Abstract
Expression of virulence-related extracellular proteases, gelatinase, and serine protease of Enterococcus faecalis is regulated by a quorum-sensing system encoded by the fsr gene cluster. In this study, a 23.9-kb chromosomal deletion containing the fsr gene cluster region was found to be present in the majority (79%) of gelatinase-negative clinical isolates of E. faecalis from urine.
Gelatinase (GelE) and serine protease (SprE) are extracellular proteases which have been shown to be related to the pathogenicity of an opportunistic pathogen, Enterococcus faecalis (2, 4, 5, 11, 13). Previous studies showed that production of these two proteases is regulated by quorum sensing, in which production starts in the late log phase and is maximal in the early stationary phase, and that this quorum sensing is mediated by a cyclic peptide termed gelatinase biosynthesis-activating pheromone (GBAP) (9, 10). GBAP is secreted from each cell, and when the concentration of GBAP that accumulates outside the cells reaches the nanomolar level, production of both proteases is triggered. GBAP is an 11-residue peptide containing a lactone ring in which the carboxyl group at the C terminus is linked to a hydroxyl group of the serine at the third residue. The amino acid sequence of GBAP corresponds to a C-terminal part of a 242-residue protein encoded by an open reading frame termed fsrB (Fig. 1) (9). It has been suggested that the GBAP signal is transduced through a two-component regulatory system comprised of a histidine protein kinase (FsrC) and a response regulator (FsrA) (9). The fsr genes and protease genes are organized as shown in Fig. 1; fsrB and fsrC are cotranscribed, gelE and sprE are also cotranscribed, and expression of both operons is positively regulated by GBAP (9, 11, 12).
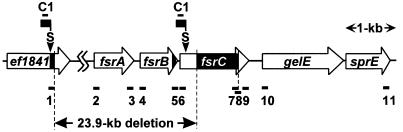
FIG. 1.
Genetic organization of the region surrounding the fsr gene cluster. The map was deduced from sequence data for the E. faecalis V583 chromosome in the TIGR database. The open arrows indicate the lengths and directions of loci. The horizontal solid bars above and below the map indicate the positions of primer annealing sites. A vertical solid bar in the fsrB locus indicates a region encoding GBAP. The sequences and directions of primers are listed in Table 2. The 23.9-kb segment was deleted in group C strains. Adapter cassettes, indicated by black flags, were ligated to the Sau3AI restriction sites indicated by arrowheads. The nested PCR with primers C1 and 8 (first step) and primers C1 and 7 (second step) amplified a 1.0-kb fragment comprised of two segments, indicated by black bars, in ef1841 and fsrC from group C strains.
It has been reported that gelatinase activity was found in approximately one-half of clinical isolates of E. faecalis and in 27% of fecal isolates from healthy volunteers (2). It has also been reported that the gelE gene was present in 89% of clinical isolates, 78% of food isolates, and 50% of starter strains of E. faecalis (3). In this study, we defined the relationship between phenotypes and genotypes related to fsr-regulated protease production in a number of clinical isolates of E. faecalis from urine.
Isolation of E. faecalis strains from patients with urinary tract infections and gelatinase activities of these strains.
The E. faecalis strains used in this study were isolated from patients with urinary tract infections at the Okayama University Hospital (Okayama, Japan) over a 5-year period from 1994 through 1998. Urine specimens were processed in parallel by cultivation on blood agar and UricultE medium (Orion Diagnostica, Espoo, Finland) for isolation and quantification of urinary tract pathogens, respectively. E. faecalis was identified with the AutoMicrobic system (Vitek Systems, Hazelwood, Mo.), and identification was confirmed by the multiplex PCR method with species-specific primers (7). From urine specimens in which Enterococcus spp. were detected at concentrations of >104 CFU/ml, a total of 149 E. faecalis isolates (one isolate per patient) were screened for production of gelatinase by the method of Su et al. (14). Production of a transparent halo around colonies after exposure to a solution saturated with ammonium sulfate on the surface of the medium was considered a gelatinase-positive (Gel+) response. Gelatinase activity was found in 76 (51%) of the 149 urine isolates. For the experiments described below, we selected 15 Gel+ and 33 Gel− strains having more than four different bands in the chromosomal restriction DNA patterns generated with SmaI by pulsed-field gel electrophoresis (15).
GBAP activity of clinical isolates.
The GBAP activity of some of the clinical isolates was measured (Table 1). Culture supernatant was collected from late-log-phase cultures (2 ml; optical density at 600 nm, 1.0) and was then loaded into a Sep-pak C18 cartridge column (100 mg; Waters Co., Milford, Mass.). The column was eluted with 2 ml of 40% acetonitrile containing 0.1% trifluoroacetic acid after it was washed with 2 ml of 20% acetonitrile containing 0.1% trifluoroacetic acid. The eluate was evaporated to dryness and was then assayed for induction of gelatinase biosynthesis in a responder strain, OG1SP (9). As a result, significant GBAP activity was detected for every Gel+ strain. This suggested that GBAP-mediated quorum sensing is common in Gel+ E. faecalis and that there is no strain specificity for GBAP activity. On the other hand, GBAP activity was not detected for any of the Gel− strains except strains OU378 and OU404.
TABLE 1.
Phenotypes and genotypes related to fsr-regulated protease production in clinical isolates of E. faecalis from urine
| Strain | Phenotype
|
Genotype
|
|||
|---|---|---|---|---|---|
| Gelatinasea | GBAPb | gelE-sprEc | fsrABCd | 23.9-kb deletione | |
| Group A strains | |||||
| OU322 | + | + | + | + | − |
| OU336 | + | + | + | + | − |
| OU341 | + | + | + | + | − |
| OU358 | + | + | + | + | − |
| OU360 | + | + | + | + | − |
| OU377 | + | + | + | + | − |
| OU397 | + | + | + | + | − |
| OU422 | + | + | + | + | − |
| OU486 | + | + | + | + | − |
| OU493 | + | + | + | + | − |
| OU502 | + | + | + | + | − |
| OU533 | + | + | + | + | − |
| OU546 | + | + | + | + | − |
| OU590 | + | + | + | + | − |
| OU592 | + | + | + | + | − |
| Group B strains | |||||
| OU378 | − | + | + | + | − |
| OU404 | − | + | + | + | − |
| OU495 | − | − | + | + | − |
| Group C strains | |||||
| OU321 | − | − | + | − | + |
| OU337 | − | − | + | − | + |
| OU350 | − | − | + | − | + |
| OU366 | − | − | + | − | + |
| OU401 | − | NT | + | − | + |
| OU417 | − | NT | + | − | + |
| OU432 | − | NT | + | − | + |
| OU442 | − | NT | + | − | + |
| OU444 | − | NT | + | − | + |
| OU461 | − | NT | + | − | + |
| OU469 | − | NT | + | − | + |
| OU477 | − | NT | + | − | + |
| OU487 | − | NT | + | − | + |
| OU492 | − | − | + | − | + |
| OU497 | − | NT | + | − | + |
| OU511 | − | NT | + | − | + |
| OU521 | − | NT | + | − | + |
| OU528 | − | NT | + | − | + |
| OU530f | − | − | + | − | + |
| OU532 | − | − | + | − | + |
| OU548 | − | − | + | − | + |
| OU579 | − | NT | + | − | + |
| OU583 | − | − | + | − | + |
| OU589 | − | − | + | − | + |
| OU598 | − | − | + | − | + |
| OU602 | − | NT | + | − | + |
| Group D strains | |||||
| OU508 | − | NT | − | − | − |
| OU540 | − | − | − | − | − |
| OU566 | − | − | − | − | − |
| OU591 | − | NT | − | − | − |
Production of gelatinase was determined by the method of Su et al. (14).
GBAP activity of partially purified culture supernatant was measured by the method of Nakayama et al. (9). +, change in optical density at 540 nm of >0.2 u; −, change in optical density at 540 nm of <0.2 u; NT, not tested.
The presence of gelE-sprE was investigated by performing PCR amplification with primers 10 and 11.
The presence of fsrA, fsrB, and fsrC was investigated by performing PCR amplification with primers 2 and 3, with primers 4 and 5, and with primers 6 and 9, respectively. +, all of these genes were detected; −, none of these genes were detected.
The presence of the 23.9-kb deletion was investigated by performing PCR amplification of the 1.0-kb DNA fragment with primers 1 and 7.
Strain used to elucidate the deleted region.
Distribution of the fsr gene cluster and the protease gene cassette among clinical isolates.
The presence of the fsr gene cluster and the protease gene cassette was examined by PCR amplification. Each PCR amplification was performed with specific primers (Table 2); fsrA, fsrB, and fsrC were amplified independently, and gelE and sprE were amplified as a fragment corresponding to a region from the 5′ end of gelE and the 3′ end of sprE (Fig. 1). Template DNA for the PCR was prepared by the method of Kariyama et al. (7). Twenty-five microliters of overnight culture broth of each strain (Todd-Hewitt broth [Oxoid, Basingstoke, United Kingdom]; optical density at 600 nm, >1.0) was mixed with 25 μl of 15% Chelex-100 (Bio-Rad Laboratories, Hercules, Calif.), and the mixture was heated for 10 min at 100°C and then centrifuged at 1,500 × g. One-half microliter of the supernatant was then added to 10 μl of a PCR solution containing 20 mM Tris-HCl (pH 8.4), 1.5 mM MgCl2, 50 mM KCl, 0.1 mM dATP, 0.1 mM dCTP, 0.1 mM dGTP, 0.1 mM dTTP, and 0.5 U of Platinum Taq DNA polymerase (Life Technologies, Paisley, Scotland) (this composition was used in other PCR experiments in this study); the PCR solution also contained each primer at a concentration of 0.2 μM. The PCR program consisted of predenaturation at 94°C for 2 min, followed by 35 cycles of amplification (denaturation at 94°C for 30 s, annealing at 48°C for 30 s, and extension at 72°C for 2 min for fsr genes or for 3 min for gelE-sprE) and then a final extension at 72°C for 6 min, in a GeneAmp PCR system 9600 (PE Biosystems, Foster City, Calif.). PCR products were analyzed by agarose gel electrophoresis followed by staining with ethidium bromide. The amplified fragments of fsrA, fsrB, fsrC, and gelE-sprE corresponding to the appropriate molecular sizes (fsrA, 0.74 kb; fsrB, 0.71 kb; fsrC, 1.3 kb; gelE-sprE, 2.4 kb) were easily recognized. In the case of the fsr genes, either fsrA, fsrB, and fsrC were all amplified from each strain or none of them was amplified.
TABLE 2.
PCR primers used in this study
| Primer | Sequence | Annealing sitea
|
||
|---|---|---|---|---|
| Gene or cassette | Direction | End | ||
| 1 | GATCAAGAAGGGAAGCCACC | ef1841 | F | |
| 2 | ATGAGTGAACAAATGGCTATTTA | fsrA | F | 5′ |
| 3 | CTAAGTAAGAAATAGTGCCTTGA | fsrA | R | 3′ |
| 4 | ATGCTAATCGATTGGATTCTAAAA | fsrB | F | 5′ |
| 5 | TCTTTTTAGGTTTTTCAGTTTGTC | fsrB | R | 3′ |
| 6 | ATGATTTTGTCGTTATTAGCTACT | fsrC | F | 5′ |
| 7 | CCAACCGTGCTCTTCTGGA | fsrC | R | |
| 8 | CATATAACAATCCCCAACCG | fsrC | R | |
| 9 | CATCGTTAACAACTTTTTTACTG | fsrC | R | 3′ |
| 10 | ATGAAGGGAAATAAAATTTTATAC | gelE | F | 5′ |
| 11 | CTGCTGGCACAGCGGATA | sprE | R | 3′ |
| C1 | GTACATATTGTCGTTAGAACGCG | Sau3AI cassette | ||
Location of primer annealing site and its direction. F, forward; R, reverse.
Table 1 shows the distribution of the fsr gene cluster (fsrABC) and the protease gene cassette (gelE-sprE). As expected, both fsrABC and gelE-sprE were amplified from every Gel+ strain (group A). Gel− strains were classified into three groups on the basis of the presence of fsrABC and gelE-sprE. Strains in group C, carrying gelE-sprE but not fsrABC, comprised the majority (79%) of the Gel− strains. Three strains in group B carried both fsrABC and gelE-sprE, even though they showed the Gel− phenotype. Strains OU378 and OU404 in this group were found to carry a point mutation in gelE (data not shown), which may cause the Gel− phenotype. Four strains in group D carried neither fsrABC nor gelE-sprE. No strain in which fsr genes were amplified but gelE-sprE was not amplified was found.
Elucidation of the deleted region in group C strains.
In order to identify the deleted region related to the Gel− phenotype of group C strains, a fragment containing a deletion junction was amplified by a nested adapter PCR (Fig. 1). Genomic DNAs were isolated from some group A and C strains by using previously described procedures (1). The genomic DNAs (0.2 μg) were digested by Sau3AI (Takara Shuzo, Tokyo, Japan), precipitated with ethanol, and then dissolved with 3 μl of H2O. The digests (2 μl) were ligated with 20 ng of a Sau3AI adapter cassette (5′ OH GTACA TATTG TCGTT AGAAC GCGTA ATACG ACTCA CTATA GGGA 3′; Takara Shuzo) by using a ligation kit (Takara Shuzo), precipitated by ethanol, dissolved with 80 μl of H2O, and then heated at 94°C for 10 min. Eighteen microliters of each ligated DNA mixture was used in a 25-μl PCR mixture with primer C1 corresponding to a portion of the Sau3AI adapter cassette and primer 8, as shown in Table 2 (10 pmol of each primer). The PCR program consisted of predenaturation at 94°C for 2 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and extension 72°C for 1.5 min, and a final extension at 72°C for 6 min. The reaction mixture containing the first PCR products was diluted 100-fold with H2O, and 1 μl of the diluted reaction mixture was used for a second PCR (total volume, 25 μl) with primers C1 and 7 (10 pmol of each primer). The second PCR program was the same as the first PCR program. A 1.0-kb amplified fragment was obtained with group C strains, while a 1.2-kb fragment, whose size coincided with the size expected from a Sau3AI restriction site predicted from fsrC sequence data (Fig. 1), was obtained with group A strains. The size difference between the amplified fragments suggested the existence of a deletion junction in the 1.0-kb fragment amplified from group C strains. The 1.0-kb fragment amplified from group C strain OU530 was extracted from the gel, cloned into the pGEM-T vector (Promega, Madison, Wis.), and then sequenced. Comparing the DNA sequence obtained with the sequence in the E. faecalis V583 genome sequence database (The Institute for Genomic Research [TIGR]) (http://www.tigr.org) revealed the deletion junction located 27 bases downstream from the 5′ end of the 1.0-kb fragment. The sequence of the 5′-end 27-base DNA (Fig. 1) was found in a locus designated ef1841 by TIGR, which is located approximately 24 kb upstream of the 5′ end of fsrC. The 3′ end of the 27-bp segment and the 5′ end of the other segment of the junction fragment correspond to positions 1789741 and 1765811, respectively, in the complete sequence of the E. faecalis V583 chromosome in the TIGR database, suggesting that strain OU530 carries a 23,930-bp deletion compared with V583 (Fig. 1). According to the locus assignment by TIGR, this region contains complete 20 loci from ef1821 to ef1840, including fsrA and fsrB, as well as a 5′-end portion of fsrC (ef1820) and a 3′-end portion of ef1841. Putative translational products of ef1829, ef1830, ef1831, ef1836, and ef1838 showed sequence similarities to proteins involved in phosphotransferase systems. The putative product of ef1826 showed sequence similarities to known alcohol dehydrogenases. ef1824 could be translated into a 206-kDa protein whose N-terminal half showed sequence similarities to some glycosyl hydrolases classified in family 31. The putative product of ef1823 showed sequence similarities to some N-acetyl muramidases.
Distribution of the 23.9-kb deletion among clinical isolates.
In order to investigate the distribution of the 23.9-kb deletion among the clinical isolates, a PCR with specific primers 1 and 7 was performed. The template DNA and PCR conditions were the same as those used for PCR amplification of fsr genes except that 52°C was the annealing temperature and extension was for 2 min. As a result, a 1.0-kb amplified DNA fragment was detected in all strains in group C. This indicated that the 23.9-kb deletion was commonly present in group C strains. No clearly amplified band was detected with strains in other groups. In the case of strains in groups A and B, it is likely that there was no big deletion between the two primer annealing sites, and the deduced amplified fragment (about 25 kb) was larger than the limit of amplification under the PCR conditions used. In the case of strains in group D, PCR with primers 1 and 11 also amplified no fragment, suggesting that the deletion extended to downstream of sprE.
In this study, we examined the relationship between phenotype and genotype related to GBAP-induced protease production in a number of clinical isolates of E. faecalis from urine and defined three groups showing different genotypes in gelatinase-negative strains in addition to group A Gel+ strains. Group C strains having fsrABC− and gelE-sprE+ genotypes comprised 79% of the Gel− strains, indicating that the Gel− phenotype is mainly determined by a defective fsr gene cluster. Eaton and Gasson have shown that there is an apparently silent gelE gene in some clinical and food isolates (3). The apparently silent gelE phenotype could also be explained by a lack of the fsr gene cluster instead of gelE-sprE genes. On the basis of a homology search of the putative translated products encoded in the deleted region, it is assumed that the 23.9-kb deletion changes some phenotypes which may or may not be related to the pathogenicity of E. faecalis in addition to gelatinase production. In our preliminary experiment, 2 of 12 Gel+ strains tested became Gel− after six generations (unpublished data). Eaton and Gasson have also reported that gelatinase activity was lost during subculturing from the original stock of a Gel+ strain (3). These observations suggest that this kind of deletion may occur at a high frequency. Recently, pathogenicity islands which are accessory genetic elements that range in size from 10 to 200 kb and contain one or more genes associated with virulence have been found in a number of species of bacteria (6). These pathogenicity islands are bordered by directly repeated sequences and can be deleted en bloc, and they may have integrase-like genes. In the 23.9-kb deleted region, neither a repeated sequence or nor an integrase-like gene was found; however, a 30-kb region located approximately 10 to 40 kb upstream of the deleted region was found to encode several transposase-like genes in the TIGR E. faecalis V583 genome sequence. The putative transposases may cause the very frequent deletion. This type of deletion mediated by an insertion element encoded in its flanking region has been reported in methicillin-resistant staphylococci (8). The reason why this deletion was frequently found in the clinical isolates of E. faecalis from urine even though the gelatinase is considered a virulence factor (2, 4, 5, 11, 13) is unclear at present; however, it is likely that the gelatinase is not essential for survival of E. faecalis in the urinary tract.
Acknowledgments
We thank Ritsuko Mitsuhata and Mizuki Ando for technical assistance. The sequence data and locus assignment used in this study were obtained from the database created by TIGR.
REFERENCES
- 1.Alander, M., R. Satokari, R. Korpela, M. Saxelin, T. Vilpponen-Salmela, T. Mattila-Sandholm, and A. von Wright. 1999. Persistence of colonization of human colonic mucosa by a probiotic strain, Lactobacillus rhamnosus GG, after oral consumption. Appl. Environ. Microbiol. 65:351-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coque, T. M., J. E. Patterson, J. M. Steckelberg, and B. E. Murray. 1995. Incidence of hemolysin, gelatinase, and aggregation substance among enterococci isolated from patients with endocarditis and other infections and from feces of hospitalized and community-based persons. J. Infect. Dis. 171:1223-1229. [DOI] [PubMed] [Google Scholar]
- 3.Eaton, T. J., and M. J. Gasson. 2001. Molecular screening of Enterococcus virulence determinants and potential for genetic exchange between food and medical isolates. Appl. Environ. Microbiol. 67:1628-1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garsin, D. A., C. D. Sifri, E. Mylonakis, X. Qin, K. V. Singh, B. E. Murray, S. B. Calderwood, and F. M. Ausubel. 2001. A simple model host for identifying gram-positive virulence factors. Proc. Natl. Acad. Sci. USA 98:10892-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gold, O. G., H. V. Jordan, and J. van Houte. 1975. The prevalence of enterococci in the human mouth and their pathogenicity in animal models. Arch. Oral Biol. 20:473-477. [DOI] [PubMed] [Google Scholar]
- 6.Hacker, J., G. Blum-Oehler, I. Muhldorfer, and H. Tschape. 1997. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol. Microbiol. 23:1089-1097. [DOI] [PubMed] [Google Scholar]
- 7.Kariyama, R., R. Mitsuhata, J. W. Chow, D. B. Clewell, and H. Kumon. 2000. Simple and reliable multiplex PCR assay for surveillance isolates of vancomycin-resistant enterococci. J. Clin. Microbiol. 38:3092-3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katayama, Y., T. Ito, and K. Hiramatsu. 2001. Genetic organization of the chromosome region surrounding mecA in clinical staphylococcal strains: role of IS431-mediated mecI deletion in expression of resistance in mecA-carrying, low-level methicillin-resistant Staphylococcus haemolyticus. Antimicrob. Agents Chemother. 45:1955-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, A. D. L. Akkermans, W. M. de Vos, and H. Nagasawa. 2001. Gelatinase biosynthesis-activating pheromone: a peptide lactone that mediates a quorum sensing in Enterococcus faecalis. Mol. Microbiol. 41:145-154. [DOI] [PubMed] [Google Scholar]
- 10.Nakayama, J., Y. Cao, T. Horii, S. Sakuda, and H. Nagasawa. 2001. Chemical synthesis and biological activity of the gelatinase biosynthesis-activating pheromone of Enterococcus faecalis and its analogs. Biosci. Biotechnol. Biochem. 65:2322-2325. [DOI] [PubMed] [Google Scholar]
- 11.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2000. Effects of Enterococcus faecalis fsr genes on production of gelatinase and a serine protease and virulence. Infect. Immun. 68:2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qin, X., K. V. Singh, G. M. Weinstock, and B. E. Murray. 2001. Characterization of fsr, a regulator controlling expression of gelatinase and serine protease in Enterococcus faecalis OG1RF. J. Bacteriol. 183:3372-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Singh, K. V., X. Qin, G. M. Weinstock, and B. E. Murray. 1998. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. J. Infect. Dis. 178:1416-1420. [DOI] [PubMed] [Google Scholar]
- 14.Su, Y. A., M. C. Sulavik, P. He, K. K. Makinen, P.-L. Makinen, S. Fiedler, R. Wirth, and D. B. Clewell. 1991. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect. Immun. 59:415-420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tenover, F. C., R. D. Arbeit, R. V. Goering, P. A. Mickelsen, B. E. Murray, D. H. Persing, and B. Swaminathan. 1995. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J. Clin. Microbiol. 33:2233-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]