | − | − | − | A1 (DUP-6626) | B | 288 | 7.8 | 27 | 85 |
| FSL-Y1-022 | − | − | − | − | A2 (DUP-6626) | C | 104 | 2.0 | 11 | 67 |
| FSL-Y1-005 | − | − | vf | + | G | G | 147 | 4.3 | 6 | 84 |
| FSL-Y1-011 | − | − | − | − | K | K | ND | ND | 43 | 90 |
| FSL-Y1-012 | − | − | − | − | L | L | ND | ND | 55 | 100g |
| FSL-Y1-003 | + | − | − | − | E | E | 120 | 0.9 | 4 | 48 |
| FSL-Y1-002 | + | − | − | − | I | UTi | 172 | 9.9 | 14 | 52 |
| FSL-Y1-006 | − | − | − | vf | H | H | 153 | 7.6 | 70 | 100g |
| FSL-Y1-010 | + | − | − | − | J | J | 72 | 1.4 | 2 | 96 |
| FSL-Y1-013 | − | − | +f | + | A1 (DUP-6626) | M | 138 | 2.6 | 6 | 100g |
| FSL-Y1-001 | − | − | v | +f | D (DUP-6621) | D | 341 | 4.9 | 57 | 100 |
| FSL-Y1-004 | − | − | − | v | F | F | 240 | 6.8 | 7 | 95 |
| FSL-Y1-008 | − | − | − | − | A2 (DUP-6626) | I | 128 | 6.2 | 3 | 14 |
+, gene present. Negative results (−) were confirmed by Southern blotting.
Cells were inoculated on TSA supplemented with 0.5% NaCl or 0.5% NaCl and 0.15% MgSO2. Positive, negative, and weak reactions are indicated as +, −, and +/−, respectively. v, variable.
Value determined by plate counts.
Value represents the average number of bacteria adhered per HeLa cell, with 20 HeLa cells per sample observed by direct microscopic observation.
Each value represents the average of two independent trials in which triplicate samples were tested in parallel.
Showed burst formation.
Value was rounded to 100.
ND, not determined.
UT, untypeable.
Preparation of DNA templates for PCR.
Two hundred fifty microliters of cultures that had been grown overnight in TSBS was centrifuged for 10 min at 13,000 × g, and then pellets were resuspended in 95 μl of 1× PCR buffer (Gibco BRL, Life Technologies, Rockville, Md.) and treated with lysozyme (2-mg/ml final concentration). After incubation at 37°C for 15 min, proteinase K (200-μg/ml final concentration) was added to the mixture. The mixture was held for 1 h at 60°C and then placed in a boiling water bath for 8 min to denature proteinase K. The resulting lysate was stored at −20°C until use.
tdh PCR.
Oligonucleotide primers were derived from five tdh sequences deposited in GenBank (accession no. S76724, D90238, M10069, S67841, and D90101). Primers tdh-F2 (5′ GCA TTA CTG TTA CTT ATA GAG T 3′) and tdh-R2 (5′ ATT ATT ATT AAT TAT TTT CAA AG 3′) were designed to amplify an ∼700-bp region comprising the tdh promoter and coding region. Primers tdh-F2 and tdh-R (5′ CAG AAT ATT CTG TGG CTT TTT A 3′) were used to sequence PCR products.
Hot Start PCR was performed in a 25-μl reaction volume in a GeneAmp PCR System 2400 (Perkin-Elmer, Foster City, Calif.). Each reaction mixture had a final concentration of 1× PCR buffer (Gibco), 1.5 mM MgCl2, 50 μM deoxynucleoside triphosphates (dNTPs), 1 μM primer set, 1 U of Taq polymerase (PE Applied Biosystems, Foster City, Calif.), and 1 μl of template DNA. The following reaction conditions produced a single band of the expected size: preincubation at 94°C for 5 min; amplification for 40 cycles of 94°C for 45 s, 45°C for 45 s, and 72°C for 90 s; and a final hold at 72°C for 7 min. PCR products were purified from primers and nucleotides with a QIAquick PCR purification kit (Qiagen, Inc., Valencia, Calif.).
ORF8 PCR.
Primers ORF8-F (5′ GTT CGC ATA CAG TTG AGG 3′) and ORF8-R (5′ AAG TAC AGC AGG AGT GAG 3′) were used to amplify an ∼700-bp fragment of the ORF8 region of phage f237 (17). The PCR conditions were as described for the tdh PCR, except that the amplification program consisted of a preincubation step at 94°C for 3 min; 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 60 s; and a final incubation at 72°C for 5 min.
Sequence determination of tdh.
Purified tdh amplicons were sequenced directly with the Perkin-Elmer cycle sequencing kit and an Applied Biosystems model 373A automated sequencer. Both strands were sequenced with tdh-F2 and tdh-R. The resulting sequences were assembled by the software SeqMan (DNAStar, Madison, Wis.) and aligned by the Clustal method in MegAlign (DNAStar).
Ribotyping.
V. parahaemolyticus isolates were characterized by automated ribotyping with the restriction enzyme EcoRI and the RiboPrinter (Qualicon Inc., Wilmington, Del.) as previously described (5).
PFGE.
Isolates were grown overnight at 35°C on TSA supplemented with 3% NaCl. Bacteria were harvested from the plates and suspended in salt-Tris-EDTA buffer (STE; 1 M NaCl, 10 mM Tris, 10 mM EDTA), with the cell density adjusted to approximately 0.9 at a fixed absorbance wavelength of 550 nm with a Microscan turbidity meter (Dade Behring, Inc., Deerfield, Ill.). Cell suspensions were mixed at 56°C with an equal volume of 1.3% InCert (FMC BioProduct, Rockland, Maine) agarose (prepared in a mixture of 10 mM Tris, 1 M NaCl, and 0.25 M EDTA) and then pipetted in 85-μl aliquots into 10-well plug molds (Bio-Rad Laboratories, Richmond, Calif.). After solidification, plugs for each sample were transferred to tubes containing 3 ml of lysis buffer (6 mM Tris, 1 M NaCl, 100 mM EDTA, 0.5% Brij-58, 0.2% sodium deoxycholate, 0.5% N-lauroyl sarcosine) supplemented with 1 mg of lysozyme per ml (Sigma Co., St. Louis, Mo.) just before use. Tubes were incubated in a 37°C water bath for 2.5 h. Following incubation, the lysis buffer was removed, replaced with 4 ml of protease solution (0.5 M EDTA, 1% N-lauroyl sarcosine) supplemented with 100 μg of proteinase K per ml (Sigma), and incubated at 50°C overnight.
Each plug to be cleaved by restriction enzyme was washed at 37°C three times for 1 h in 1 ml of TE (10 mM Tris, 1 mM EDTA) and a fourth time overnight and finally washed for 1 h in low-EDTA-TE (10 mM Tris, 0.1 mM EDTA). Whole plugs were cleaved for at least 6 h at 37°C with 30 U of the restriction enzyme NotI (New England BioLabs, Inc., Beverly, Mass.) in 250-μl reaction volumes. Plugs were sliced, and approximately one-fourth-plug sections were loaded directly onto a 15-tooth comb, which was then positioned in the gel tray for electrophoresis. A molten 100-ml gel of 1% FastLane agarose (FMC BioProducts) in 0.5× TBE buffer (1× TBE is 0.089 M Tris, 0.089 M boric acid, 0.002 M EDTA) was poured and allowed to solidify for at least 45 min. PFGE was performed with a contour-clamped homogeneous electric field instrument (Bio-Rad CHEF DRII). The running conditions were as follows: 6 V/cm at 14°C for 18 h; initial and final switch times of 2 and 40 s, respectively; and 0.5× TBE running buffer. A control strain, nyc#98-2379, from a 1998 Vibrio parahaemolyticus outbreak associated with shellfish from New York (6), was subject to electrophoresis for comparison in three lanes of the gel. Following electrophoresis, gels were stained with ethidium bromide and photographed under UV transillumination. Dendrograms were plotted by BioNumerics (Applied Maths BVBA, Sint-Martens-Latem, Belgium). PFGE patterns were clustered by numerical analysis by the unweighted pair group method with arithmetic averages and the Dice similarity coefficient.
Southern blot hybridization.
Genomic DNA was prepared with the QIAamp DNA mini kit (Qiagen) from cultures that had been grown overnight in TSBS. DNA purity and concentrations were determined with a Beckman DU 640 spectrophotometer (Beckman Coulter, Fullerton, Calif.). Southern blot hybridization was carried out as described previously (1). The probes for tdh and ORF8 were synthesized with PCR as described above and by incorporating digoxigenin-11-dUTP (Roche Diagnostics Corp., Indianapolis, Ind.) as a label into PCR products. Genomic DNA (2.5 μg) was digested with HindIII, and the resulting DNA fragments were separated in agarose gels and transferred to nylon membranes. Probes were hybridized at 55°C in the presence of blocking reagent (Roche Diagnostics). Anti-digoxigenin-AP Fab fragment (Roche Diagnostics) and CDP-Star (Roche Diagnostics) were used to detect the presence of target sequences.
Adherence.
HeLa cells (human epithelial cells) were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum and 1% nonessential amino acids to form monolayers on 12-mm-diameter untreated glass coverslips, which were maintained in a humidified atmosphere of 5% CO2 in air at 37°C. V. parahaemolyticus isolates were grown overnight in TSBS, and 250 μl of these cultures was centrifuged and resuspended with 250 μl of phosphate-buffered saline (PBS). Bacterial cell suspensions were then used to infect HeLa cells at a multiplicity of infection (MOI) of about 100:1. Extracellular nonadherent bacterial cells were removed 2 h after the infection by three washes with PBS. Immediately after washing, monolayers were lysed and cell suspensions were plated onto TSAS plates to determine the number of V. parahaemolyticus cells that had adhered to the host cells. To lyse host cells, triplicate coverslips were aseptically removed from the wells, deposited into 4-ml sterile 0.1% Triton X-100 in PBS and vigorously vortexed. In addition, one coverslip was stained with modified Giemsa's solution, and monolayers were observed microscopically.
Cytotoxicity.
Cytotoxicity was assessed by measurement of released host cell lactate dehydrogenase (LDH) with the CytoTox 96 nonradioactive cytotoxicity assay (Promega, Madison, Wis.). Briefly, HeLa cells were grown in 1 ml of DMEM in 24-well plates to form monolayers. After infection with V. parahaemolyticus at an MOI of about 100:1, 10 μl of culture supernatants was removed at various time points and pipetted into 96-well plates. After incubation for 30 min at room temperature with 50 μl of substrate mix, the enzymatic reaction was terminated by the addition of 50 μl of stop solution. Plates were read at 490 nm with a μQuant platereader (Bio-Tek Instruments, Inc., Winooski, Vt.). Cytotoxicity calculations were based on the formula % cytotoxicity = 100 × [(Asample − Aspontaneous)/(Atotal − Aspontaneous)], where Asample is the absorbance of infected cells, Aspontaneous is the absorbance of uninfected cells, and Atotal is the absorbance of cells lysed for maximal LDH release after freezing cells at −80°C for at least 30 min. The average absorbance of blank wells was subtracted from all other readings.
Statistical analyses.
Results from the adherence and cytotoxicity tests for the O3:K6 and the non-O3:K6 groups were compared by using the two-sample t test (parametric) and the Mann-Whitney U test (nonparametric) to evaluate the significance of the differences observed between the groups. The relationship between adherence and cytotoxicity was evaluated by correlation. All statistical analyses were performed with SAS version 8.0 (SAS Institute, Inc., Cary, N.C.) or Minitab version 12.0 (Minitab, Inc., State College, Pa.).
Nucleotide sequence accession number.
The tdh sequence data reported in this work were submitted to GenBank under accession no. AY044107, AY044108, AY044109, AY044110, AY044111, AY044112, AY044113, and AY044114.
RESULTS
We assembled a collection of 25 V. parahaemolyticus isolates to allow a comparative characterization of O3:K6 and non-O3:K6 isolates. Isolates were characterized by ribotyping, PFGE, tdh sequencing, and PCR and Southern blot screening for ORF8, an ORF that appears to be unique to strains closely related to recent O3:K6 isolates (17). Isolates also were tested for their swarming phenotype and for tissue culture cytopathogenicity and adherence characteristics.
Ribotyping.
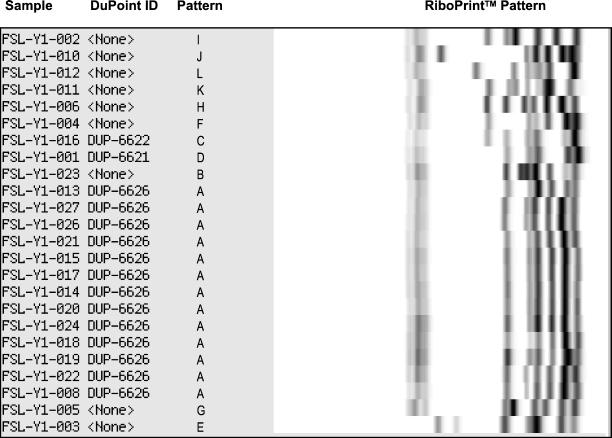
EcoRI ribotyping yielded 13 different patterns among the 25 V. parahaemolyticus isolates characterized (Table 2 and Fig. 1). Fourteen isolates, which included 10 recent (i.e., isolated in and after 1996) and “old” (i.e., isolated before 1996) O3:K6 isolates, one O4:K68 isolate, one O1 isolate, one O6:K18 isolate, and one unknown isolate, shared an almost identical ribotype (patterns A1 and A2; Table 2 and Fig. 1). One “old” O3:K6 isolate (FSL-Y1-021) was characterized as pattern A1, which was indistinguishable from recent O3:K6 isolates (isolated in 1996 and later). Overall, three different ribotypes (A1, B, and C) were found among the 12 isolates in the O3:K6 group. Eleven different ribotypes were identified among the 13 isolates in the non-O3:K6 group.
FIG. 1.
Ribotypes of V. parahaemolyticus isolates. Ribotype patterns are arbitrarily defined such that identical patterns are denoted by the same letter. DuPoint designations for ribotypes already present in the RiboPrinter database are also indicated.
PFGE.
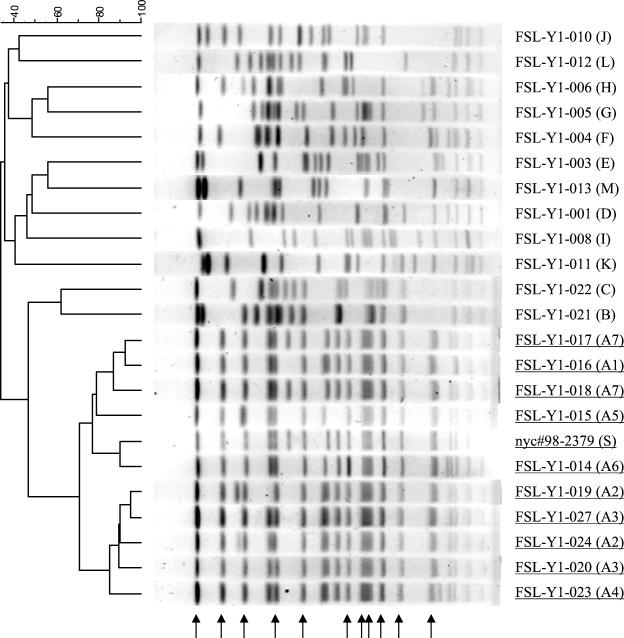
NotI PFGE differentiated 19 subtypes among the 22 isolates characterized. In contrast to EcoRI ribotyping, NotI PFGE reliably discriminated between recent and “old” O3:K6 isolates. To illustrate, although FSL-Y1-016 and FSL-Y1-023, both recent O3:K6 isolates, had different ribotypes, their PFGE patterns were not only very similar to each other, but were also clearly distinct from those of the “old” O3:K6 isolates (Table 2 and Fig. 2). Isolates in the O3:K6 group shared 11 common bands (pattern A). Pattern A could be further divided into seven subpatterns, which differed by up to four bands. FSL-Y1-017 (New York isolate) and FSL-Y1-018 (Connecticut isolate) shared an identical PFGE pattern, but were discernible from FSL-Y1-016 (Texas isolate) by three bands (Fig. 2). In contrast to the very similar PFGE patterns shared by isolates in the O3:K6 group, 12 distinct patterns were identified for 12 isolates in the non-O3:K6 group.
FIG. 2.
DNA restriction patterns of V. parahaemolyticus isolates by PFGE after digestion with NotI. Running conditions involved a switching time of 2 to 40 s, 6 V/cm, and running time of 18 h. The standard (S) was NotI-restricted DNA of nyc#98-2379, which is a V. parahaemolyticus isolate associated with the 1998 New York outbreak. PFGE patterns were generated successfully for 22 isolates. The dendrogram groups PFGE patterns by percent similarity (shown above the dendrogram). Isolates in the O3:K6 group are underlined. Pattern designations are given in parentheses. Common bands displayed by isolates in the O3: K6 group are indicated by arrows.
Sequence determination of the tdh gene.
The presence of the tdh gene was assessed in all 25 isolates by PCR with a set of primers flanking the entire tdh coding region. Among these isolates, 16 bore the tdh gene (Table 2). The nine isolates that were negative for tdh by PCR were also negative by Southern blot hybridization. Of the 16 isolates positive for tdh, 15 were clinical isolates, and 1 was a food isolate. Of the nine tdh-negative isolates, six were food isolates and three were clinical isolates.
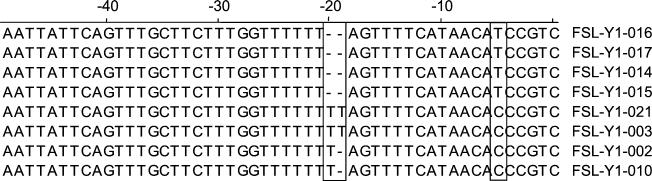
Complete tdh DNA sequences were determined for eight V. parahaemolyticus isolates (four from the O3:K6 group and four from the non-O3:K6 group). These sequences were highly similar to each other (99.4 to 100% identity), as well as to previously published tdh sequences. Overall, isolates in the O3:K6 group (FSL-Y1-014, FSL-Y1-015, FSL-Y1-016, and FSL-Y1-017) showed few, but distinct, sequence differences from those of the non-O3:K6 group (FSL-Y1-002, FSL-Y1-003, FSL-Y1-010, and FSL-Y1-021). The predicted Tdh protein was comprised of 165 amino acids (aa), preceded by a putative signal peptide sequence of 24 aa (18). The predicted amino acid sequences encoded by tdh in all eight V. parahaemolyticus isolates were identical, except that the Gly109 present in the O3:K6 group was predicted to be an Asp109 in the non-O3:K6 group. In addition, FSL-Y1-003 had a deletion of 11 nucleotides near the carboxyl terminus. Sequence differences also were found in the putative tdh promoter region, specifically in the number of thymidine (T) residues present in this region (Fig. 3). While isolates in the O3:K6 group had six T residues in this region, sequences for non-O3:K6 isolates had seven (FSL-Y1-002 and FSL-Y1-010) or eight (FSL-Y1-003 and FSL-Y1-021) T residues. Isolates in the O3:K6 group also had a thymidine at position −5 instead of the cytidine present in the non-O3:K6 isolates (Fig. 3).
FIG. 3.
Nucleotide sequence variation of the putative tdh promoter region among isolates in the V. parahaemolyticus O3: K6 group (FSL-Y1-014, FSL-Y1-015, FSL-Y1-016, and FSL-Y1-017), and in the non-O3: K6 group (FSL-Y1-002, FSL-Y1-003, FSL-Y1-010 and FSL-Y1-021). Sequence differences among isolates are boxed.
Screening for the presence of ORF8.
The discovery and characterization of a new filamentous phage, f237, in recent V. parahaemolyticus O3:K6 isolates suggests the possibility that the enhanced virulence of this clone might be associated with the presence of newly introduced virulence-related genes. The eighth ORF in f237, referred to as ORF8, was found by colony hybridization analysis to be unique to V. parahaemolyticus O3:K6 strains isolated after 1996 (17). We thus screened by PCR for ORF8 in 25 V. parahaemolyticus isolates and found that this gene was exclusively present in isolates of the O3:K6 group (Table 2). Results were confirmed by Southern blot hybridization and sequence analysis.
Swarming.
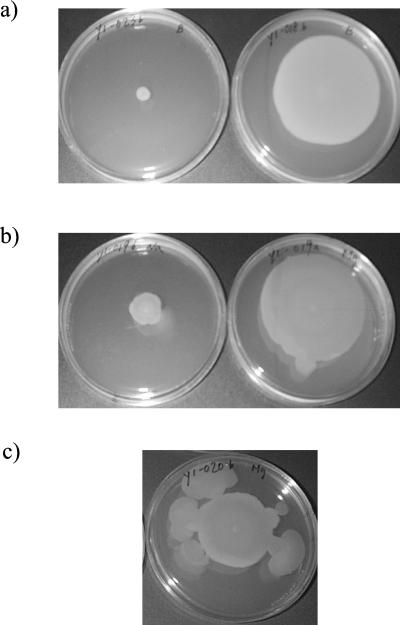
To determine if swarming behavior was associated with specific serotypes, we characterized the swarming phenotypes of our 25 V. parahaemolyticus isolates. Swarming was determined on TSA supplemented with sodium chloride or sodium chloride and magnesium. Results are summarized in Table 2. Of 12 isolates in the O3:K6 group, 4 (33%) did not exhibit swarming. Of 13 isolates in the non-O3:K6 group, 10 (77%) did not exhibit swarming. In general, isolates in the O3:K6 group appeared more likely to have a swarming phenotype than non-O3:K6 isolates (P = 0.028). Nine of 11 isolates (82%) with a swarming phenotype were characterized by a “burst formation,” which was an irregular outgrowth from a discrete locus within the colony, as shown in Fig. 4. For eight isolates (FSL-Y1-001, FSL-Y1-004, FSL-Y1-005, FSL-Y1-006, FSL-Y1-016, FSL-Y1-018, FSL-Y1-025, and FSL-Y1-026), swarming was induced by the presence of magnesium.
FIG. 4.
Representative swarming phenotypes of different V. parahaemolyticus isolates. (a) Negative (left) and positive (right) swarming phenotypes. (b) Swarming was stimulated by magnesium (right) for comparison with swarming on medium without magnesium (left). (c) Burst formation.
Adherence.
Because ORF8 of f237 is predicted to encode an adhesion molecule (17), we hypothesized that the recent O3:K6 clone might have enhanced adherence capabilities relative to other strains and that this property may contribute to pathogenesis. We thus assessed adherence of V. parahaemolyticus to human host cells in a tissue culture model (Table 2). The numbers of bacterial cells that adhered to HeLa cells ranged from 13 × 106 to 47 × 106 per coverslip and 72 × 105 to 34 × 106 per coverslip for isolates in the O3:K6 and non-O3:K6 groups, respectively, as determined by plate counts. The average number of adhered bacterial cells per HeLa cell as determined by microscopic counts ranged from 1.8 to 14.5 (mean ± standard deviation = 8.5 ± 3.7) and 0.9 to 9.9 (mean ± standard deviation = 5.4 ± 2.9) for the O3:K6 and non-O3:K6 groups, respectively. Statistically, isolates in the O3:K6 group exhibited higher levels of adherence to HeLa cells than the non-O3:K6 isolates (P <0.05) by both measurements.
Cytotoxicity.
LDH release from host cells was measured to reflect cytotoxicity resulting from interactions with bacterial pathogens (20). LDH release from HeLa cells following 3 h of bacterial exposure differed by up to 40-fold among the tested isolates. Seventy-five percent of isolates in the O3:K6 group exhibited cytotoxicities greater than 50%, compared to 23% of those in the non-O3:K6 group. As shown in Table 2, cytotoxicities ranged from 15 to 83% and 2 to 70% for isolates in the O3:K6 and non-O3:K6 groups, respectively. After 6 h of exposure, host cell LDH release increased for all isolates in comparison to values measured after 3 h of exposure.
In general, isolates in the O3:K6 group appear to have an enhanced ability to elicit cytotoxic effects on human epithelial cells within 3 h relative to the non-O3:K6 isolates. A statistically significant relationship was found between the number of adhered V. parahaemolyticus cells per coverslip and cytotoxicity after 3 h of exposure (P = 0.009). A smaller, but also significant, correlation was determined between the number of adhered V. parahaemolyticus cells per HeLa cell and cytotoxicity after 3 h of exposure (P = 0.084).
DISCUSSION
The objectives of this study were to evaluate the abilities of molecular methods to differentiate recent V. parahaemolyticus O3:K6 isolates from unrelated serotypes and isolates and to perform a comparative genetic and phenotypic characterization of these O3:K6 isolates and of unrelated isolates to better understand the apparent enhanced virulence of the O3:K6 group. To these ends, we characterized a collection of genetically diverse V. parahaemolyticus isolates by using molecular subtyping strategies and phenotypic methods and tissue culture models.
Molecular subtyping and genetic characterization.
Molecular subtyping methods can be applied to identify different strains within a species; hence, these methods are useful in taxonomic and epidemiologic studies. The application of molecular subtyping methods has become integral to food-borne outbreak investigations (15). While some previous studies have compared various subtyping methods (PFGE, ribotyping, fla locus restriction fragment length polymorphism analysis, and enterobacterial repetitive intergenic consensus [ERIC] PCR) for differentiation of V. parahaemolyticus isolates (3, 15, 21), none of these studies specifically focused on V. parahaemolyticus serotype O3:K6. PFGE was shown to most effectively discriminate among 60 isolates from a 1997 outbreak in British Columbia, Canada (15). Marshall et al. (15) suggested that the utility of PFGE for typing V. parahaemolyticus might be limited by a high proportion of untypeable isolates; however, only 1 isolate was untypeable among the 23 that we tested.
Our EcoRI ribotyping and NotI PFGE results showed significant genetic diversity of non-O3:K6 isolates in our collection (12 PFGE patterns and 11 ribotypes), while isolates categorized into the O3:K6 group represented a more genetically homogeneous group (three ribotypes and seven closely related PFGE patterns). The NotI PFGE patterns found among the O3:K6 group were unique; thus, PFGE allowed differentiation of recent O3:K6 isolates from non-O3:K6 isolates, as well as from “old” O3:K6 isolates. Similarly, Bag et al. (3) showed that a nearly identical PFGE pattern was shared by five O3:K6 isolates isolated between August 1996 and March 1998, whereas several distinct patterns were identified for four O3:K6 isolates that had been obtained between February and August 1996. Our results support the hypothesis that strains bearing serotypes O4:K68 and O1:KUT, which have been responsible for gastroenteritis in India and other Southeast Asian countries, were likely to have originated from the pandemic O3:K6 clone (8). The O4:K68 and O1:KUT isolates included in our study had the same ribotype, as well as closely related PFGE patterns, as the majority of recent O3:K6 isolates. Our results are consistent with reports by Gendel et al. (11) and by Chowdhury et al. (8), who reported that 35 pandemic isolates of V. parahaemolyticus (21 O3:K6, 10 O4:K68 and 4 O1:KUT) isolated primarily from Asian countries between 1996 to 1999 all showed highly similar BglI ribotypes. In these studies, 33 isolates exhibited an identical pattern with 11 fragments. Two isolates—from Japan and the United States—differed from the consensus pattern by a single band. However, in another study, five different BglI ribotypes were found among 30 O3:K6 isolates isolated in Calcutta between 1996 and 1998 (3). In our study, EcoRI ribotyping did not allow consistent discrimination of the recent O3:K6 isolates from other isolates, because one “old” O3:K6 isolate and one O6:K18 isolate showed the same patterns as the most recent O3:K6 isolates.
Our results indicate some genetic diversity among isolates classified into the O3:K6 group, including the presence of multiple ribotypes, multiple closely related PFGE patterns, and multiple serotypes. We found that three O3:K6 isolates from the United States had NotI PFGE patterns very similar to those of three Calcutta O3:K6 isolates. Interestingly, the NotI pattern obtained from an isolate associated with the New York V. parahaemolyticus outbreak differed by three bands from the pattern generated by an isolate from a Texas (Galveston Bay) outbreak. This finding suggests the possibility that different V. parahaemolyticus subtypes may have been responsible for the 1998 U.S. outbreaks. Alternatively, it is possible that the New York outbreak isolate is a genetic variant of the Texas outbreak isolate.
Our DNA sequence comparisons of tdh genes from both the O3:K6 group and the non-O3:K6 group are consistent with previous reports of high levels of conservation in the tdh sequence among different strains (18). Our analyses revealed few, but potentially significant, differences between these groups. Specifically, we identified one amino acid polymorphism in the tdh ORF that appears to differentiate isolates in the O3:K6 group (Gly109) from non-O3:K6 isolates (Asp109). Interestingly, we also observed sequence polymorphisms in the putative tdh promoter regions of these isolates. We found that the non-O3:K6 isolates have one or two additional T residues in this region relative to the O3:K6 isolates (Fig. 3). Different numbers of nucleotide residues in a promoter region may alter the spacing between the −10 and −35 promoter binding sites. Therefore, we speculate that the binding affinity of RNA polymerase to the −10 and −35 promoter regions of tdh may differ between strains in the O3:K6 and non-O3:K6 groups. Further experiments are necessary to determine possible effects of these additional T residues on tdh transcription initiation.
We hypothesized that the O3:K6 group might have emerged as a result of the transfer of genetic elements. Nasu et al. (17) recently reported the presence of the unique filamentous phage f237 in the V. parahaemolyticus O3:K6 Calcutta clone. The genome of this phage included ORF8, which has been proposed as a reliable marker for the O3:K6 Calcutta clone. Nasu et al. (17) screened only isolates collected between January and August 1996 for the presence of ORF8. In this study, we used PCR to screen for the presence of ORF8 in 25 isolates and found that not only the recent O3:K6 isolate, but also the O4:K68 and O1:KUT isolates, possessed ORF8. These findings are consistent with those of Iida et al. (14), who reported that O4:K68 and O1:KUT strains are closely related to the recent O3:K6 strains. The presence of tdh was not always associated with ORF8, because ORF8 was absent in several tdh-positive, non-O3:K6 isolates. Thus, our results confirm the specificity of ORF8 to the O3:K6 clonal group (including O4:K68 and O1:KUT strains). Thus, ORF8 may provide a unique genetic target for development of a PCR-based detection strategy for the V. parahaemolyticus O3:K6 clonal group.
Virulence-associated characteristics of O3:K6 isolates.
Virulence-related characteristics (including cytopathogenicity, host cell adherence, and motility) of the isolates in our collection were determined to further define the unique features of recent V. parahaemolyticus O3:K6 isolates and to better understand their apparent emergence as a particularly virulent group. Various V. parahaemolyticus strains previously had been shown to induce cytotoxicity in a number of cell lines, including Caco-2 cells (20). By using HeLa cells, we found that the mean cytotoxicity and adherence were greater for the O3:K6 group than for the non-O3:K6 group. Importantly, these findings establish quantitative evidence for enhanced virulence-associated characteristics of O3:K6 clonal group isolates compared to those of other V. parahaemolyticus strains.
Furthermore, our findings have established a statistically significant relationship between adherence and cytotoxicity, suggesting that cytotoxicity of some isolates may be associated with stronger adherence to epithelial cells. An association between adherence and cytotoxicity also has been reported by Baffone et al. (2), who suggested that a cytotoxic distortion of host cell epithelial structures might facilitate adhesion by additional bacterial cells. Alternatively, enhanced adherence might enable a greater number of cells to attach to the host cell surface, which may collectively produce a stronger cytotoxic effect.
Based on predicted amino acid sequence, Nasu et al. (17) speculated that ORF8 encodes an adherence protein. Adherence of pathogenic bacteria to the host cell may be related to virulence, because adherence is thought to be a prerequisite for causing disease. Hackney et al. (12) proposed using adherence as a method for differentiating virulent and avirulent V. parahaemolyticus strains, because intensity of bacterial adherence to human fetal epithelial cells appeared to be associated with a strain's ability to cause food-borne illness. Additional experimentation on the role of ORF8 in host cell adherence may provide further insight into virulence mechanisms as well as the evolution of the O3:K6 clonal group.
The motility phenotype is correlated with virulence in V. cholera (10). Specifically, V. cholera mutants with greater motility showed increased expression of virulence-associated fucose-sensitive hemagglutination and HEp-2 adhesins. V. parahaemolyticus possess lateral flagella, which enable migration across semisolid surfaces in a phenomenon called swarming. As an enhanced swarming ability might allow bacterial cells to more rapidly and effectively reach a target host, we tested the ability of the isolates in our collection to swarm. In general, isolates in the O3:K6 group demonstrated an enhanced ability to swarm over agar surfaces. The presence of magnesium appeared to further stimulate swarming in eight isolates. None of the four non-O3:K6 isolates that were tdh positive exhibited any swarming capability, nor were they especially effective at adherence or cytotoxicity. While the mechanism triggering swarming and its importance for virulence are not well understood, further characterization of this behavior may contribute to an understanding of its role, if any, in V. parahaemolyticus pathogenesis.
Conclusions.
In this study, we have confirmed the close genetic relationships among isolates in the O3:K6 clonal group, including serotypes O4:K68 and O1:KUT, by using three different molecular subtyping approaches (EcoRI ribotyping, NotI PFGE, and tdh sequencing). In general, isolates in the O3:K6 clonal group had enhanced cytopathogenicity, host cell adherence, and motility compared to non-O3:K6 isolates. We hypothesize that the unique pathogenic potential of the O3:K6 clonal group may be related to these phenotypic characteristics. Future studies on the effects of ORF8 and of sequence variation in the tdh promoter region may provide additional insight into the evolution and virulence characteristics of this recently emerged group.
Acknowledgments
We are grateful to Martin Wiedmann of Cornell University for providing technical advice. We also thank Suzanne Barth of the Texas Department of Health for Texas isolates, Tim Root of the New York State Department of Health for New York isolates, Bob Howard of the Connecticut Department of Public Health for Connecticut isolates, and Mitsuaki Nishibuchi of Kyoto University for Asian isolates.
This work was supported by the Cooperative State Research, Education, and Extension Service, National Research Initiative Competitive Grants Program (NRI Proposal 199902756) of the United States Department of Agriculture.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1995. Current protocols in molecular biology. John Wiley & Sons, Inc., New York, N.Y.
- 2.Baffone, W., A. Pianetti, F. Bruscolini, E. Barbieri, and B. Citterio. 2000. Occurrence and expression of virulence-related properties of Vibrio species isolated from widely consumed seafood products. Int. J. Food Microbiol. 54:9-18. [DOI] [PubMed] [Google Scholar]
- 3.Bag, P. K., S. Nandi, R. K. Bhadra, T. Ramamurthy, S. K. Bhattacharya, M. Nishibuchi, T. Hamabata, S. Yamasaki, Y. Takeda, and G. B. Nair. 1999. Clonal diversity among recently emerged strains of Vibrio parahaemolyticus O3:K6 associated with pandemic spread. J. Clin. Microbiol. 37:2354-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blake, P. A., R. E. Weaver, and D. G. Hollis. 1980. Diseases of humans (other than cholera) caused by vibrios. Annu. Rev. Microbiol. 34:341-367. [DOI] [PubMed] [Google Scholar]
- 5.Bruce, J. 1996. Automated system rapidly identifies and characterizes microorganisms in food. Food Technol. 50:77-81. [Google Scholar]
- 6.Centers for Disease Control and Prevention. 1999. Outbreak of Vibrio parahaemolyticus infection associated with eating raw oysters and clams harvested from Long Island Sound—Connecticut, New Jersey, and New York, 1998. Morb. Mortal. Wkly. Rep. 48:48-51. [PubMed] [Google Scholar]
- 7.Chiou, C.-S., S.-Y. Hsu, S.-I. Chiu, T.-K. Wang, and C.-S. Chao. 2000. Vibrio parahaemolyticus serovar O3:K6 as cause of unusually high incidence of food-borne disease outbreaks in Taiwan from 1996 to 1999. J. Clin. Microbiol. 38:4621-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowdhury, N. R., S. Chakraborty, T. Ramamurthy, M. Nishibuchi, S. Yamasaki, Y. Takeda, and G. B. Nair. 2000. Molecular evidence of clonal Vibrio parahaemolyticus pandemic strains. Emerg. Infect. Dis. 6:631-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Food and Drug Administration. January2001, posting date. Bacteriological analytical manual. [Online.] Food and Drug Administration, Rockville, Md. http://www.cfsan.fda.gov/∼ebam/bam-9.html.
- 10.Gardel, C. L., and J. J. Mekalanos. 1996. Alterations in Vibrio cholerae motility phenotypes correlate with changes in virulence factor expression. Infect. Immun. 64:2246-2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gendel, S. M., J. Ulaszek, M. Nishibuchi, and A. DePaola. 2001. Automated ribotyping differentiates Vibrio parahaemolyticus O3:K6 strains associated with a Texas outbreak from other clinical strains. J. Food Prot. 64:1617-1620. [DOI] [PubMed] [Google Scholar]
- 12.Hackney, C. R., E. G. Kleeman, B. Ray, and M. L. Speck. 1980. Adherence as a method of differentiating virulent and avirulent strains of Vibrio parahaemolyticus. Appl. Environ. Microbiol. 40:652-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hlady, W. G., and K. C. Klontz. 1996. The epidemiology of Vibrio infections in Florida, 1981-1993. J. Infect. Dis. 173:1176-1183. [DOI] [PubMed] [Google Scholar]
- 14.Iida, T., A. Hattori, T. Kenichi, H. Nasu, R. Naim, and T. Honda. 2001. Filamentous phage associated with recent pandemic strains of Vibrio parahaemolyticus. Emerg. Infect. Dis. 7:477-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marshall, S., C. G. Clark, G. Wang, M. Mulvey, M. T. Kelly, and W. M. Johnson. 1999. Comparison of molecular methods for typing Vibrio parahaemolyticus. J. Clin. Microbiol. 37:2473-2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto, C., J. Okuda, M. Ishibashi, M. Iwanaga, P. Garg, T. Rammamurthy, H. C. Wong, A. Depaola, Y. B. Kim, M. J. Albert, and M. Nishibuchi. 2000. Pandemic spread of an O3:K6 clone of Vibrio parahaemolyticus and emergence of related strains evidenced by arbitrarily primed PCR and toxRS sequence analyses. J. Clin. Microbiol. 38:578-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nasu, H., T. Iida, T. Sugahara, Y. Yamaichi, K.-S. Park, K. Yokoyama, K. Makino, H. Shinagawa, and T. Honda. 2000. A filamentous phage associated with recent pandemic Vibrio parahaemolyticus O3:K6 strains. J. Clin. Microbiol. 38:2156-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishibuchi, M., and J. B. Kaper. 1985. Nucleotide sequence of the thermostable direct hemolysin gene of Vibrio parahaemolyticus. J. Bacteriol. 162:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okuda, J., M. Ishibashi, E. Hayakawa, T. Nishino, Y. Takeda, A. K. Mukhopadhyay, S. Garg, S. K. Bhattacharya, G. B. Nair, and M. Nishibuchi. 1997. Emergence of a unique O3:K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 35:3150-3155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raimondi, F., J. P. Y. Kao, C. Fiorentini, A. Fabbri, G. Donelli, N. Gasparini, A. Rubino, and A. Fasano. 2000. Enterotoxicity and cytotoxicity of Vibrio parahaemolyticus thermostable direct hemolysin in in vitro systems. Infect. Immun. 68:3180-3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wong, H. C., M. C. Chen, S. H. Liu, and D. P. Liu. 1999. Incidence of highly genetically diversified Vibrio parahaemolyticus in seafood imported from Asian countries. Int. J. Food Microbiol. 52:181-188. [DOI] [PubMed] [Google Scholar]