Abstract
Six mercury-resistant environmental proteobacterial isolates and one genetically modified mercury-resistant Pseudomonas putida strain were analyzed for physiological traits of adaptive relevance in an environment of packed-bed bioreactors designed for the decontamination of mercury-polluted chlor-alkali wastewater. The strains displayed characteristic differences in each trait (i.e., biofilm formation capability, growth rate in mercury contaminated wastewaters, and mercury reduction efficiency). Subsequently, they were immobilized either as a monoculture or as a mixed culture on porous carrier material in packed-bed bioreactors through which different batches of filter-sterilized industrial chlor-alkali wastewater were pumped. In monospecies bioreactors, the mercury retention efficiency was sensitive to rapidly increasing mercury concentrations in the wastewater. Mixed culture biofilms displayed a high mercury retention efficiency that was not affected by rapid increases in mercury or continuously high mercury concentrations. The dynamic in the community composition of the mixed culture bioreactors was determined by ribosomal intergenic spacer polymorphism analysis. Mercury-mediated selective pressure decreased the number of prevalent strains. Microbial diversity was completely restored after easing of the selective pressure. Microbial diversity provides a reservoir of strains with complementary ecological niches that results in a superior bioreactor performance under changing environmental conditions.
Mercury cycles through the environment as a result of both natural and human activities. The human activities that are most responsible for mercury emissions are (i) the incineration of mercury-containing fuels and materials and (ii) industrial processes such as those utilized in the mercury cell chlor-alkali industry. Without appropriate retention devices, mercury is released into the environment in substantial amounts (6, 19, 31). Once mercury enters waters, either directly or through air deposition, inorganic mercury can be methylated abiotically or biotically to its most toxic form, methylmercury (1, 24). Methylmercury biomagnifies readily in the food chain, endangering ecosystems and public health. In the United States it is estimated that ca. 60,000 babies per year are born with neurological damage caused by mercury poisoning of their mothers upon consuming mercury-contaminated fish during pregnancy (29). In freshwater ecosystems, methylmercury bioaccumulation is more common than in salinic environments (4, 13). Hence, it is of great importance for environment and public health to stop mercury dumping into river ecosystems.
In previous experiments we demonstrated a new, cost-effective, and environmentally friendly end-of-pipe technology: the efficient retention of mercury from chemical wastewater by mercury-resistant bacteria in packed-bed bioreactors in laboratory and technical scale (37, 40). The basic principle of this process is the enzymatic reduction of ionic mercury Hg(II) to metallic mercury Hg(0) by mercury-resistant bacteria (16, 30, 33). The metallic mercury is retained in the matrix of the bioreactor in the form of mercury droplets (2, 39). The decontamination of mercury polluted wastewater can be done both in pure culture and in mixed communities in a bioreactor. In open systems, mixed cultures developed by invasion of ubiquitous mercury-resistant strains into the bioreactors (38, 39). Here, we excluded invasion completely by sterilizing the wastewater inflow. This allowed us to compare the performance of monospecies bioreactors of highly adapted and active strains with that of multispecies bioreactors consisting of the same strains. The seven investigated strains showed clear differences in adaptive physiological traits, e.g., biofilm formation capability, growth rate, and mercury volatilization rate. The community composition at the strain level and its response to increasing and decreasing selective pressure of mercury stress was analyzed by 16S-23S ribosomal DNA intergenic spacer polymorphism analysis (RISA). A changing selective pressure was imposed by using wastewater batches with different mercury concentrations. We sought to investigate the effect of microbial diversity on mercury reduction efficiency and the stability of mercury-reducing bioreactors.
MATERIALS AND METHODS
Wastewater.
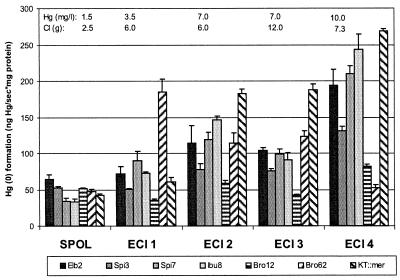
The bioreactors were run with process wastewater from two mercury cell chlor-alkali plants (Elektrochemie Ibbenbüren [ECI], Ibbenbüren, Germany, and SPOL Chemicals [SPOL], Usti nad Labem, Czech Republic), provided in batches of 800 liters (ECI) or 25 liters (SPOL) in polyethylene containers. The wastewater had a pH of 2.1 to 2.6 (ECI) or 13.0 (SPOL) and was neutralized by the addition of NaOH (5 M) or HCl (30% [wt/vol]), respectively. Oxygen saturation was obtained by bubbling with compressed air for 2 to 5 h. The wastewater batches differed with respect to mercury concentration (1.5 to 10.0 mg/liter) and chloride concentration (2.5 to 12.0 g/liter), as indicated in Fig. 2. The original batches of ECI 1 and ECI 3 were slightly diluted with deionized water to reach the mercury concentrations shown in Fig. 2. The chloride concentrations of these two batches were adjusted by adding NaCl.
FIG. 2.
Maximum Hg(0) formation rates in chlor-alkali wastewaters. Initial wastewater conditions are shown in the top lines. The columns in each data set are, from left to right: Elb2, Spi3, Spi7, Ibu8, Bro12, Bro62, and KT2442::mer-73.
Biofilm formation assay.
The ability to form biofilms was tested in glass test tubes (internal diameter, 15 mm; length, 18 cm; Schott Glaswerke, Mainz, Germany) according to the method of O'Toole et al. (23). Growth was started by mixing 0.5 ml of an overnight test tube culture with 4.5 ml of fresh medium (ECI 1 wastewater [pH 7.0], sucrose [1.0 g/liter], yeast extract [0.2 g/liter]). After 24 h of incubation at room temperature without agitation, 250 μl of 0.1% (wt/vol) safranin O (Sigma, St. Louis, Mo.) was added. Tubes were incubated for 10 min at room temperature and then rinsed thoroughly with water to remove unattached cells and residual dye. The dye was solubilized in 5 ml of ethanol (absolute) for 10 min, and 200 μl was transferred into 96-well polystyrene tissue culture test plates (Techno Plastic Products, Trasadingen, Switzerland). The absorbance of safranin O was measured at 490 nm in an Emax Precision microplate reader (Molecular Devices, Munich, Germany). Uninoculated medium was used to determine the background. Biofilm formation assays were done in triplicate.
Growth in batch culture.
The growth in batches of chlor-alkali wastewater was tested in 96-well polystyrene tissue culture test plates (Techno Plastic Products). The nutrient concentration of the bioreactor medium was too low to observe a detailed growth curve in multiwell plates (data not shown). Therefore, the growth experiments in batch cultures were performed with a 10-fold-higher nutrient concentration. Growth was started by mixing 20 μl of an overnight test tube culture with 180 μl of fresh medium (ECI 1, 2, 3, and 4; SPOL wastewater [pH 7.0]; sucrose [1.0 g/liter]; yeast extract [0.2 g/liter]). The optical density (650 nm) was determined in the Emax microplate reader (Molecular Devices). Uninoculated medium was used to determine the background. Growth measurements were done in triplicate.
Online measurement of mercury reduction.
Mercury reduction rates were analyzed by connecting three periodically aerated reaction vessels (containing the microbial culture and the mercury solution) to an atomic absorption spectrophotometer (AAS) (AAS 2100; Perkin-Elmer, Überlingen, Germany) (28). Hg(II) was transformed after injection of 600 μl of cell suspension (grown in ECI 1, 2, 3, or 4 or SPOL wastewater [pH 7.0]-sucrose [1.0 g/liter]-yeast extract [0.2 g/liter] until early stationary phase) into a reaction vessel (Nalgene polycarbonate, 50 ml) containing 5.4 ml of the wastewater (ECI 1, 2, 3, or 4 or SPOL; sucrose [0.1 g/liter]; yeast extract [0.02 g/liter]). Prior to injection, abiotically produced Hg(0) was removed in a maximum of five aeration cycles. Three assays were run in parallel in cycles of 3 min. Within one cycle, each vessel was aerated for 30 s, and the produced Hg(0) was blown into the AAS, where the amount of Hg(0) was detected. In the next 30 s, the tubes and the AAS were aerated with Hg-free air to remove excess Hg(0). The assays were mixed by a magnetic stirrer at 300 rpm. In every cycle the assays were incubated without aeration for 2.5 min, and then the produced Hg(0) was blown by compressed air into the AAS within 0.5 min. The amount of volatilized Hg(0) was highest in the first or second cycle. After the fourth cycle, the reaction was stopped. The amount of microbially produced Hg(0) was calculated for each cycle on the basis of a calibration curve obtained by the chemical reduction of HgNO3 standard solutions by SnCl2.
Determination of protein content.
After the stop of the Hg(II) transformation reaction, 2 ml of the assay was stored at −20°C for protein analysis according to the method of Lowry et al. (20). Samples were centrifuged for 10 min (10,000 × g), and cells were resuspended in 500 μl of NaOH (0.5 M) and lysed for 1 h.
Bioreactor setup and operation.
For each type of biofilm, three packed-bed bioreactors were operated in parallel. The bioreactors had an internal diameter of 60 mm and a height of 120 mm (W. O. Schmidt, Braunschweig, Germany). A refined steel grid located 50 mm above the bottom ensured an even inflow distribution. The columns were filled with 160 cm3 of pumice chips (diameter, 4 to 6 mm; Raab GmbH, Luckenau, Germany). Columns and tubes were sterilized by autoclaving (121°C, 20 min). Neutralized, oxygen-saturated chlor-alkali wastewater was pumped into the bioreactors in upflow mode by using a peristaltic pump. Sterile medium (sucrose [2.0 g/liter], yeast extract [0.4 g/liter], and NaCl [2.5, 6.0, 7.3, or 12.0 g of chloride/liter in accordance with the respective wastewater batch]) was added with a second peristaltic pump. Before entering the bioreactors, the nutrient amended wastewater passed through a filter (pore size, 0.2 μm) to prevent contamination. The total volume entering the column was 160 ml/h, consisting of nutrient medium (8 ml/h) and wastewater (152 ml/h), resulting in a final concentration of 0.1 g of sucrose and 0.02 g of yeast extract/liter. For inoculation, a second inlet between filter and bioreactor inflow was used. The monospecies bioreactors of Pseudomonas putida KT2442::mer-73 and Pseudomonas aeruginosa Bro12 and the multispecies bioreactors were started simultaneously. At day 10, in the P. putida KT2442::mer-73 bioreactors the nutrient concentration was increased to a final concentration of 20 g of sucrose and yeast extract/liter, respectively, for 24 h. They were stopped at day 21, cleaned, autoclaved, and restarted with Citrobacter freundii Bro62. Since the amount of wastewater was limited, the Bro62 monospecies bioreactors had slightly shorter periods for each wastewater batch. Between days 53 and 74, the bioreactors were run with ECI wastewater with Hg at 10.0 mg/liter (days 53 to 64), 7.0 mg/liter (days 64 to 69), and 8.9 mg/liter (days 69 to 75). From day 75 on, the bioreactors were run with SPOL wastewaters at 1.5 to 2.6 mg of Hg/liter (average, 2.0 mg/liter).
Strains.
The strains P. putida Spi3, P. putida Elb2, and Sphingomonas sp. strain Spi7 are environmental isolates from river sediments (38). The strains Pseudomonas stutzeri Ibu8 and C. freundii Bro62 were isolated from laboratory bioreactor effluents, and the strain P. aeruginosa Bro12 was isolated from the technical scale bioreactor effluent. The genetically engineered strain P. putida KT2442::mer-73 has a high and constitutive expression of the mer genes and was shown to have the highest specific activity of MerA compared to other genetically engineered microorganisms (GEMs) and a natural isolate (2, 17).
Inoculation.
For cultivation of the inoculum, one colony of each strain was picked from a plate [sucrose, 5 g/liter; yeast extract, 5 g/liter; Hg(II), 1 mg/liter] and suspended in 25 ml of growth medium (ECI 1 wastewater [pH 7.0], 0.2 M Na2HPO4/NaH2PO4 [pH 7.0], sucrose and yeast extract [15 g of each/liter]) and grown at 20°C on a rotary shaker for 1 day. The preculture was added to 600 ml (KT2442::mer-73 and Bro12) or 200 ml (Spi3, Spi7, Elb2, Bro62, and Ibu8) of fresh growth medium and grown for another day. Thereafter, the cell density of each culture was determined microscopically. Each bioreactor was inoculated with 200 ml of culture (and, if necessary, filled up with fresh growth medium) containing 1.5 × 1012 cells. For the inoculum of the community bioreactors, cultures of equal cell numbers were combined. Monospecies and multispecies cultures were divided into three parts and pumped through the bioreactors at 160 ml/h. After inoculation, wastewater was pumped through the bioreactors. During the first 16 h the nutrient supply was increased to improve growth of the biofilm on the carrier material (sucrose and yeast extract, 1.0 g of each/liter).
Determination of mercury and chloride concentrations.
The amount of total mercury was determined by flameless cold vapor atomic absorption spectroscopy with a flow injection system (FIAS 200 [FIAS]; Perkin-Elmer) which was connected to an AAS. Daily bioreactor effluent samples of ca. 20 ml were stabilized with 400 μl of HNO3 (65%). Subsamples of 1 ml were oxidized with 150 μl of KMNO4 (5%), 10 μl of H2SO4 (96%), and 10 μl of HNO3 (65%). After 2 min, 100 μl of K2S2O8 (potassium peroxodisulfate, 4%) was added, followed by mixing for further oxidation. After 10 min, 230 μl of H3NOHCl (hydroxylammoniumchloride, 10%) was added, followed by mixing until decolorization of the sample occurred. By addition of deionized water the sample was diluted to Hg concentrations of <100 μg/liter (maximum value of calibration). Subsequently, the sample was injected into the FIAS, and ionic mercury was reduced by the addition of NaBH4 (4 g/liter) to metallic mercury, which was volatilized by the carrier gas argon and detected at 253.7 nm by the AAS.
Chloride concentrations were determined photometrically with Dr. Lange kits (LCK 311; Dr. Lange, Düsseldorf, Germany).
Determination of culturable cell number in bioreactor effluent.
Densities of culturable cells were determined in the bioreactor effluent by counting the CFU on complex, Hg-free medium. A representative sample (50 ml) was collected within ca. 20 min. The sample was mixed well, and an aliquot (100 μl) was diluted serially in a sterile salt solution (NaCl, 15 g/liter). Then, 50 μl of the expected appropriate dilution (minimum of 10 and maximum of 100 CFU per plate) were plated in triplicate on agar plates (NaCl [15 g/liter], sucrose [4 g/liter], yeast extract [2 g/liter]) immediately after sampling. Plates were incubated at room temperature for 3 days before counting.
Community analysis. (i) DNA extraction.
Approximately 50 ml of bioreactor effluent was sampled and centrifuged (30 min, 5,000 × g). Samples were taken at least twice, at the beginning and at the end of each wastewater batch period. The pellet was resuspended in 100 μl of Tris-EDTA buffer. Total cell DNA was extracted by using guanidium thiocyanate as described by Pitcher et al. (25).
(ii) Amplification primers for the 16S-23S ISR.
The primers G1 and L1 were described by Jensen et al. (18). The primer G1 (5′-GAAGTCGTAACAAGG-3′) binds to a highly conserved region of the 16S rRNA gene adjacent to the 16S-23S interspacer region (ISR). The primer L1 (5′-CAAGGCATCCACCGT-3′) binds to a conserved region of the 23S rRNA gene close to the 16S-23S spacer. The primer R4c (5′-CTCATTTCTGAGCTTTCAGCG-3′) was selected from an alignment of five 16S-23S ISR sequences (unpublished data) of P. stutzeri Ibu8 and two different P. fulva and two different P. fluorescens strains. It binds specifically within the 16S-23S ISR of P. stutzeri Ibu8, 300 nucleotides upstream of the 16S gene.
(iii) PCR conditions.
The amplification assay (20 μl) contained 250 μM concentrations of each deoxynucleoside triphosphate (MBI Fermentas, Vilnius, Lithuania); 10% (vol/vol) dimethyl sulfoxide (Fluka, p.A.); 1× PCR buffer (Qiagen, Hilden, Germany); 1 U of Taq DNA polymerase (Qiagen); 0.5 μM concentrations of the primers G1, L1, and R4c (Gibco-BRL, Karlsruhe, Germany); and 0.5 μl of template DNA of various concentrations. To prevent evaporation, the mixture was covered with a drop of Nujol mineral oil (Perkin-Elmer). The PCR was performed in a thermal cycler (Landgraf; TC Varius V) with a temperature program consisting of a first DNA denaturation step of 120 s at 94°C; followed by 35 cycles of 30 s at 94°C, 60 s at 45°C, and 120 s at 72°C; a final elongation step of 600 s at 72°C; and subsequent cooling at 4°C. The ramping rate between the different temperatures was set to the fastest available.
(iv) Electrophoresis.
After the success of DNA amplification had been monitored in an agarose gel, the PCR products were separated in 4 to 20% TBE (Tris base, boric acid, EDTA) acrylamide gradient minigels (Novex, Frankfurt/Mainz, Germany) as described by Hirsch and Sigmund (15). Portions (3 μl) of the PCR products were mixed with 0.5 μl of TBE sample buffer (Novex) and loaded into the gel. For strain discrimination within the community, a community standard consisting of the PCR products of the six different strains (amplified separately and then mixed in equal amounts) was used. The template DNA concentration of each separately amplified strain was 10 μg/ml. The DNA was separated by electrophoresis at 200 V for 100 min in 1× TBE buffer (Novex). The gel was stained for 10 min in ethidium bromide (5 mg/liter) and then destained for 5 min in 1× TBE buffer. Bands were visualized with UV light (254 nm; UV Transilluminator 200/2.0; UVP Inc., San Gabriel, Calif.) and digitalized with the Enhanced Analysis System (EASY; Herolab, Wiesloch, Germany).
(v) Analysis of the community fingerprint.
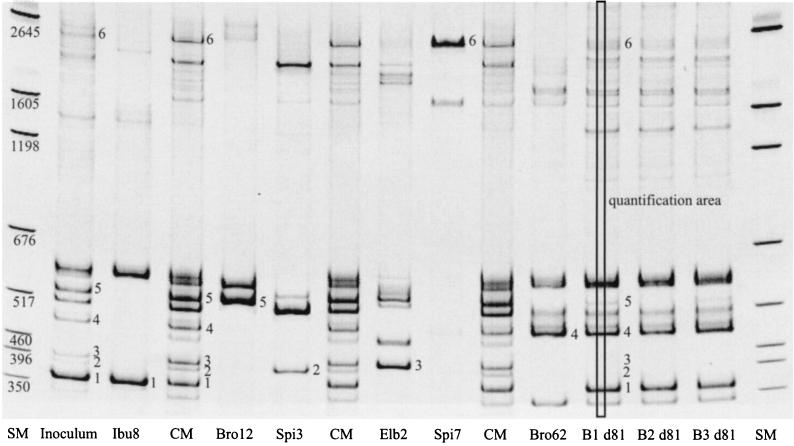
The light intensity (i.e., the number of pixels) of the quantification area (see Fig. 5) of each diagnostic band was calculated with Scion Image (version Beta3b; Scion Corp., Frederick, Md.). For the determination of the community composition, only one diagnostic band per strain was used. The relative intensities of the diagnostic bands of different gels were normalized in relation to the intensities of the size markers of the different gels.
FIG. 5.
ISR fingerprints of the inoculum strains, the inoculum, and the final bioreactor effluent samples. The community marker (CM) bands consists of a mixture of the ISR-PCR products of the six inoculum strains (separately amplified). The numbers at the fingerprint bands indicate the diagnostic bands and their position in the community marker. The numbers at the size marker (SM) bands indicate their sizes in base pairs. B1 d81, bioreactor 1 (day 81); B2 d81, bioreactor 2 (day 81); B3 d81, bioreactor 3 (day 81).
RESULTS
Biofilm formation capability.
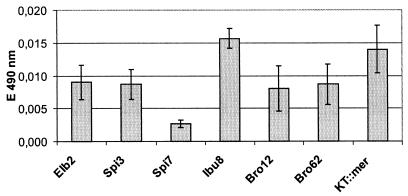
The ability of each strain to form biofilms was tested in the first wastewater batch of the bioreactor run, ECI 1. The relative amounts of biomass bound in biofilms on glass testtube walls were deduced from the amount of retained safranin O. The absorption of the dye is shown in Fig. 1. All tested strains were able to form biofilms within 24 h. The biofilm formation capacity was similar for strains Elb2, Spi3, Bro12, and Bro62. Ibu8 and KT::mer clearly produced a higher biofilm mass, and Spi7 produced a distinctly smaller biofilm mass.
FIG. 1.
Biofilm formation in ECI 1 wastewater after 24 h in glass test tubes by the multispecies and pure culture bioreactor strains.
Mercury reduction rates in five wastewater batches.
All strains were able to transform soluble Hg(II) from the chlor-alkali wastewaters to volatile Hg(0), as shown in Fig. 2. The transformation rates of P. putida strains Elb2, Spi3, and KT2442::mer-73, P. stutzeri strain Ibu8, and Sphingomonas sp. strain Spi7 showed a similar trend of increasing transformation rates at increasing Hg(II) concentrations. Only P. aeruginosa Bro12 showed no general trend, with a low transformation rate throughout. In contrast to all other strains, C. freundii Bro62's transformation rate was highest at ECI 1 and decreased with increasing Hg(II) concentrations.
No strain had superior reduction rates in all wastewater batches. In SPOL wastewater, all strains had similar low reduction rates. In ECI 1 wastewater, Bro62 had the highest reduction rates. In ECI batches 2, 3, and 4, the GEM ranked first. ECI 2 and ECI 3 had the same mercury level but different chloride concentrations. No clear effect of the higher chloride concentration in ECI 3 on the mercury reduction rate was observed.
Growth in wastewater batch cultures.
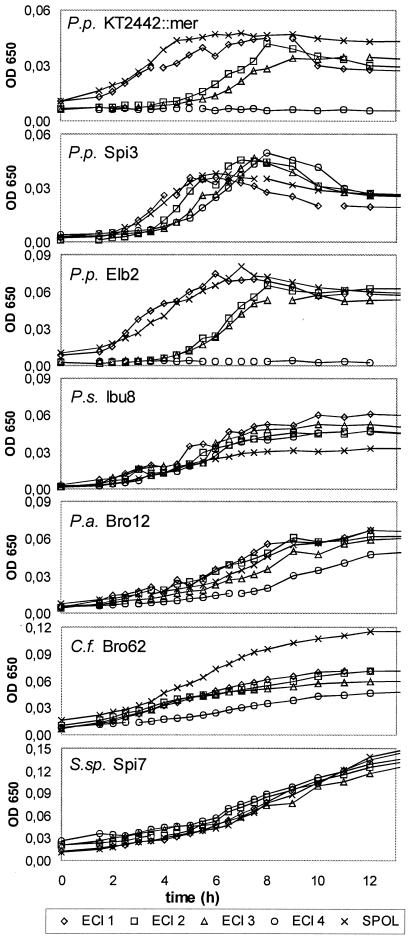
Fig. 3 shows the growth of the seven investigated strains in batch cultures of the five different wastewaters. All strains were able to grow in SPOL and ECI 1, 2, and 3 wastewater. In ECI 4 wastewater at 10 mg of Hg(II)/liter, two P. putida strains (KT2442::mer-73 and Elb2) did not grow within 24 h. The strains differed in lag time, duration of growth phase, and transition of growth to stationary phase. The P. putida strains showed a distinct lag phase at high Hg(II) concentrations, a short growth phase, and an early death phase. All other strains displayed continuous growth without a clear lag, growth, stationary, or death phase. The α-proteobacterium Sphingomonas sp. strain Spi7 differed from the other six γ-proteobacteria by its long growth phase, which lasted at least 35 h (data not shown), and its high maximum optical density, which was two- to threefold higher than that of the next best strain, Bro62. However, since the Spi7 cells were much larger than those of the other six strains, its cell density was smaller (data not shown). Upon comparison with the growth seen in ECI 2 and ECI 3 wastewater, we observed no significant effect of the doubled chloride concentration, but only a slightly delayed growth in a few ECI 3 cultures. While most strains grew best in the low-mercury wastewaters SPOL and ECI 1 and showed a longer lag phase or slower growth rate at increasing mercury concentrations, strains Ibu8 and Spi7 showed no effect of the mercury concentration on their growth curves, i.e., they grew similarly in all wastewater batches.
FIG. 3.
Growth of chlor-alkali wastewater batch cultures. The data are mean values of three parallel experiments.
Mercury retention efficiency.
The mercury retention efficiency of the bioreactors is shown in Fig. 4. The number of culturable cells in the effluent was determined as an indication of the growth activity of the biofilms and is also shown in Fig. 4. For inoculation of the monospecies bioreactors, P. aeruginosa Bro12 was chosen because it dominated the biofilm community in a technical scale bioreactor operated on site at a factory for 8 months. C. freundii Bro62 was chosen because it was the dominant strain in laboratory scale bioreactors operated for 15 months. P. putida KT2442::mer-73 was chosen because of its genetically modified constitutive high Hg(II) resistance.
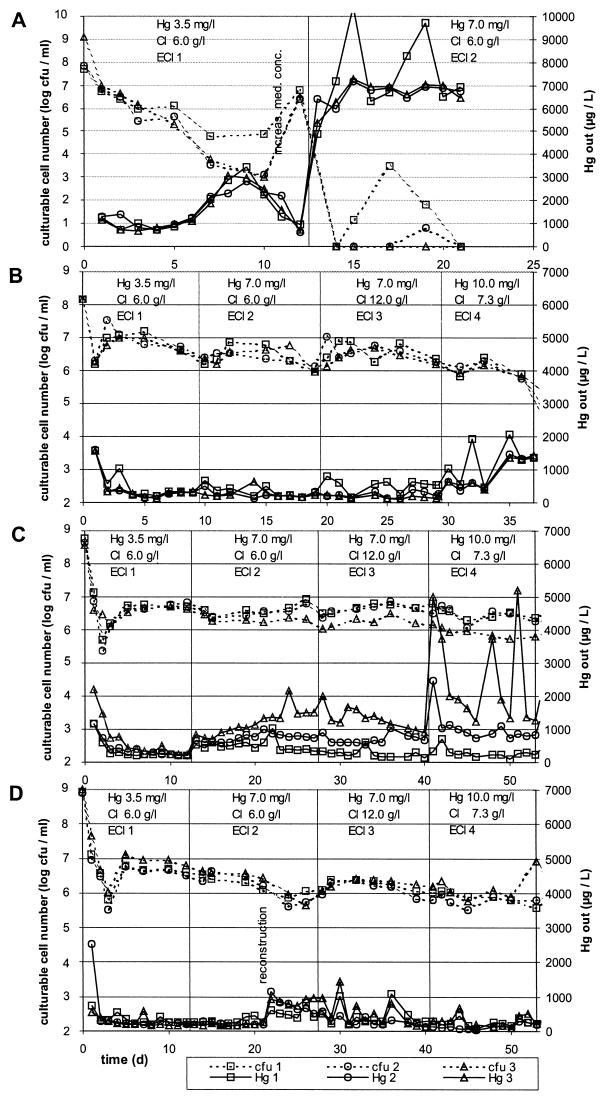
FIG. 4.
Culturable cell numbers and mercury concentration in the bioreactor effluent. Solid lines, total mercury in bioreactor effluent; dotted lines, CFU in bioreactor effluent. Squares, bioreactor 1; circles, bioreactor 2; triangles, bioreactor 3. (A) P. putida KT2442::mer-73 monospecies bioreactors; (B) C. freundii Bro62 monospecies bioreactors; (C) P. aeruginosa Bro12 monospecies bioreactors; (D) multispecies bioreactors.
Monospecies mercury-reducing biofilms.
The three parallel bioreactors inoculated with P. putida KT2442::mer-73 showed a permanent decrease in effluent cell numbers in the first 10 days (Fig. 4A). This correlated with an increase in the effluent Hg concentration up to a near breakthrough of mercury. Both trends were reversed at day 10 by increasing the nutrient concentration in the feeding solution. However, after the switch to ECI 2 wastewater with a higher mercury concentration and a regular nutrient concentration, the number of culturable cells in the effluent dropped to zero within a few days. Correspondingly, no mercury was retained. Thus, P. putida KT2442::mer-73 was not able to establish an active biofilm under these conditions.
The three parallel bioreactors inoculated with C. freundii Bro62 had a high and stable Hg(II) retention efficiency and low Hg effluent concentrations with the wastewater batches ECI 1, 2, and 3, as shown in Fig. 4B. Minimum Hg effluent concentrations were 110 μg/liter at a 7.0-mg/liter inflow concentration, which corresponded to a 98% Hg retention. The effluent's culturable cell numbers were also stable. After the switch to ECI 4 wastewater, Hg effluent concentration and effluent cell densities remained stable for a few more days. The mercury retention efficiency and effluent cell density then decreased strongly.
The three parallel bioreactors inoculated with P. aeruginosa Bro12 drifted apart during the course of the experiment with respect to performance and effluent cell densities, as shown in Fig. 4C. While bioreactor 1 reached mercury effluent concentrations of 100 μg/liter when operated with wastewater at 10.0 mg/liter, bioreactor 3 reached only a level of ca. 1,200 μg/liter in the effluent when operated with the same wastewater. These differences in performance were probably caused by differences in the biofilm thickness, which were visible to the eye on the glass walls of the bioreactors between the inlet and the packed bed. The mercury retention efficiency of the Bro12 bioreactors was impaired by increasing mercury concentrations, as demonstrated by the increased mercury effluent concentrations at the inflow mercury shifts of from 3.5 to 7 mg/liter (day 12) and 7.0 to 10.0 mg/liter (day 40), although the bioreactors recovered spontaneously within a few days.
Multispecies mercury-reducing biofilms.
The multispecies bioreactors had extremely low mercury effluent concentrations as low as 40 μg/liter at 10 mg of Hg/liter in the inflow, as shown in Fig. 4D, corresponding to more than 99% Hg retention. Only due to the reconstruction of adjacent bioreactors, combined with mechanical perturbations and an 8-h stop of nutrient amendment to the wastewater, was the performance of the multispecies bioreactors decreased temporarily, but it recovered spontaneously within several days. Apart from this artifact, the community bioreactors had a very high and stable Hg retention efficiency in all wastewater batches and were not impaired by changes in the mercury inflow concentrations.
Composition of mercury-reducing biofilm communities and selection pressure.
Figure 5 shows the clearly discriminable “fingerprints” of the six inoculum strains and their identification in the inoculum community fingerprint and in the bioreactor effluent fingerprints from day 81. The gel image shows that all inoculum strains were still present in all bioreactors at the end of the run (the bands from Elb2 and Spi3 were visible in bright close-ups) and that Ibu8, Bro12, and Bro62 were the prevalent strains.
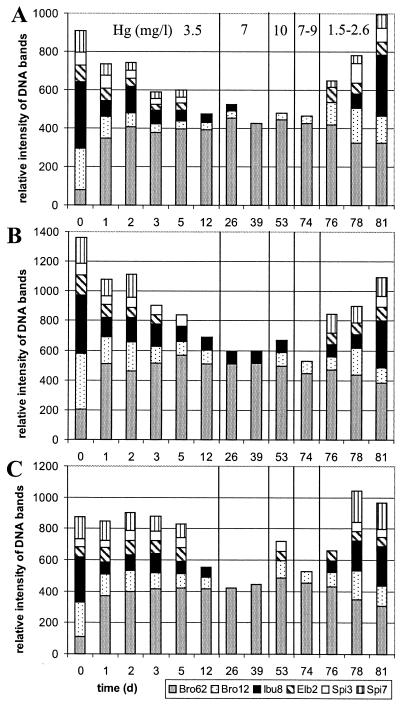
Figure 6 shows the time course of relative strain abundance in the effluents of the three bioreactors for a period of 81 days. The dynamics were similar in all three bioreactor communities. The bioreactors started with a moderate selection pressure of 3.5 mg of Hg(II)/liter. As a result, at the end of the first wastewater batch the concentrations of Sphingomonas sp. strain Spi7 and P. putida strains Spi3 and Elb2 had dropped below the detection limit by the fingerprint method. An increase in inflow mercury concentration to 7 mg of Hg(II)/liter after day 12 resulted in a further decrease of diversity, because the strains P. stutzeri Ibu8 and P. aeruginosa Bro12 became undetectable in bioreactor 3 and in bioreactors 2 and 3, respectively. Bro62 was by far the dominant strain during remediation of highly concentrated wastewater. Interestingly, a further increase of the selection pressure to 10 mg of Hg(II)/liter at day 40 resulted in an increase of diversity, since a few strains became detectable again. By reducing the selection pressure to 2.0 mg of Hg(II)/liter after day 74, the initial diversity was completely restored. The community fingerprints of the inoculum and of the effluent after 81 days of operation were similar, except for Bro62, which had a higher abundance at the end of the experiment. Strains Elb2, Spi3, and Spi7 were undetectable for ca. 70 days. However, their reappearance at day 81 showed that these strains did not die out but only dropped below the limit of detection under unfavorable conditions. According to agarose gel images, the fingerprints from days 7, 14, 28, 42, and 67 were identical to the fingerprints from days 5, 12, 26, 39, and 74, respectively, and therefore not used for polyacrylamide gel electrophoresis fingerprinting. The relative abundances of the strains as shown by RISA were consistent with plate counts of the respective colony morphotypes, which were performed randomly (data not shown).
FIG. 6.
Time course analyses of relative strain abundances in the multispecies bioreactor effluents. The presence of the inoculum strains in the effluxes of bioreactors 1 (A), 2 (B), and 3 (C) was based on the intensity of the diagnostic band of each strain within the community fingerprint relative to the size marker intensity. The mercury concentration of the wastewater is indicated in panel A.
DISCUSSION
Mercury resistance: mercury volatilization and growth.
Growth in wastewaters containing ionic mercury depends on the Hg(II) bioavailability and toxicity. Nutrients containing sulfhydryl groups (e.g., yeast extract), as well as negatively charged ions (e.g., chloride), bind to ionic mercury and thereby alter its bioavailability and toxicity (8, 9). Therefore, the same wastewater batches were used for mercury volatilization, for growth in batch culture, and in the bioreactor runs. The ability to grow in the presence of mercury (a process requiring several hours) has been shown here to be a more important feature of mercury-resistant cells than short-term volatilization rates (a reaction of several minutes length) for the application in a long-term bioremediation process. High concentrations of ionic mercury can kill even mercury-resistant bacteria after a few hours (3). Accordingly, high Hg(0) volatilization rates, as observed during the first 12 min of Hg(II) exposure, were no guarantee for growth, as has been shown for P. putida Elb2 and the GEM at 10 mg of Hg(II) per liter.
Settling in the bioreactors: growth strategy and biofilm formation capability.
The patterns of growth curves represented different growth strategies. The P. putida strains had long lag phases, which were presumably used for the removal of inhibitory mercury (3), short growth phases, and early starts of the death phase. Community analysis of the bioreactor effluent revealed that the P. putida growth strategy was not successful for establishing growth in high abundance at medium and high mercury concentrations. However, the opposite growth strategy of Sphingomonas sp. Spi7—no lag phase and a long growth phase—was also not successful. The growth phases of Ibu8, Bro12, and Bro62 ranged between those of the P. putida strains and strain Spi7, and no lag phase even at high mercury concentrations was detected. This intermediate strategy may be the reason why Ibu8, Bro12, and Bro62 had the highest abundance in the community fingerprints. However, an explanation of the community composition on this basis is not possible, since there were no data to suggest that Bro62 would dominate over Bro12 and Ibu8. The presence of all strains at the end of the run confirmed, however, that all strains' capabilities were sufficient for a long-term existence in the bioreactors at various relative abundances.
Growth strategy and growth rate in batch cultures of planktonic cells are important, but they are not crucial for high abundance in biofilms. Biofilms consist of nonmotile, sessile bacteria and a large fraction of planktonic cells, as demonstrated by Tolker-Nielsen et al. for Pseudomonas biofilms (36). All strains tested here had the general ability to form biofilms in glass test tubes as monocultures, although to a different extent. However, conditions may be different in multispecies biofilms. Stewart et al. demonstrated that a Klebsiella pneumoniae strain, although growing at twice the rate of a P. aeruginosa strain under planktonic conditions, did not outcompete the slower-growing strain in a biofilm (32). On the contrary, P. aeruginosa cells formed the main parts of the biofilm, and K. pneumoniae cells persisted in the biofilm only by virtue of a higher growth rate and an ability to attach to the P. aeruginosa biofilm.
RISA.
The variety in numbers, lengths, and sequences of the ISRs, along with a flanking by the highly conserved 16S and 23S rRNA genes, makes the ISR a sensitive marker for the discrimination of different bacterial species and strains (14, 18). The patterns produced by RISA have been shown to be highly reproducible and sufficient to discriminate both closely and distantly related species and subspecies (11). For community fingerprinting, the diversity which can be described is limited by the amount of length variation of the spacer region (12), i.e., the presence of unique, diagnostic bands. It was previously shown that the tested strains provide a unique fingerprint and were distinguishable in community fingerprints (38). However, RISA allows only a semiquantitative analysis of the relative proportions of dominant strains. An absolute quantification is impossible due to the bias inherent in any PCR amplification approach, such as preferential amplification of particular sequences (34) and multicopy target sequences. Bacteria may possess more than one rrn operon with identical or multiple spacer lengths (14). In the case of identical operons, this results in comparably strong bands, and in the case of diverse operons, RISA profiles exhibit several bands of the same strain.
Whiteley et al. reported that the relative concentration of each strain in a biofilm was not identical to its relative abundance in the liquid phase of planktonic cells and biofilm fragments (41), but in this case the community was in principle stable (no invader, no fader) and thus comparison of the effluent banding patterns allowed a conclusion about changes in the relative strain abundances. By analyzing the effluent community, we avoided intrusive sampling within the bioreactor.
Osborn et al. demonstrated the effect of decreasing DNA concentrations in the PCR on terminal restriction fragment length polymorphism profiles. Diluting the initial template concentration resulted in a decline in both the number of peaks and the height of each peak present in a profile (22). In our study, the amount of template DNA in each sample was basically the same, with only minor variations. Only in the samples of the inoculum and of day 1 was the DNA concentration much higher, due to the high cell numbers in the bioreactor effluent.
Monospecies versus multispecies mercury-reducing biofilms.
It is an old question whether highly specialized pure cultures are more effective in food utilization or pollutant degradation than mixed cultures of diverse species. In plant ecosystems, greater diversity leads to greater productivity in plant communities, greater nutrient retention, and greater ecosystem stability (35). In a glucose-limited chemostat incubated with up to 20 bacterial strains, the biofilm population size increased with increasing species diversity (41). However, what happens if species of high productivity but low competitiveness (i.e., low capability to settle, grow, and maintain high abundance in competition with other community members) are combined with superior competitors of lower productivity? In the present study, the productivity (in terms of mercury reduction) of highly specialized pure cultures was compared with a multispecies community's performance. Although the monoculture strains were selected due to their superior traits (the GEM P. putida KT2442::mer-73 had a high and constitutive expression of the mercury resistance genes) or dominance in previous bioreactor runs (C. freundii Bro62 and P. aeruginosa Bro12), no monospecies biofilm matched the highly efficient and stable performance of the multispecies biofilms. The GEM was clearly not adapted to these wastewater conditions and was gradually washed out, resulting in a breakthrough of the mercury load. Since only increased feeding stopped the washout, we assume that the growth rate at a regular nutrient concentration was too low to replace washed-out cells. In previous runs with nonsterile wastewater, P. putida KT2442::mer-73 was shown to disappear quickly and to be replaced by ubiquitous mercury-reducing bacteria (10). The performance of the Bro12 bioreactors drifted apart during operation, and they were sensitive to shifts in the mercury concentration. We assume that the diverse performances of the three bioreactors were caused by incidental differences in biofilm formation at the bioreactor inflow. Although the multispecies biofilms were dominated by Bro62, its monospecies biofilms were less effective. The decrease in mercury retention of Bro62 monocultures at 10 mg of Hg/liter may have been caused by a slower growth, resulting in a decreased number of active cells. The better and more stable performance of the multispecies bioreactors clearly showed that the presence of the other community members, although of very low abundance, improved biofilm activity. Because all strains had higher mercury reduction rates than Bro62 in ECI4 wastewater, their higher activity protected Bro62 cells from mercury concentrations toxic for this strain and thereby maintained the activity of the biofilm as a whole. Thus, the “species pool” increases the chance that the species that are better adapted to handle particular conditions are present, which is particularly important under changing conditions. This is consistent with the observations of Erb et al. and Whiteley et al. that even very low concentrations of well-adapted and resistant bacteria can protect less-resistant strains and thus maintain community activity (7, 41).
Microbial diversity provides a reservoir of strains with different physiological traits that are selectively promoted under changing environmental conditions, resulting in a surplus benefit above the performance and stability of the best-adapted strain alone. As a consequence, it is favorable to operate mercury-reducing bioreactors in an open system. The chance for additional mercury-resistant strains to enter the bioreactor will increase the pool of active strains.
Microbial diversity and selective pressure.
Most studies report a decrease in diversity as result of a pollutant's selection pressure (21, 26, 27, 41). Eventually, a few tolerant species increase in cell number. Little is known about the ease of selection pressure and reconstitution of the former diversity. Eichner et al. reported that activated sludge microbial communities lacking bioprotective community members decreased drastically in diversity after spiking with pollutants. But microbial diversity was partially or almost completely restored within 7 to 18 days after degradation of the pollutant. However, the community composition shifted, and a different community, although of similar diversity, evolved (5). In the present study it was demonstrated that a bacterial community composition decreased substantially in diversity as the selection pressure of ionic mercury increased, but the initial diversity was completely restored after the selection pressure was decreased.
Stewart et al. observed that a binary biofilm consisted of K. pneumoniae microcolonies residing on top of, or intermixing with, a base film of well-attaching P. aeruginosa cells (32). Such a structural differentiation and direct interaction of the cells results in nutrient and toxicant concentration gradients, i.e., different ecological niches and selection pressure. The abundance of a biofilm member is therefore not only governed by growth and death rates (and attachment and detachment rates) but also by the interaction with other community members and the mass transport limitations of nutrients, as well as toxicants in its spatial and ecological niche.
Acknowledgments
Wanda Fehr is gratefully acknowledged for her calibration of the Hg(II) transformation assay. We are very obliged to Dieter Sass, Hans Hardemann, Karen Bartel, and Werner Janot of ECI and to Jaroslav Novotny, Karel Cipra, and Ladislav Dycka of SPOL for their kind supply of wastewater.
This work was supported by European Union grants LIFE LIFE97-ENV/D/000463 and BIOTECH BIO4-CT-98-0168.
REFERENCES
- 1.Akagi, H., Y. Fujita, and E. Takabatake. 1977. Methylmercury: photochemical transformation of mercuric sulfide into methylmercury in aqueous solution. Photochem. Photobiol. 26:363-370. [Google Scholar]
- 2.Brunke, M., W.-D. Deckwer, A. Frischmuth, J. M. Horn, H. Lünsdorf, M. Rhode, M. Rohricht, K. N. Timmis, and P. Weppen. 1993. Microbial retention of mercury from waste streams in a laboratory column containing merA gene bacteria. FEMS Microbiol. Rev. 11:145-152. [DOI] [PubMed] [Google Scholar]
- 3.Chang, J. S., and J. Hong. 1995. Estimation of kinetics of mercury detoxification from low-inoculum batch cultures of Pseudomonas aeruginosa PU21 (Rip64). J. Biotechnol. 42:85-90. [DOI] [PubMed] [Google Scholar]
- 4.Compeau, G. C., and R. Bartha. 1987. Effect of salinity on mercury-methylating activity of sulfate-reducing bacteria in estuarine sediments. Appl. Environ. Microbiol. 53:261-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eichner, C. A., R. W. Erb, K. N. Timmis, and I. Wagner-Döbler. 1999. Thermal gradient gel electrophoresis analysis of bioprotection from pollutant shocks in the activated sludge microbial community. Appl. Environ. Microbiol. 65:102-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Environmental Protection Agency. 1997. Mercury study report to Congress (volume I) EPA-452/R-97-003. Office of Air Quality Planning and Standards and Office of Research and Development, Environmental Protection Agency, Washington, D.C.
- 7.Erb, R. W., C. A. Eichner, I. Wagner-Döbler, and K. N. Timmis. 1997. Bioprotection of microbial communities from toxic phenol mixtures by a genetically designed pseudomonad. Nat. Biotechnol. 15:378-382. [DOI] [PubMed] [Google Scholar]
- 8.Farrell, R. E., J. J. Germida, and P. M. Huang. 1990. Biotoxicity of mercury as influenced by mercury(II) speciation. Appl. Environ. Microbiol. 56:3006-3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Farrell, R. E., J. J. Germida, and P. M. Huang. 1993. Effects of chemical speciation in growth media on the toxicity of mercury(II). Appl. Environ. Microbiol. 59:1507-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Felske, A., B. V. Pauling, H. F. von Canstein, Y. Li, J. Lauber, J. Buer, and I. Wagner-Döbler. 2001. Detection of small sequence differences using competitive PCR: molecular monitoring of genetically improved, mercury-reducing bacteria. BioTechniques 30:142-148. [DOI] [PubMed] [Google Scholar]
- 11.Fisher, M. M., and E. W. Triplett. 1999. Automated approach for ribosomal intergenic spacer analysis of microbial diversity and its application to freshwater bacterial communities. Appl. Environ. Microbiol. 65:4630-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garcia-Martinez, J., S. G. Acinas, A. I. Anton, and F. Rodriguez-Valera. 1999. Use of the 16S-23S ribosomal genes spacer region in studies of prokaryotic diversity. J. Microbiol. Methods 36:55-64. [DOI] [PubMed] [Google Scholar]
- 13.Gilmour, C. C., E. A. Henry, and R. Mitchell. 1992. Sulfate stimulation of mercury methylation in freshwater sediments. Environ. Sci. Technol. 26:2281-2287. [Google Scholar]
- 14.Gurtler, V., and V. A. Stanisich. 1996. New approaches to typing and identification of bacteria using the 16S-23S rDNA spacer region. Microbiology 142:3-16. [DOI] [PubMed] [Google Scholar]
- 15.Hirsch, C. F., and J. M. Sigmund. 1995. Use of polymerase chain reaction (PCR) fingerprinting to differentiate bacteria for microbial products screening. J. Ind. Microbiol. 15:85-93. [DOI] [PubMed] [Google Scholar]
- 16.Hobman, J. L., and N. L. Brown. 1997. Bacterial mercury-resistance genes, p. 503-568. In A. Sigel and H. Sigel (ed.), Metal ions in biological systems: mercury and its effects on environment and biology, vol. 34. Marcel Dekker, Inc., New York, N.Y. [PubMed] [Google Scholar]
- 17.Horn, J. M., M. Brunke, W.-D. Deckwer, and K. N. Timmis. 1993. Pseudomonas putida strains which constitutively overexpress mercury resistance for biodetoxification of organomercurial pollutants. Appl. Environ. Microbiol. 60:357-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen, M. A., J. A. Webster, and N. Straus. 1993. Rapid identification of bacteria on the basis of polymerase chain reaction-amplified ribosomal DNA spacer polymorphisms. Appl. Environ. Microbiol. 59:945-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lacerda, L. D. 1997. Evolution of mercury contamination in Brazil. Water Air Soil Pollut. 97:247-255. [Google Scholar]
- 20.Lowry, O. H., A. L. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 21.Nyman, J. A. 1999. Effect of crude oil and chemical additives on metabolic activity of mixed microbial populations in fresh marsh soils. Microb. Ecol. 37:152-162. [DOI] [PubMed] [Google Scholar]
- 22.Osborn, A. M., E. R. Moore, and K. N. Timmis. 2000. An evaluation of terminal-restriction fragment length polymorphism (T-RFLP) analysis for the study of microbial community structure and dynamics. Environ. Microbiol. 2:39-50. [DOI] [PubMed] [Google Scholar]
- 23.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 24.Pak, K., and R. Bartha. 1998. Mercury methylation by interspecies hydrogen and acetate transfer between sulfidogens and methanogens. Appl. Environ. Microbiol. 64:1987-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pitcher, D. G., N. A. Saunders, and R. J. Owen. 1989. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 8:151-156. [Google Scholar]
- 26.Ramirez-Saad, H. C., A. Sessitsch, W. M. de Vos, and A. D. Akkermans. 2000. Bacterial community changes and enrichment of Burkholderia-like bacteria induced by chlorinated benzoates in a peat-forest soil-microcosm. Syst. Appl. Microbiol. 23:591-598. [DOI] [PubMed] [Google Scholar]
- 27.Sandaa, R., V. V. Torsvik, Enger, F. L. Daae, T. Castberg, and D. Hahn. 1999. Analysis of bacterial communities in heavy metal-contaminated soils at different levels of resolution. FEMS Microbiol. Ecol. 30:237-251. [DOI] [PubMed] [Google Scholar]
- 28.Schneider, M. 1994. Ph.D. dissertation. TU Carola-Wilhelmina, Braunschweig, Germany.
- 29.Schrope, M. 2001. US to take temperature of mercury threat. Nature 409:124. [DOI] [PubMed] [Google Scholar]
- 30.Silver, S., and L. T. Phung. 1996. Bacterial heavy metal resistance: new surprises. Annu. Rev. Microbiol. 50:753-789. [DOI] [PubMed] [Google Scholar]
- 31.Smith, J. N., and C. T. Schafer. 1999. Sedimentation, bioturbation, and Hg uptake in the sediments of the estuary and Gulf of St. Lawrence. Limnol. Oceanogr. 44:207-219. [Google Scholar]
- 32.Stewart, P. S., A. K. Camper, S. D. Handran, C. Huang, and M. Warnecke. 1997. Spatial distribution and coexistence of Klebsiella pneumoniae and Pseudomonas aeruginosa in biofilms. Microb. Ecol. 33:2-10. [DOI] [PubMed] [Google Scholar]
- 33.Summers, A. O. 1986. Organization, expression, and evolution of genes for mercury resistance. Annu. Rev. Microbiol. 40:607-634. [DOI] [PubMed] [Google Scholar]
- 34.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tilman, D. 2000. Causes, consequences and ethics of biodiversity. Nature 405:208-211. [DOI] [PubMed] [Google Scholar]
- 36.Tolker-Nielsen, T., U. C. Brinch, P. C. Ragas, J. B. Andersen, C. S. Jacobsen, and S. Molin. 2000. Development and detachment of Pseudomonas sp. biofilms. J. Bacteriol. 182:6482-6489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Canstein, H., Y. Li, K. N. Timmis, W.-D. Deckwer, and I. Wagner-Döbler. 1999. Removal of mercury from chlor-alkali electrolysis wastewater by a mercury-resistant Pseudomonas putida strain. Appl. Environ. Microbiol. 65:5279-5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Canstein, H., Y. Li, and I. Wagner-Döbler. 2001. Long-term stability of mercury reducing microbial biofilm communities analysed by 16S-23S rDNA interspacer region polymorphism. Microb. Ecol. 42:624-634. [DOI] [PubMed] [Google Scholar]
- 39.Wagner-Döbler, I., H. Lünsdorf, T. Lübbehüsen, H. F. von Canstein, and Y. Li. 2000. Structure and species composition of mercury-reducing biofilms. Appl. Environ. Microbiol. 66:4559-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wagner-Döbler, I., H. von Canstein, Y. Li, K. N. Timmis, and W.-D. Deckwer. 2000. Removal of mercury from chemical wastewater by microorganisms in technical scale. Environ. Sci. Technol. 34:4628-4634. [Google Scholar]
- 41.Whiteley, M., J. R. Ott, E. A. Weaver, and R. J. McLean. 2001. Effects of community composition and growth rate on aquifer biofilm bacteria and their susceptibility to betadine disinfection. Environ. Microbiol. 3:43-52. [DOI] [PubMed] [Google Scholar]