Abstract
Purified bile salt hydrolase from bile-adapted Xanthomonas maltophilia displays Michaelis-Menten kinetics on cholylglycine and cholyltaurine and hydrolyzes bile salts also in crude bovine bile. The protein is a dimer and is resistant to proteinases and to heating at 55 to 60°C for up to 60 min, in agreement with calorimetric data.
Recently, we examined the metabolism of bile salts by the bile-adapted strain Xanthomonas maltophilia CBS 827.97, which hydrolyzes bile salt conjugates, rearranging the steroid nucleus (1, 5). Bile salts have multiple functions in digestion of lipids, solubilization, and excretion of cholesterol (2, 19, 23). Ursodeoxycholic acid is particularly interesting in this regard, solubilizing gallbladder cholesterol stones after oral administration (20). Ursodeoxycholic acid is currently prepared by alkaline hydrolysis of bile conjugates, inversion of the hydroxyl configuration at carbon 7, and resolution of the racemic mixture (reviewed in reference 1). Procedures for enzymatic synthesis would be useful for controlling stereochemistry, increasing yields, and avoiding toxic or aggressive chemicals. In this paper we describe a solution to the hydrolysis step, in which purified bile salt hydrolase (cholylglycine hydrolase [CGH], EC 3.5.1.24) from our bacterial strain is used.
Procedures for purification of CGH and of other enzymes of bile salt metabolism are covered by an Italian patent (1). For purification of CGH, the enzyme was isolated from 10 g of bacteria grown as described previously (5) and lysed by overnight stirring in 10 volumes of 20 mM sodium phosphate-1 mM EDTA-2 mM mercaptoethanol (pH 7.5) (NaP buffer) containing 0.3 mg of egg lysozyme per ml and 1 mM phenylmethylsulfonyl fluoride. Cell debris was removed by centrifugation (200,000 × g, 20 min), and nucleic acids were removed by precipitation with protamine sulfate (2 mg/ml). The supernatant, adjusted to pH 8.3, was chromatographed on a DEAE-Sepharose column (30 ml) and eluted with a linear 0 to 0.25 M NaCl gradient in NaP buffer (pH 8.3). The second protein peak, displaying hydrolase activity, was fractionated with ammonium sulfate in order to collect proteins insoluble between 45 and 75% saturation. After dialysis against NaP buffer, the phosphate concentration of the solution was adjusted to 200 mM at pH 7.5 by adding solid salts, and the solution was heated to 55°C with continuous swirling in a water bath at 70°C. After the solution was cooled on ice, denatured aggregated proteins were removed by high-speed centrifugation, and the supernatant, diluted with 1 volume of water, was loaded onto a phenyl-Sepharose column (2 ml) equilibrated in 100 mM phosphate buffer (pH 7.5). Hydrolase activity was eluted with NaP buffer. The CGH activity assay was performed by measuring glycine release from 5 mM cholylglycine in NaP buffer by a cadmium-ninhydrin procedure (6). For the cholyltaurine hydrolysis assay, we employed a different ninhydrin formulation (17). The protein concentration was determined as described previously (18).
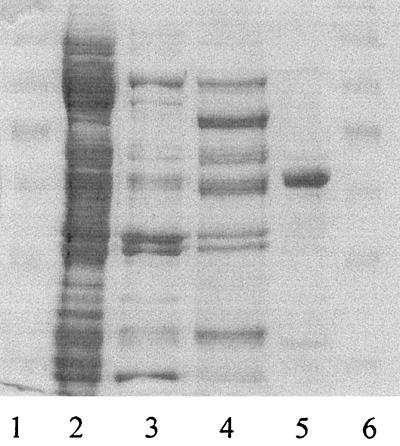
The yield was 0.4 mg of CGH from 10 g of bacteria, with 20% recovery, a specific activity of 100 U/mg, and a purification factor of 168-fold (Table 1). The activity was stable for 2 weeks at 4°C and resisted prolonged heating at temperatures below 55°C. The purified hydrolase (minor contaminants were occasionally present, as shown in Fig. 1) had an Mr of 52,000 as determined by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (16) and an Mr of 100,000 as determined by native size exclusion chromatography, suggesting that the enzyme is a dimer of identical subunits.
TABLE 1.
Purification of CGH from X. maltophilia CBS 827.97a
| Step | Vol (ml) | Protein (mg) | Activity (U) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Extract | 89 | 356 | 213 | 0.6 | 1 | 100 |
| DEAE-Sepharose | 35 | 12.9 | 209 | 16.2 | 27 | 98 |
| Ammonium sulfate (45-70%) | 7 | 7.7 | 140 | 18.2 | 30 | 65 |
| Heat treatment | 7 | 4 | 132.6 | 34 | 56 | 63 |
| Hydrophobic chromatography | 2.5 | 0.33 | 33.5 | 101 | 168 | 16 |
Results of a typical purification in which the starting material was 8.9 g of X. maltophilia. The activity and specific activity values for the ammonium sulfate step were calculated after extensive dialysis.
FIG. 1.
SDS-PAGE of X. maltophilia CGH at different stages of purification. Lanes 1 and 6, Mr standards (Mrs, 172,600, 111,400, 79,600, 61,300, 49,000, 36,400, and 24,700); lane 2, extract; lane 3, DEAE-Sepharose eluate; lane 4, heated ammonium sulfate fraction; lane 5, representative fraction (protein load, 3 μg) eluted by hydrophobic interaction chromatography with NaP buffer (pH 7.5). The gel was stained with Coomassie blue R-250 and destained with aqueous isopropanol-acetic acid.
The enzyme was inactivated slowly by thiol (iodoacetamide and N-ethylmaleimide), histidyl (diethylpyrocarbonate), carboxyl (ethyl-dimethyl-aminopropyl-carbodiimide), and arginyl (hydroxyphenylglyoxal) reagents and rapidly (15 min) by 1 mM o-phthalaldehyde, indicating the importance of closely spaced thiol and lysine residues for the catalytic activity. Heavy metals were inhibitory, and there was complete inactivation by 10 mM copper(II) salts.
The purified protein was resistant to narrow-specificity proteinases (trypsin, chymotrypsin, and V8 proteinase) but was slowly digested by the broad-specificity proteases elastase and pronase, with a half-life of 1.25 h at a hydrolase/protease ratio of 30:1 (wt/wt). No proteolytic intermediate that was detectable by SDS-PAGE accumulated, indicating the tight structure of the protein (13), and there was rapid digestion into small peptides after the initial cleavage. Proteolysis increased under mildly denaturing conditions (0.1% SDS). Automated Edman degradation (21) identified the N-terminal sequence AEGN, which was different from the N-terminal CTXY sequence (where X is Gly or Ala and Y is Ile, Leu, or Val) of other bacterial bile salt hydrolases (3, 4, 7, 22).
The enzyme hydrolyzed cholylglycine with Michaelis-Menten kinetics (Km, 1.1 ± 0.15 mM), and there was competitive inhibition by cholyltaurine (Ki, 2.5 mM), as if both conjugated acids were hydrolyzed at a single site. The rate of inactivation by 5 mM iodoacetate was the same with both substrates. The products taurine and cholic acid inhibited uncompetitively the hydrolysis of cholylglycine, with Ki values of 2.5 and 4.7 mM. A set of parallel straight lines in double-reciprocal plots indicated that there was an inhibitory mechanism involving formation of a rate-limiting acyl intermediate or abortive ternary complexes with cholylglycine and taurine (data not shown). The pH optimum for hydrolysis was 7.9 to 8.5 with two apparent pK values (pK 7.2 and 9), which was consistent with catalytic roles for cysteine and lysine residues.
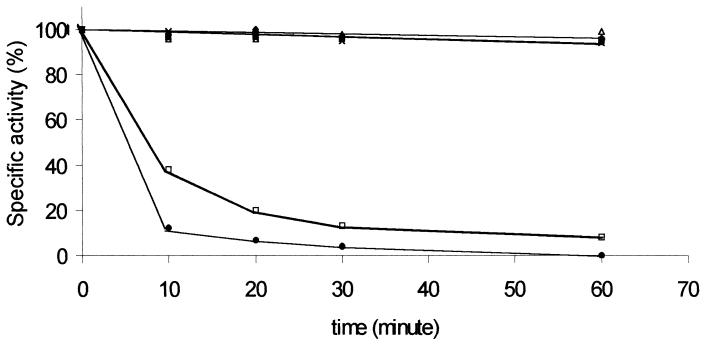
The enzyme was remarkably thermostable, with total preservation of activity during incubation for 75 min at 55°C (Fig. 2); however, it was rapidly inactivated at slightly higher temperatures, with half-lives of 9 and 5 min at 60 and 65°C, respectively. Differential scanning calorimetry revealed a transition at 64.5°C, with exothermic heat exchange and massive aggregation at temperatures above 80°C (data not shown). The thermal unfolding was clearly irreversible and complex, and deconvolution did not fit a two-state model, as shown by the large difference between the calorimetric (82.5 kJ/mol) and Van't Hoff (2.02 kJ/mol) enthalpies.
FIG. 2.
Thermal stability of CGH activity. The purified enzyme was incubated at 25°C (⧫), 40°C (▪), 50°C (▵), 55°C (×), 60°C (□), and 65°C (•), and the activity was determined in the presence of 5 mM cholylglycine at 25°C at different times.
Numerous enteric bacteria express hydrolases for conjugated bile salts as protection against the toxicity of bile acids (9, 10, 15), and there are differences in cellular distribution (4, 14) and molecular structure (trimeric or tetrameric proteins with Mrs of 150,000 to 250,000) (3, 7, 8, 22). These hydrolases display high affinity for conjugated bile salts, have a slightly acid pH optimum, moderate thermostability, and peptide chains (314 to 329 amino acids) shorter than the peptide chain of our hydrolase (52 kDa), and are homologous to penicillin V acylase (22). The N-terminal sequence of our enzyme is different from those reported previously (3, 22), indicating that there has been appreciable divergence in these enzymes and a loss of the N-terminal cysteine, which is thought to be crucial for catalysis.
Some properties of Xanthomonas CGH (broad pH optimum, high hydrophobicity, appreciable thermal stability, and favorable substrate affinity) seem to be useful for carrying out biotransformations at high temperatures for prolonged times, with advantages in terms of the product yield. We examined this possibility by analyzing hydrolysis of conjugated salts in crude bovine bile (containing 35, 33, 10, and 7% taurocholic, glycocholic, taurochenodeoxycholic, and cholic acids, respectively, and smaller amounts of glycodeoxycholic, glycochenodeoxycholic, and taurodeoxycholic acids). At a concentration of 6 mM (close to the reported critical micellar concentration) (11, 12), conjugated bile salts were hydrolyzed completely by X. maltophilia CGH (0.04 U/ml) within 40, 22, and 12 min at 25, 37, and 50°C, respectively, without interference by other bile components (cholesterol, phospholipids) or reaction products (taurine, glycine, cholic acid).
All these features make X. maltophilia CGH attractive from theoretical and applicative points of view, and we are cloning this enzyme to obtain large amounts of it for complete physicochemical characterization.
Acknowledgments
This research was supported by grants from the University of Ferrara to C.M.B. and from Industria Chimica Emiliana, Reggio Emilia to A.M.
Footnotes
This report is dedicated to the memory of our teacher, Mario Rippa, who suddenly died on 27 March 2001.
REFERENCES
- 1.Bartoli, E., A. Medici, and C. M. Bergamini. June 1999. Italian patent MI99A 001383.
- 1a.Bortolini, O., A. Medici, and S. Poli. 1997. Biotransformations on steroid nucleus of bile acids. Steroids 62:564-577. [DOI] [PubMed] [Google Scholar]
- 2.Carey, M. C., D. M. Small, and C. M. Bliss. 1983. Lipid digestion and absorption. Annu. Rev. Physiol. 45:651-677. [DOI] [PubMed] [Google Scholar]
- 3.Christiaens, H., R. J. Leer, P. H. Pouwels, and W. Verstraete. 1992. Cloning and expression of a conjugated bile acid hydrolase gene from Lactobacillus plantarum by using a direct plate assay. Appl. Environ. Microbiol. 58:3792-3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman, J. P., and L. L. Hudson. 1995. Cloning and characterization of a conjugated bile acid hydrolase gene from Clostridium perfringens. Appl. Environ. Microbiol. 61:2514-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dean, M., G. Fantin, M. Fogagnolo, A. Medici, P. Pedrini, and S. Poli. 1999. Microbial 7-OH epimerization of bile acids. Chem. Lett. 1999:693-694. [Google Scholar]
- 6.Doi, E., D. Shiobata, and T. Matoba. 1981. Modified colorimetric ninhydrin methods for peptidase assay. Anal. Biochem. 118:173-184. [DOI] [PubMed] [Google Scholar]
- 7.Elkins, C. A., and D. C. Savage. 1998. Identification of genes encoding conjugated bile salt hydrolase and transport in Lactobacillus johnsonii 100-100. J. Bacteriol. 180:4344-4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gopal-Srivastava, R., and P. B. Hylemon. 1988. Purification and characterization of bile salt hydrolase from Clostridium perfringens. J. Lipid Res. 29:1079-1085. [PubMed] [Google Scholar]
- 9.Grill, J. P., F. Schneider, J. Crociani, and J. Ballongue. 1995. Purification and characterization of conjugated bile salt hydrolase from Bifidobacterium longum BB536. Appl. Environ. Microbiol. 61:2577-2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grill, J. P., S. Perrin, and F. Schneider. 2000. Bile salt toxicity to some bifidobacteria strains: role of conjugated bile salt hydrolase and pH. Can. J. Biochem. 46:878-884. [DOI] [PubMed] [Google Scholar]
- 11.Helenius, A., D. R. McCaslin, E. Fries, and C. Tanford. 1979. Properties of detergents. Methods Enzymol. 56:734-749. [DOI] [PubMed] [Google Scholar]
- 12.Hofmann, A. F., and K. J. Mysels. 1992. Bile acid solubility and precipitation in vitro and in vivo: the role of conjugation, pH, and Ca2+ ions. J. Lipid Res. 33:617-626. [PubMed] [Google Scholar]
- 13.Hubbard, S. J. 1998. The structural aspects of limited proteolysis of native proteins. Biochim. Biophys. Acta 1382:191-206. [DOI] [PubMed] [Google Scholar]
- 14.Kishinaka, M., A. Umeda, and S. Kuroki. 1994. High concentrations of conjugated bile acids inhibit bacterial growth of Clostridium perfringens and induce its extracellular cholylglycine hydrolase. Steroids 59:485-489. [DOI] [PubMed] [Google Scholar]
- 15.Kurdi, P., H. W. van Veen, H. Tanaka, I. Mierau, W. N. Konings, G. W. Tannock, F. Tomita, and A. Yokota. 2000. Cholic acid is accumulated spontaneously, driven by membrane pH, in many lactobacilli. J. Bacteriol. 182:6525-6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 17.Lee, Y. P., and T. Takahashi. 1966. An improved colorimetric determination of amino acids with use of ninhydrin. Anal. Biochem. 14:71-77. [Google Scholar]
- 18.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 19.Reddy, B. S. 1981. Diet and excretion of bile acids. Cancer Res. 41:3766-3768. [PubMed] [Google Scholar]
- 20.Salen, G., A. Colalillo, D. Verga, E. Bargan, G. S. Tint, and S. Shefer. 1980. Effect of high and low doses of ursodeoxycholic acid on gallstone dissolution in humans. Gastroenterology 78:1412-1418. [PubMed] [Google Scholar]
- 21.Soundar, S., and R. F. Colman. 1993. Identification of metal-isocitrate binding site of pig heart NADP-specific isocitrate dehydrogenase by affinity cleavage of the enzyme by Fe(2+)-isocitrate. J. Biol. Chem. 268:5264-5271. [PubMed] [Google Scholar]
- 22.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum—biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tint, G. S., G. Salen, and S. Shefer. 1986. Effect of ursodeoxycholic acid and chenodeoxycholic acid on cholesterol and bile acid metabolism. Gastroenterology 91:1007-1018. [DOI] [PubMed] [Google Scholar]