Abstract
A novel antifungal protein (SAP) was found in the culture supernatant of a marine bacterium, Streptomyces sp. strain AP77, and was purified. This protein was characterized by chemical, biochemical, and biological analyses. By using gel filtration, the molecular mass of SAP was estimated to be 160 kDa. Structural analysis of SAP by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and matrix-assisted laser desorption ionization-time of flight mass spectrometry suggested that SAP is composed of three heterologous protein subunits of 41.7 kDa (SAP1), 21.7 kDa (SAP2), and 18.7 kDa (SAP3) at a molar ratio of 1:1:5 (or 1:1:6). N-terminal amino acid sequence analysis and a homology search revealed that SAP1, SAP2, and SAP3 exhibit 64.3, 68.4, and 86.7% similarity to three Streptomyces coelicolor polypeptides, puromycin resistance protein (Pur8), a conserved hypothetical protein, and bacterioferritin, respectively. The MIC of purified SAP against Pythium porphyrae was determined to be 1.6 μg/disk, whereas no inhibitory effect was observed at concentrations up to 100 μg/disk against most of the fungal and bacterial strains tested; the only exception was relatively strong antifungal activity against Pythium ultimum (MIC, 6.3 μg/disk). In vitro and in vivo toxicity tests demonstrated that SAP showed no toxicity against Porphyra yezoensis cells, human normal dermal fibroblasts, and mice at doses up to 700 μg/ml (for 24 h), 250 μg/ml (for 12 h), and 75 mg/kg (for 35 days), respectively. SAP was labile when it was subjected to a heated-air drying treatment, which is a great advantage in food production procedures. These results indicated that Streptomyces sp. strain AP77 might be useful as a gene source for safe transgenic Porphyra breeding for tolerance to Pythium infection.
A marine red alga, Porphyra (Nori in Japanese), is extensively cultivated in Japan, Korea, and other oriental countries and is one of the popular and edible seaweeds. In Japan, the annual production of Porphyra spp. is 10 billion sheets, which earns 800 to 900 million United States dollars per year. However, Porphyra farming has frequently suffered great economic damage due to various diseases, such as red rot disease (50), chytrid blight disease (34), and green spot disease (47), caused by pathogenic microorganisms and unusual weather. Red rot disease (Akagusare-byo in Japanese) is one of the most serious problems in Porphyra farming, and this disease is prevalent on Porphyra farms throughout Japan. It causes losses of about 40 to 60 million United States dollars every year.
Red rot disease was first reported by Arasaki (5) in 1947. The causative organism of this disease is a member of the fungal genus Pythium, an oomycete (6, 57), and the pathogen was designated a new species, Pythium porphyrae, by Takahashi in 1970 (52). This disease spreads by means of motile biflagellate zoospores which are released into seawater (13), and the zoospores then infect Porphyra cells via the following process: adhesion of a zoospore to a host cell surface, encystment on the surface, appressorium formation beside the cyst formed, gemmation of the cyst, and penetration of the germ tube into the host cell (56). The spread of zoospores in Porphyra farms is too rapid for serious damage to be prevented in many cases, because on Porphyra farms simple communities consisting of a single species are cultivated. This pathogen severely infects Porphyra thalli, causing death of the host within a few days (13, 18). Moreover, infection by this pathogen reduces the quality and market value of Porphyra products even if they narrowly escape critical damage before harvest, because the sheet products obtained from infected thalli appear to be less lustrous, uneven, and discolored (17).
In the face of this situation, several countermeasures are employed on Porphyra farms. These include immersing the Porphyra cultivating nets into an organic acid, such as citric or phosphoric acid (at a pH of about 2), for 5 to 10 min, early harvesting of infected Porphyra prior to critical damage, exposing the Porphyra cultivating nets to the air, and short-term freezing at around −20°C for several days (48, 49). However, these treatments are effective only when they are implemented at the early stages of infection. Furthermore, the use of organic acids may be problematic with regard to possible environmental pollution in Porphyra cultivating areas.
Farmers have also made a constant effort to obtain Porphyra strains with beneficial properties by means of natural mutation and artificial selection (22, 36). This conventional method, however, requires a great deal of time and effort. Moreover, crossbreeding seems to be useless because the foliose thalli of Porphyra yezoensis possess a haploid nuclear phase (41). Therefore, biotechnological approaches in which protoplast fusion and regeneration techniques were used were recently attempted to improve breeding properties, such as disease tolerance (1, 4, 14, 15, 16, 31), growth rate (14, 22, 25), and resistance to higher temperatures (32). Although there have been a few reports of transient expression of a reporter gene in a red alga, Porphyra miniata (29), transgenic techniques for stable gene expression have not been established yet for seaweeds.
On the other hand, several crops which exhibit tolerance to fungal pathogens have been developed among the higher terrestrial plants by using the transgenic technique (2, 9, 33, 35, 38, 40, 59). Many of the resistance genes used for transformation of these plants encode polysaccharolytic enzymes, such as chitinase (8, 9, 23, 51) and β-1,3-glucanase (7, 23, 45, 51, 60), which should degrade the cell walls of invading fungi.
Given this situation, we have focused on marine bacteria indigenous to Porphyra cultivation environments in which the P. porphyrae concentration should be high. We previously screened these bacteria for anti-Pythium activity and Pythium cell wall-degrading activity and stored the bacteria showing either activity so that they could be used as possible sources of anti-Pythium genes to be introduced into Porphyra cells by means of transgenic techniques. At this time, 33 bacterial isolates that show anti-Pythium activity and four bacterial isolates that show Pythium cell wall-degrading activity have been isolated (28; S. Kawaguchi and Y. Kamei, Abstr. Meet. Jpn. Soc. Fish. Sci., p. 231, 1995). One of these 37 isolates, AP77, exhibited the greatest anti-Pythium activity and produced an extracellular anti-Pythium protein (Kawaguchi and Kamei, Abstr. Meet. Jpn. Soc. Fish. Sci., 1995). The anti-Pythium protein, designated SAP, had neither Pythium cell wall-degrading activity nor any other polysaccharolytic activity, including chitinase or β-1,3-glucanase activity (unpublished data), implying that AP77 may produce a unique antifungal protein with a function different from the functions of the polysaccharolytic enzymes used in the higher terrestrial plants.
In the present study, we first identified the SAP-producing bacterium, strain AP77, to the genus level. SAP was then purified from bacterial culture supernatant and characterized. We determined its molecular mass and structure, its stability in the presence of heat, different pH values, and drying conditions, its N-terminal amino acid sequences, and its antimicrobial spectrum. We also evaluated its in vitro and in vivo toxicities. Below we discuss the availability of the genes encoding SAP to produce transgenic Porphyra.
MATERIALS AND METHODS
Bacterial strains.
An anti-Pythium bacterium, strain AP77, was isolated by enrichment culture in September 1994 from the seawater of a P. yezoensis cultivation facility near the estuary of the Rokkaku River, Saga Prefecture, Japan (S. Kawaguchi and Y. Kamei, Abstr. Meet. Jpn. Soc. Fish. Sci., p. 231, 1995). Bacillus subtilis IFO 14419 , Pseudomonas aeruginosa IFO 13736, Staphylococcus aureus IFO 15036, and Vibrio parahaemolyticus IFO 12711 were purchased from the Institute for Fermentation, Osaka, Japan. TSB medium (Difco) and ZoBell 2216E broth (42) were used as basal media for storage and culture of these bacteria (see Table 2). The basal medium was supplemented with 20% glycerol and with 1.5% agar for storage of bacteria at −80°C and for MIC determinations, respectively.
TABLE 2.
MICs of SAP against bacterial and fungal strains and culture media used in the tests
| Microorganism | Medium used | MIC (μg/disk) |
|---|---|---|
| Bacteria | ||
| Bacillus subtilis IFO 14419 | TSB | >100 |
| Pseudomonas aeruginosa IFO 13736 | TSB | >100 |
| Staphylococcus aureus IFO 15036 | TSB | >100 |
| Vibrio parahaemolyticus IFO 12711 | ZoBell | >100 |
| Fungi | ||
| Alternaria alternata IFO 31189 | PDB | >100 |
| Aspergillus fumigatus TIMM 0063 | Malt extract | >100 |
| Aspergillus versicolor IFO 31223 | PDB | >100 |
| Botrytis cinerea IFO 33008 | PDB | >100 |
| Candida albicans IFO 1594 | 108 | >100 |
| Fusarium graminearum IFO 9462 | PDB | >100 |
| Mucor circinelloides IFO 31398 | PDB | >100 |
| Penicillium citrinum IFO 7784 | PDB | >100 |
| Phytophthora nicotianae IFO 33193 | PDB | >100 |
| Pythium aphanidermatum IFO 32440 | PDB | >100 |
| Pythium oligandrum IFO 32559 | PDB | >100 |
| Pythium porphyrae IFO 30347 | CSL | 1.6 |
| Pythium ultimum IFO 32612 | PDB | 6.3 |
| Trichophyton rubrum TIMM 2659 | Sabouraud | >100 |
| Ustilago maydis IFO 6907 | PDB | >100 |
Fungal strains.
Alternaria alternata IFO 31189, Aspergillus versicolor IFO 31223, Botrytis cinerea IFO 33008, Candida albicans IFO 1594, Fusarium graminearum IFO 9462, Mucor circinelloides IFO 31398, Penicillium citrinum IFO 7784, Phytophthora nicotianae IFO 33193, Pythium aphanidermatum IFO 32440, Pythium oligandrum IFO 32559, Pythium porphyrae IFO 30347, Pythium ultimum IFO 32612, and Ustilago maydis IFO 6907 were provided by the Institute for Fermentation, Osaka, Japan, and Aspergillus fumigatus TIMM 0063 and Trichophyton rubrum TIMM 2659 were provided by Teikyo University Institute of Medical Mycology, Tokyo, Japan. The following basal media were used for storage and culture of mycelia of these fungi: PDB medium (20% potato extract, 2% dextrose [Eiken Chemical, Tokyo, Japan]; pH 5.6), malt extract broth (2% malt extract, 2% glucose, 0.1% peptone; pH 6.0), 108 broth (1% glucose [Katayama Chemical, Osaka, Japan], 0.5% peptone [Katayama Chemical], 0.3% yeast extract [Nihon Seiyaku, Tokyo, Japan], 0.3% malt extract [Difco]; pH 5.6), CSL broth (0.2% corn steep liquor [Wako Pure Chemical, Osaka, Japan], 0.1% yeast extract, 75% artificial seawater [Jamarin Laboratory, Osaka, Japan]; pH 7.5), and Sabouraud broth medium (4% glucose, 1% peptone; pH 6.0). These basal media were supplemented with 10% dimethyl sulfoxide and with 1.5% agar for storage of fungal mycelia at −80°C and for MIC determinations, respectively.
Cells and animals.
Dried P. yezoensis thalli (ca. 5 by 25 mm) attached to synthetic rope were kindly provided by Y. Kawamura, Saga Prefectural Ariake Fisheries Research and Development Center, Saga, Japan, and were stored in our laboratory at −20°C until they were used. After the thallus cells were thawed at 15°C, they were cultured at 15°C for 3 days in a 300-ml flask with SWM-III medium (11) with aeration (10 ml/min) by using cool white fluorescent light (50 μmol·m−2·s−1; 10 h of light and 14 h of darkness per day). Human normal dermal fibroblasts (HDF) were provided by Morinaga Institute of Biological Science, Yokohama, Japan, and were maintained at 37°C in a 5% CO2 humidified incubator by using E-RDF medium (Kyokuto Pharmaceutical Industrial Inc., Tokyo, Japan) containing 10% fetal bovine serum (Sigma, St. Louis, Mo.). Four-week-old BALB/c mice (Nihon Clea Inc., Osaka, Japan) were reared at 25°C until the experiment was initiated.
Characterization and identification of strain AP77.
Strain AP77 was identified by using the methods described by Williams et al. (58). The bacterium was cultured at 25°C for 3 days on a marine agar plate (Difco) and then used for morphological tests performed with a light microscope or a scanning electron microscope (model JSM-840A; JEOL Ltd., Tokyo, Japan). For scanning electron microscopy, the bacterium was fixed on a specimen mount with 2% glutaraldehyde and then with 2% osmic acid solution vapor for 4 h. Then the sample was dehydrated in a graduated ethanol series (30 to 100%) and critical point dried in the presence of CO2. After the preparation was sputter coated (model JPC-1100; JEOL Ltd.) with gold for 1 min, it was observed at 10 kV. For genetic characterization of strain AP77, the bacterial DNA was extracted by the method of Hopwood et al. (19). To hydrolyze DNA into nucleotides, 0.02 U of nuclease P1 (Yamasa Shoyu Co., Choshi, Japan) was added to denature a DNA solution (10 μg) that was incubated at 50°C for 60 min. After centrifugation at 10,000 × g for 10 min, the mixture was then filtered through a Millipore filter (DISMIC-3; pore size, 0.45 μm; Millipore Corp., Bedford, Mass.), and the hydrolysate containing nucleotides was analyzed with a high-performance liquid chromatography (HPLC) system (JASCO Corporation, Tokyo, Japan) equipped with a reversed-phase HPLC column (Pack AQ312; YMC, Kyoto, Japan). Nucleotides were eluted with a mixture of 10 mM H3PO4 and 10 mM KH2PO4 (pH 3.0) at a flow rate of 1.5 ml/min (24). A standard mixture of nucleotides (Yamasa Shoyu Co.) was used as a reference to calibrate the G+C content. PCR amplification of 16S ribosomal DNA (rDNA) fragments was carried out by using three sets of primers as described by Takeuchi et al. (53). Both DNA strands were sequenced directly by using the PRISM Big Dye terminator cycle sequencing reagent (PE Biosystems Japan, Tokyo, Japan) and an ABI PRISM 310 genetic analyzer (PE Biosystems Japan). The 16S rDNA fragments were analyzed separately in triplicate, which resulted in a reproducible sequence. The sequence determined was compared with 16S rDNA sequences obtained from the EMBL, GenBank, and DDBJ databases. Multiple alignment of sequences, calculation of nucleotide substitution rates (Knuc values) (27), construction of a neighbor-joining phylogenetic tree (46), and a bootstrap analysis with 1,000 replicates for evaluation of phylogenetic tree topology (12) were performed by using the CLUSTAL W (version 1.5) program (55).
Determination of antimicrobial activity.
The antimicrobial activity of SAP was determined in triplicate by using disk diffusion on a double-layer agar plate (diameter, 90 mm) as previously described (20). P. porphyrae was inoculated into CSL broth and cultured at 25°C with agitation at 230 rpm until it reached the turbidity of the 0.5 McFarland standard. A 1-ml aliquot of this culture was added to 10 ml of CSL broth with 0.8% agar and overlaid onto a CSL broth plate with 1.5% agar. Sterile paper disks (diameter, 8 mm; Advantec, Tokyo, Japan) were permeated with 50 μl of serially diluted SAP samples and placed onto the agar plate immediately after the medium had solidified. The plate was then incubated at 25°C for 48 h, and the inhibitory zones around the paper disks were observed. One unit of anti-Pythium activity was defined as the activity exhibited in 1 mm of the inhibitory zone. The approximate MICs of SAP against fungi and bacteria also were determined by using the procedure described above and were defined as the lowest concentrations of SAP at which inhibitory zones were still observed around the disks. The bacterial inocula were prepared to give a density of 106 cells/ml. Most fungal and bacterial strains were incubated at 37°C; the only exception was V. parahaemolyticus, which was incubated at 25°C. The incubation times were 24 h for C. albicans, B. subtilis, P. aeruginosa, and S. aureus, 48 h for V. parahaemolyticus, and 72 h for the other fungi.
Purification of SAP.
Strain AP77 was precultured in ZoBell 2216E broth at 25°C for 2 days to obtain a seed culture. Five milliliters of the seed culture was inoculated into 10 liters of ZoBell 2216E broth in a 15-liter jar fermentor (model TS-AL15L; Takasugi Seisakusho Ltd., Tokyo, Japan) and cultured at 25°C for 4 days with aeration at a rate of 1 liter/min and with agitation at 150 rpm. The culture supernatant collected by centrifugation at 7,500 × g for 15 min at 4°C was concentrated to 100 ml by using a polysulfone ultrafiltration membrane (30,000 normal molecular weight limits; filter area, 60 cm2; Millipore) and was mixed with 400 ml of distilled water. The resulting solution (500 ml) was then saturated (95%) with solid ammonium sulfate and kept overnight at 4°C. The precipitate was collected by centrifugation at 18,000 × g for 30 min at 4°C and dissolved in 10 ml of 10 mM Tris-HCl buffer (pH 7.5). The protein solution was dialyzed against Tris-HCl buffer at 4°C for 24 h by using a cellulose dialysis membrane (molecular weight cutoff, 3,500; Spectrum Laboratories Inc., Rancho Dominguez, Calif.). The dialyzed solution (20 mg of protein) was applied to a DEAE-cellulose column (diameter, 2.5 cm; length, 30 cm; Bio-Rad, Hercules, Calif.) preequilibrated with the Tris-HCl buffer. After the column was washed with 2 bed volumes of the Tris-HCl buffer, proteins were eluted with 5 bed volumes of a linear NaCl gradient (0 to 1.0 M) at a flow rate of 1 ml/min. The elute was collected (5 ml per test tube), and the anti-Pythium activity of each fraction and the absorbance at 280 nm were determined. The active fractions were pooled and concentrated by ultrafiltration (filter area, 0.2 cm2; Millipore). Then the concentrated SAP solution (3 mg of protein) was applied to a Protein KW-803 column (diameter, 0.8; length, 30 cm; Shodex, Tokyo, Japan) and eluted with the Tris-HCl buffer at a flow rate of 0.5 ml/min. The active fractions were again pooled and concentrated by using the same ultrafilter and then loaded onto a native 5% polyacrylamide gel electrophoresis (PAGE) gel for final purification. To identify the active protein band, the resulting PAGE gel was placed on a CSL agar plate on which P. porphyrae had been precultured at 25°C for 24 h, and the preparation was incubated at 25°C for 24 h. The active band which produced a growth inhibition zone for P. porphyrae was extracted from the gel as purified SAP. The protein assay in each step of SAP sample preparation was performed by using a bicinchoninic acid protein assay kit (Pierce, Rockford, Ill.) with bovine serum albumin as the standard.
Preparation of antiserum.
To prepare an anti-SAP serum, 100 μl (8 μg) of the purified SAP was mixed with an equal volume of an adjuvant (Titer Max GOLD; CytRx Corp., Atlanta, Ga.), and the mixture was subcutaneously injected to the femoral regions of a 5-week-old female mouse (weight, 20 g). The same inoculum was used for two subsequent booster doses at 2 week-intervals. Two weeks after the last booster dose, the mouse was sacrificed, and the blood serum was collected.
Elucidation of molecular mass and structure of SAP.
The molecular mass of SAP in its native state was estimated by gel filtration on an HPLC column (Protein KW-803) with molecular mass standard proteins (Gel Filtration Standard; Bio-Rad). The molecular mass of SAP was also determined by sodium dodecyl sulfate (SDS)-PAGE in the presence and absence of β-mercaptoethanol by using a 12.5% polyacrylamide gel, molecular mass standards (Prestained Protein Markers; New England Biolabs Inc., Beverly, Mass.), and the method of Laemmli (30). After electrophoresis, proteins were stained with 1% Coomassie brilliant blue R-250. The stained proteins in the gel were then analyzed with NIH Image software (version 1.58), and the molar ratio of the subunits of SAP was calculated based on the molecular masses and protein amounts determined in five independent experiments. Western blot analysis also was performed by electroblotting at 200 mA for 90 min onto a nitrocellulose membrane (Immobilon; Millipore). After the electroblotting, SAP was detected by using the anti-SAP mouse serum (dilution, 1:20,000) described above as the primary antibody, anti-mouse immunoglobulin G goat serum (dilution, 1:2,000; Funakoshi Inc., Tokyo, Japan) as the secondary antibody, and an immunostaining kit (HRP-1000; Konica, Tokyo, Japan). To confirm the molecular mass of SAP or its subunits, the purified SAP was desalted with a microtip column (ZipTip C18; Millipore) and subjected to matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) (Voyager-Elite BioSpectrometry; PerSeptive Biosystems, Framingham, Mass.). The acceleration voltage was 20 kV, and α-cyano-4-hydroxycinnamic acid (Sigma) was used as the matrix.
Stability of SAP.
The stability of purified SAP at temperatures ranging from 4 to 100°C was determined by incubating SAP in 10 mM Tris-HCl buffer (pH 7.5) for 20 min. After the solutions were cooled to 4°C, the remaining anti-Pythium activities were determined. The stability of SAP under acid and alkaline conditions was tested by using McIlvaine buffer (pH 2.5 to 8.0) and 10 mM Tris-HCl buffer (pH 7.5 to 10.5). SAP was incubated in each buffer at various pHs at 4°C for 120 min. The remaining anti-Pythium activities were assayed after the pH of each SAP solution was readjusted to 7.5. A stability test imitating the heated-air drying conditions in the manufacturing process used to produce Porphyra sheets also was conducted. Fifty-microliter portions of SAP in sterile artificial seawater were added to capped or uncapped 200-μl microtubes and incubated under a vacuum (60 hPa) at 43°C for 120 min or at 33°C for 150 min. After incubation, each tube was filled to the initial level with ice-cold distilled water, and the remaining anti-Pythium activity was determined.
N-terminal amino acid sequencing of subunit proteins of SAP.
To analyze the N-terminal amino acid sequences of the subunit proteins of SAP, purified SAP was subjected to SDS-PAGE under reducing conditions and then electroblotted onto a polyvinylidene difluoride membrane (ProBlott; PE Biosystems Japan). After the membrane was stained with 0.1% Coomassie brilliant blue R-250, the protein bands on the membrane were cut out and rinsed with 50 and 100% methanol (three times each) to eliminate the staining reagent. Each protein band was analyzed to determine the N-terminal amino acid sequence by using the Edman degradation method and an automated protein sequencer (model 491; PE Biosystems Japan), and the resulting N-terminal amino acid sequences were compared with the protein sequences in the DAD, PDB, PIR, and SWISS-PROT databases.
Toxicity of SAP.
In vitro cytotoxicity of purified SAP was evaluated by using P. yezoensis cells and HDF. Thalli of P. yezoensis were cut into pieces that were 5 mm square and treated with 700 μg of purified SAP per ml in a dish containing SWM-III medium at 25°C for 1 to 24 h. After treatment, the thalli were washed three times with SWM-III medium and stained with 0.01% Evans blue (Merck, Darmstadt, Germany) for 10 min. The percentages of surviving cells were then calculated by counting the numbers of viable and dead cells with a microscope. HDF were seeded into a 96-well plate to give 5 × 103 cells/100 μl in each well and immediately treated with serial dilutions (0.98 to 500 μg/ml) of SAP in E-RDF medium at 37°C for 12 h. After treatment, the cells were washed three times with E-RDF medium, and the MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (Sigma) assay (37) was performed. The growth rate of HDF was determined by comparing the absorbance of the treated group with that of the untreated control, and the cytotoxicity of SAP was evaluated. To examine the toxicity of SAP in vivo, 75 mg of SAP/kg of mouse in 10 mM Tris-HCl buffer (pH 7.5) was intraperitoneally injected into 5-week-old mice (weight, 18 to 22 g; n = 5) three times at 2-week intervals. For the control, the Tris-HCl buffer was inoculated into mice (n = 2) by using the same schedule. The pathological changes were then observed daily for 35 days after the first administration.
Nucleotide and amino acid sequence accession numbers.
The nucleotide sequence of 16S rDNA from strain AP77 has been deposited in DDBJ database under accession number AB052845. The amino acid sequences of puromycin resistance protein (Pur8), a conserved hypothetical protein, and bacterioferritin from Streptomyces coelicolor have been deposited in the DDBJ database under accession numbers AL391541, AL035161, and AL109661, respectively.
RESULTS AND DISCUSSION
Characterization and identification of strain AP77.
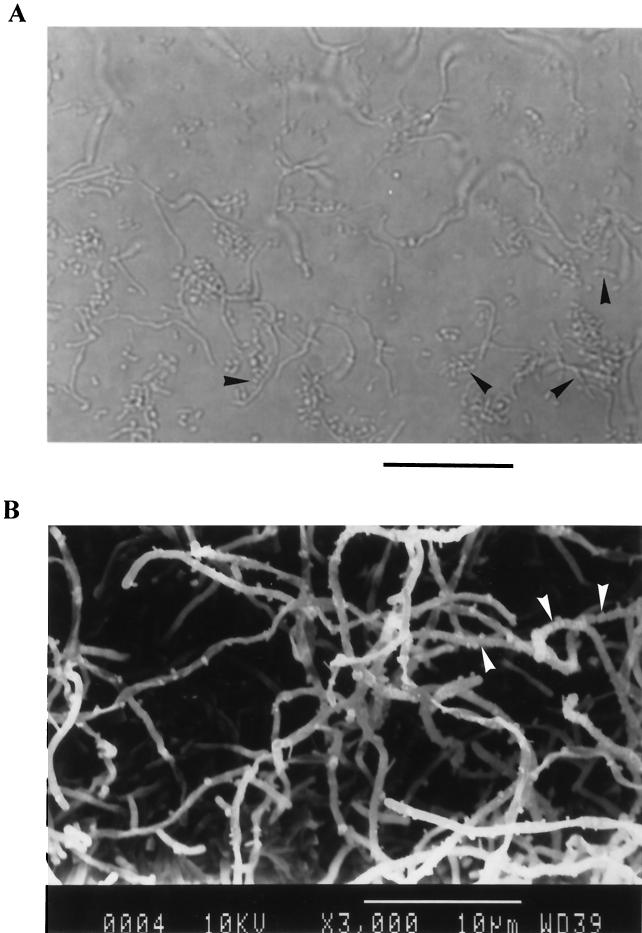
Strain AP77 grew well on a marine agar plate and formed white to ash gray, wrinkled colonies, while no growth was observed on potato dextrose agar (Eiken Chemical) supplemented with 50 μg of chloramphenicol per ml. During observation with light and electron microscopes, this strain was found to be a gram-positive filamentous bacterium with extensively branched aerial mycelia (diameter, 0.5 to 1 μm), on which chains of arthrospores were observed (Fig. 1). No sporangia or motile spores were found. The G+C content of this strain was 74.4%. These results indicated that strain AP77 is a bacterium belonging to the genus Steptomyces (58).
FIG. 1.
Light micrograph (A) and scanning electron micrograph (B) of strain AP77 cultured at 25°C for 3 days on a marine agar plate. The arrowheads indicate chains of arthrospores formed on branched aerial mycelia. (A) Bar = 20 μm; (B) bar = 10 μm.
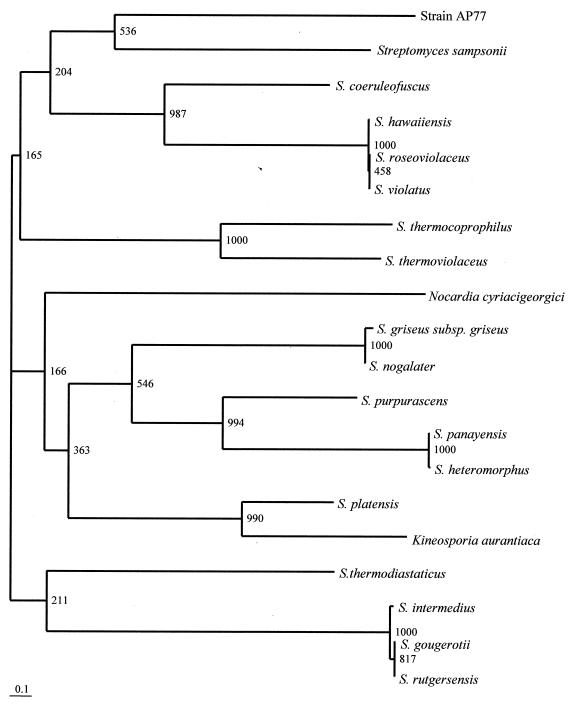
The results described above were obviously supported by the 16S rDNA sequence analysis of strain AP77. The phylogenetic tree determined by the neighbor-joining method revealed that Steptomyces sampsonii was the bacterial species most closely related to strain AP77 (Fig. 2), and the sequence similarity of the two bacteria was 99.8% (1,440 of 1,443 nucleotides were identical). However, further investigations that include DNA-DNA hybridization are required to identify the bacterium to the species level.
FIG. 2.
Phylogenetic position of strain AP77 within the genus Streptomyces and allied bacteria. The branching pattern was generated by the neighbor-joining method. The numbers at the nodes are bootstrap values. Bar = 0.1 nucleotide substitution per site (Knuc value).
Purification of SAP.
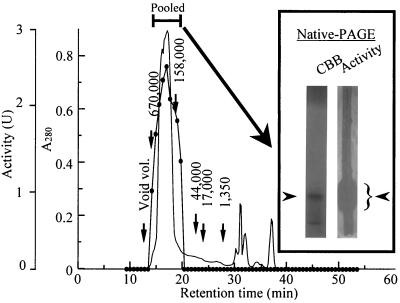
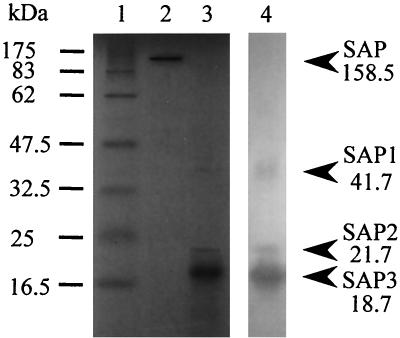
The yields SAP in the different purification steps are summarized in Table 1. In the first step, SAP was purified 10.5-fold by ultrafiltration. The yield of SAP was, however, decreased to 5.9% after the subsequent steps involving ammonium sulfate precipitation and dialysis. This loss of protein appeared to be due to removal of another antifungal substance through the dialysis membrane. This antifungal substance, which had a low molecular mass, was extracted by chloroform, was not colored by the ninhydrin reaction, and retained its activity even after it was boiled for 20 min (data not shown). Therefore, we concluded that this substance could not be used as an anti-Pythium protein to confer disease tolerance and continued the purification process. Ion-exchange chromatography with DEAE-cellulose resulted in effective separation of SAP from other proteins. The active protein was eluted with 0.8 M NaCl, and then SAP was purified 13.7-fold. Subsequent purification was performed by gel filtration with HPLC by using a Protein KW-803 column, which resulted in a single peak at a retention time of 16 min (Fig. 3). By means of these chromatography steps, SAP was purified 23.1-fold. The active fractions were pooled and subjected to native PAGE, which revealed two protein bands on the gel stained with Coomassie brilliant blue R-250 (Fig. 3). To evaluate which band had antifungal activity, the gel was placed on an agar plate on which P. porphyrae had been precultured. After incubation, the upper band was found to exhibit antifungal activity against P. porphyrae (Fig. 3). Therefore, the upper protein band extracted from the gel was subjected to SDS-PAGE without β-mercaptoethanol, which resulted in a single protein band (Fig. 4, lane 2). Finally, the specific activity of purified SAP was increased 33.5-fold, and the yield was 0.9%.
TABLE 1.
Purification of SAP from the culture supernatant of Streptomyces sp. strain AP77
| Purification step | Vol (ml) | Protein (mg) | Total activity (103 U) | Sp act (U/mg) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Culture supernatant | 10,000 | 3,410 | 250 | 73 | 1.0 | 100 |
| Ultrafiltration | 500 | 234 | 180 | 769 | 10.5 | 72 |
| (NH4)2SO4 precipitation and dialysis | 20 | 191 | 14.7 | 77 | 1.1 | 5.9 |
| Ion exchange | 10 | 13.0 | 13.0 | 1,000 | 13.7 | 5.2 |
| Gel filtration | 10 | 3.2 | 5.4 | 1,688 | 23.1 | 2.2 |
| Native PAGE | 10 | 0.9 | 2.2 | 2,444 | 33.5 | 0.9 |
FIG. 3.
Gel filtration chromatography of SAP with Protein KW-803 and the native PAGE profile. SAP (3 mg) was applied to the column (diameter, 0.8; length, 30 cm) and was eluted with 10 mM Tris-HCl buffer (pH 7.5) at a flow rate of 0.5 ml/min. The solid circles indicate anti-Pythium activity, and the line without circles shows the absorbance at 280 nm. The numbers above the vertical arrows indicate the retention times of the molecular mass standards eluted in the same column. The active fractions were subjected to native PAGE and were stained with Coomassie brilliant blue (CBB) or visualized on an agar plate with precultured P. porphyrae (Activity). The fungal growth on the agar plate is evident in the white border on either side of the native PAGE gel on the right. The arrowheads next to the native PAGE gels indicate the positions of SAP (left gel) and anti-Pythium activity (right gel).
FIG. 4.
SDS-PAGE and Western blot analysis profiles of SAP. Lane 1, standard protein markers; lane 2, preparation not treated with β-mercaptoethanol; lane 3, preparation treated with β-mercaptoethanol; lane 4, Western blot.
Molecular mass and structure of SAP.
The molecular mass of native SAP was estimated to be approximately 160 kDa by gel filtration (Fig. 3) and SDS-PAGE without β-mercaptoethanol (Fig. 4, lane 2), while the results of SDS-PAGE under reducing conditions gave three protein bands at 41.7, 21.7, and 18.7 kDa; these bands were designated SAP1, SAP2, and SAP3, respectively (Fig. 4, lane 3). The immunoreactivities of these three proteins with an anti-SAP serum were shown by Western blot analysis (Fig. 4, lane 4). The results indicated that SAP consists of three protein subunits. The apparent discrepancy between the molecular mass of native SAP and the molecular masses of the three subunits was resolved by analysis with NIH Image software. The numbers of subunits per mole of SAP were calculated to be 0.61 (± 0.24), 1.25 (± 0.53), and 5.29 (± 0.88) for SAP1, SAP2, and SAP3, respectively, suggesting that SAP consists of the three subunits at a molar ratio of 1:1:5 (or 1:1:6). Furthermore, at least five peaks were observed in a MALDI-TOF MS profile of SAP, at molecular masses of 18,852, 37,704, 56,693, 75,450, and 94,623 Da. These MALDI-TOF MS data may support the results of the NIH Image software analysis indicating that five or six SAP3 molecules are included in a SAP molecule. Neither SAP1 nor SAP2 was ionized under the conditions tested.
Stability of SAP.
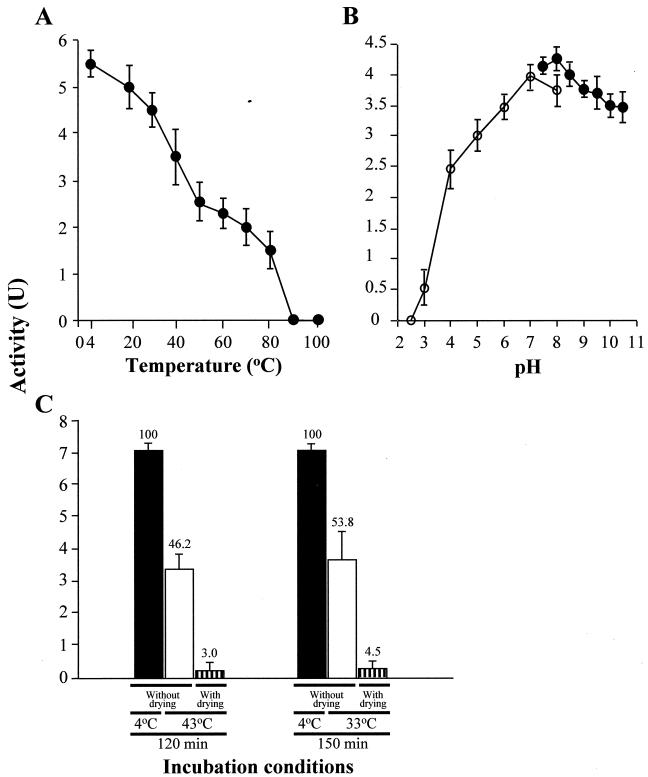
The anti-Pythium activities of purified SAP after exposure to various temperatures and pH conditions are summarized in Fig. 5. The activity of SAP decreased with increasing temperature, and SAP was completely inactivated by heating at 90°C for 20 min in 10 mM Tris-HCl buffer at pH 7.5 (Fig. 5A). SAP was stable to pHs ranging from 6.0 to 10.5, and the maximal activity was observed at pH 7.0 to 8.0 in Tris-HCl or McIlvaine buffer for 120 min of exposure (Fig. 5B). Since having an expressed foreign protein that remains in the crop and shows bioactivity after manufacturing is undesirable and affects food safety, we investigated the stability of SAP under conditions similar to the heated-air drying process used to manufacture Porphyra sheets. After the SAP solution was treated at 43°C for 120 min and at 33°C for 150 min in artificial seawater at pH 8.0, the activities declined 46.2 and 53.8%, respectively, compared to the activities of the controls kept at 4°C (Fig. 5C). Moreover, it is remarkable that the activities of the dehydrated protein in uncapped tubes decreased to 3.0 and 4.5% of the control values, respectively (Fig. 5C), and most of the activities were lost under these practical conditions.
FIG. 5.
Heat stability, pH stability, and stability under drying conditions for purified SAP. SAP was exposed to various temperatures for 20 min (A), to various pH values at 4°C for 2 h (B), and to heated-air drying conditions imitating the conditions used in the Porphyra manufacturing process (C). (A and B) Symbols: •, anti-Pythium activities in 10 mM Tris-HCl buffer; ○, anti-Pythium activities in McIlvaine buffer. (C) The bars indicate the activities of SAP incubated under different conditions (see Materials and Methods). The numbers above the bars show the percentages of activity compared with the activity of the untreated control (solid bars). The data and error bars represent means and standard deviations for triplicate assays.
MICs of SAP.
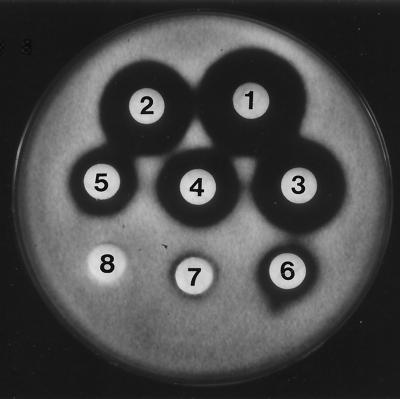
Approximate MICs of purified SAP were determined by using bacterial and fungal strains and concentrations ranging from 0.8 to 100 μg/disk. The MICs of SAP for P. porphyrae and P. ultimum were determined to be 1.6 and 6.3 μg/disk, respectively (Fig. 6 and Table 2), while no antimicrobial activity was observed even at a concentration of 100 μg/disk against gram-positive and gram-negative bacteria and the other fungi tested. These results indicate that the activity of SAP may be specific to P. porphyrae and P. ultimum.
FIG. 6.
Inhibitory activity of purified SAP against P. porphyrae as determined by the disk diffusion susceptibility test. Disk 1, 100 μg of purified SAP/disk; disk 2, 50 μg/disk; disk 3, 25 μg/disk; disk 4, 12.5 μg/disk; disk 5, 6.3 μg/disk; disk 6, 3.2 μg/disk; disk 7, 1.6 μg/disk; disk 8, 0.8 μg/disk. The approximate MIC was determined to be 1.6 μg/disk.
N-terminal amino acid sequences of SAP subunits.
The N-terminal amino acid sequences of the subunit proteins of SAP were determined to be QVFLRDRSGRRGGHLDVPGVLLGSGGLVAL (SAP1), VEGDRLLAAGALDPEGVVVGEGAPAVQLRD (SAP2), and MQGDPEVLEFLNEQLTAELTAINQYWLHYR (SAP3). Based on DAD, PDB, PIR, and SWISS-PROT database data, 30 N-terminal amino acid residues of SAP1, SAP2, and SAP3 showed 64.3, 68.4, and 86.7% similarity to the residues of puromycin resistance protein Pur8 (FALLHDRPGHADARLDVPGVVLGCGGLVAL), a conserved hypothetical protein (GRFVAVSGQLALDEDGKVVGEGDPAAQARQ), and bacterioferritin (MQGDPEVIEFLNEQLTAELTAINQYFLHAK) from S. coelicolor (44), respectively. These results suggest that SAP1 is associated with a transport system in the host bacterial cells (54), while SAP3 participates in bacterial iron storage with its possible heme structure (3, 10, 21, 43).
There have been no reports on the proteins active against Pythium spp., but two proteins active against Phytophthora, a chitin-negative fungal genus related to Pythium, have been reported (26, 39). These 14- and 34-kDa proteins were identified as basic pathogenesis-related protein 1 from tomato (Lycopersicon esculentum Mill. cv. Baby) and β-1,3-glucanase from pepper (Capsicum annuum L. cv. Hanbyul), respectively. The subunits of SAP do not show similarity to either pathogenesis-related protein 1 or β-1,3-glucanase, and there have been no reports showing antifungal activity of puromycin resistance proteins or bacterioferritins in vitro and in vivo. The contribution of SAP2 to the antifungal activity of SAP is cannot be predicted at this time.
Toxicity of SAP in vitro and in vivo.
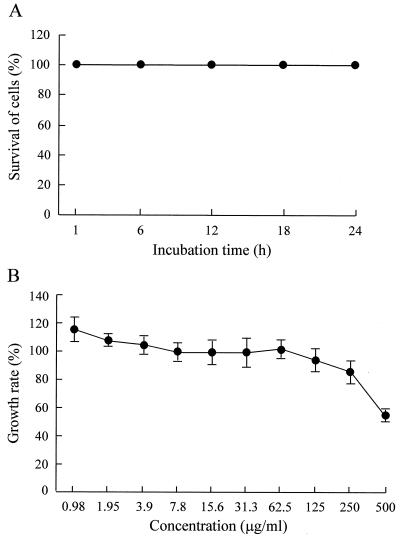
In a preliminary test performed at 25°C for 1 h, purified SAP exhibited no cytotoxcity against Porphyra cells at concentrations ranging from 1.4 to 700 μg/ml (data not shown). Therefore, the incubation time was increased to 24 h, and cytoxicity at a concentration of 700 μg/ml was examined. However, no cytotoxicity against Porphyra cells was observed at this concentration, even in the cells incubated for 24 h (Fig. 7A). When the cytotoxicity of SAP against HDF was tested at 37°C for 12 h at concentrations ranging from 0.98 to 500 μg/ml, no significant decrease in the growth rate was found in the cells treated with SAP at concentrations up to 250 μg/ml, although the 500-μg/ml SAP treatment decreased the growth rate 54.9% compared with that of the untreated control (Fig. 7B). Based on these results, the minimum cytotoxic concentrations of SAP in vitro were estimated to be >700 and 500 μg/ml against Porphyra cells and HDF, respectively. The low toxicity of SAP in vivo also was shown by an acute toxicity test performed with BALB/c mice. During the 35-day observation period, neither mortality nor pathological symptoms were found in mice injected with 75 mg of SAP per kg three times at 2-week intervals.
FIG. 7.
Cytotoxocity of purified SAP against P. yezoensis cells and HDF. (A) After incubation with 700 μg of SAP per ml at 25°C, thallus cells of P. yezoensis were stained with Evans blue, and the percentage of surviving cells was determined. (B) HDF cells were incubated with each concentration of SAP at 37°C for 12 h, and the growth rate of the cells was evaluated by the MTT assay. The data points and error bars represent means and standard deviations for triplicate experiments.
This is the first report showing that a protein has activity against Pythium species. Considering that the MIC of SAP against P. porphyrae was 1.6 μg/disk (Fig. 6 and Table 2), we suggest that SAP exhibits selective inhibition of growth of P. porphyrae and that the range of effective concentrations is wide. Moreover, most of the antifungal activity was eliminated under the conditions used for heated-air drying during Porphyra manufacturing (Fig. 5C), suggesting that the genes encoding SAP or its subunits are safe and excellent candidates for use as resistant genes to establish Porphyra strains tolerant to red rot disease by means of transgenic techniques. However, we have not determined which subunits are essential to generate this activity. Detailed studies focusing on the anti-Pythium mechanisms of SAP, the sequences and expression of SAP genes, and determination of the SAP active domain are in progress.
REFERENCES
- 1.Achiha, H. 1995. Crossbreeding of Porphyra tenuipedalis by electroporation. Kaiyo Monthly 27:671-677. [Google Scholar]
- 2.Alexander, D., R. M. Goodman, M. Gut-Rella, C. Glascock, K. Weymann, L. Friedrich, D. Maddox, P. Ahl-Goy, T. Luntz, E. Ward, and J. Ryals. 1993. Increased tolerance to two oomycete pathogens in transgenic tobacco expressing pathogenesis-related protein 1a. Proc. Natl. Acad. Sci. USA 90:7327-7331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews, S. C., J. M. Smith, J. R. Guest, and P. M. Harrison. 1989. Amino acid sequence of the bacterioferritin gene (cytochrome b1) of Escherichia coli K-12. Biochem. Biophys. Res. Commun. 158:489-496. [DOI] [PubMed] [Google Scholar]
- 4.Araki, T., and T. Morishita. 1990. Fusion of protoplasts from wild type Porphyra yezoensis and green type P. tenera thalli (Rhodophyta). Nippon Suisan Gakkaishi 56:1161. [Google Scholar]
- 5.Arasaki, S. 1947. Studies on the rot of Porphyra tenera by a Pythium. Bull. Jpn. Soc. Sci. Fish. 13:74-90. [Google Scholar]
- 6.Arasaki, S., K. Akino, and T. Tomiyama. 1968. A comparison of some physiological aspects in a marine Pythium on the host and on the artificial medium. Bull. Misaki Marine Biol. Inst. Tokyo Univ. 12:203-206. [Google Scholar]
- 7.Benhamou, N., J. Grenier, A. Asselin, and M. Legrand. 1989. Immunogold localization of β-1,3-glucanases in two plants infected by vascular wilt fungi. Plant Cell 1:1209-1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bishop, J. C., A. M. Dean, and T. Mitchell-Olds. 2000. Rapid evolution in plant chitinases: molecular targets of selection in plant-pathogen coevolution. Proc. Natl. Acad. Sci. USA 97:5322-5327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broglie, K., I. Chet, M. Holliday, R. Cressman, P. Biddle, S. Knowlton, C. J. Mauvais, and R. Broglie. 1991. Transgenic plants with enhanced resistance to the fungal pathogen Rhizoctonia solani. Science 254:1194-1197. [DOI] [PubMed] [Google Scholar]
- 10.Brooks, B. W., N. M. Young, D. C. Watson, R. H. Robertson, E. A. Sugden, K. H. Nielsen, and S. A. Becker. 1991. Mycobacterium paratuberculosis antigen D: characterization and evidence that it is a bacterioferritin. J. Clin. Microbiol. 29:1652-1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen, L. C., M. T. Edelstein, E. Ogata, and J. McLachlan. 1970. The life history of Porphyra miniata. Can. J. Bot. 48:385-389. [Google Scholar]
- 12.Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the boostrap. Evolution 39:783-791. [DOI] [PubMed] [Google Scholar]
- 13.Fujita, Y. 1978. Studies on pathogenic Pythium of laver red rot in Ariake Sea farm. V. Germination of Pythium porphyrae oospores. Bull. Jpn. Soc. Sci. Fish. 44:15-19. [Google Scholar]
- 14.Fujita, Y. 1993. Crossbreeding of green and red algae by cell fusion. Kaiyo Monthly 281:690-695. [Google Scholar]
- 15.Fujita, Y., and S. Migita. 1987. Fusion of protoplasts from thalli of two different color types in Porphyra yezoensis UEDA and development of fusion products. J. Phycol. 35:201-208. [Google Scholar]
- 16.Fujita, Y., and M. Saito. 1990. Protoplast isolation and fusion in Porphyra (Bangiales, Rhodophyta). Hydrobiologia 204:161-166. [Google Scholar]
- 17.Fujita, Y., and S. R. Uppalapati. 1997. Genetic improvement of Porphyra through cell culture techniques: present status and future prospects. Nat. Hist. Res. 3:71-81. [Google Scholar]
- 18.Fuller, M. S., B. Lewis, and P. Cook. 1966. Occurrence of Pythium sp. on the marine alga Porphyra. Mycologia 58:313-318. [Google Scholar]
- 19.Hopwood, D. A., M. J. Bibb, K. F. Chater, T. Kieser, C. J. Bruton, H. M. Kieser, D. J. Lydiate, C. P. Smith, J. M. Ward, and H. Schrempf. 1985. Genetic manipulation of Streptomyces: a laboratory manual. The John Innes Foundation, Norwich, United Kingdom.
- 20.Horikawa, M., T. Noro, and Y. Kamei. 1999. In vitro anti-methicillin-resistant Staphylococcus aureus activity found in extract of marine algae indigenous to the coastline of Japan. J. Antibiot. 52:186-189. [DOI] [PubMed] [Google Scholar]
- 21.Inglis, N. F., K. Stevenson, A. H. Hosie, and J. M. Sharp. 1994. Complete sequence of the gene encoding the bacterioferritin subunit of Mycobacterium avium subspecies silvaticum. Gene 150:205-206. [DOI] [PubMed] [Google Scholar]
- 22.Iwabuchi, M. 1995. Establishment of stress-tolerant Porphyra using mutated cell. Kaiyo Monthly 27:666-670. [Google Scholar]
- 23.Jongedijk, E., H. Tigelaar, J. S. C. Van Roekel, S. A. Bress-Vloemans, I. Dekker, P. J. M. Van den Elzen, B. J. C. Cornelissen, and L. S. Melchers. 1995. Synergistic activity of chitinases and β-1,3-glucanases enhances fungal resistance in transgenic tomato plants. Euphytica 85:173-180. [Google Scholar]
- 24.Katoh, K., A. Suzuki-Onzaki, T. Ohta, H. Ebine, M. Kumagai M. Fujimoto, and A. Kuninaka. 1983. Microbiological identification of single cell proteins on DNA-GC contents. Part II. Chemical determination of DNA-GC contents. Rep. Natl. Food Res. Inst. 43:79-89. [Google Scholar]
- 25.Kawamura, Y., and I. Aoto. 1995. Cultivation of Porphyra protoplast and its application in breeding. Kaiyo Monthly 27:661-665. [Google Scholar]
- 26.Kim, Y. J., and B. K. Hwang. 1997. Isolation of a basic 34 kilodalton β-1,3-glucanase with inhibitory activity against Phytophthora capsici from pepper stems. Physiol. Mol. Plant Pathol. 50:103-115. [Google Scholar]
- 27.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 28.Kitamura, E., H. Myouga, and Y. Kamei. 2002. Polysaccharolytic activities of bacterial enzymes which degrade the cell walls of Pythium porphyrae, a causative fungus of red rot disease in Porphyra yezoensis. Fish. Sci. 68:436-445. [Google Scholar]
- 29.Kubler, J. E., S. C. Minocha, and A. C. Mathieson. 1994. Transient expression of the GUS reporter gene in protoplasts of Porphyra miniata (Rhodophyta). J. Mar. Biotechnol. 1:165-169. [Google Scholar]
- 30.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 31.Matsumoto, M., Y. Kawashima, H. Tokuda, E. Fukui, T. Aoyama, and H. Kageyama. 1991. Intrageneric protoplast fusion in Porphyra. J. Phycol. 27(Suppl.):48. [Google Scholar]
- 32.Matsumoto, M., Y. Kawashima, and E. Fukui. 1995. Transduction of high temperature-tolerant to Porphyra by cell fusion. Kaiyo Monthly 27:677-682. [Google Scholar]
- 33.Michelmore, R. 2000. Genomic approaches to plant disease resistance. Curr. Opin. Plant Biol. 3:125-131. [DOI] [PubMed] [Google Scholar]
- 34.Migita, S. 1973. Studies on chytrid blight disease of Porphyra, p. 12-20. In Japanese Society of Fisheries Science (ed.), The disease of cultivated Porphyra. Koseisha Koseikaku, Tokyo, Japan.
- 35.Mitsuhara, I., H. Matsufuru, M. Ohshima, H. Kaku, Y. Nakajima, N. Murai, S. Natori, and Y. Ohashi. 2000. Induced expression of sarcotoxin IA enhanced host resistance against both bacterial and fungal pathogens in transgenic tobacco. Mol. Plant-Microbe Interact. 13:860-868. [DOI] [PubMed] [Google Scholar]
- 36.Miura, A. 1984. A new variety and new form of Porphyra (Bangiales, Rhodophyta) from Japan: Porphyra tenera Kjellman var. tamatsuensis Miura, var. nov. and P. yezoensis Ueda form narawaensis Miura form nov. J. Tokyo Univ. Fish. 71:1-4. [Google Scholar]
- 37.Mosmann, T. 1983. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. 65:55-63. [DOI] [PubMed] [Google Scholar]
- 38.Nakajima, H., T. Muranaka, F. Ishige, K. Akutsu, and K. Oeda. 1994. Fungal and bacterial disease resistance in transgenic plants expressing human lysozyme. Plant Cell Rep. 16:674-679. [DOI] [PubMed] [Google Scholar]
- 39.Niderman, T., I. Genetet, T. Bruyere, R. Gees, A. Stintzi, M. Legrand, B. Fritig, and E. Mosinger. 1995. Pathogenesis-related PR-1 proteins are antifungal. Plant Physiol. 108:17-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishizawa, Y., M. Suzuki, and T. Hibi. 1999. Current status of the disease-resistant transgenic plants. KASEAA 37:385-392. [Google Scholar]
- 41.Notoya, M. 1997. Diversity of life history in the genus Porphyra. Nat. Hist. Res. 3:47-56. [Google Scholar]
- 42.Oppenheimer, C. H., and C. E. ZoBell. 1952. The growth and viability of sixty-three species of marine bacteria as influenced by hydrostatic pressure. J. Mar. Res. 11:10-18. [Google Scholar]
- 43.Pessolini, M. C., D. R. Smith, B. Rivorire, J. McCormick, S. A. Hefta, S. T. Cole, and P. J. Brennan. 1994. Purification, characterization, gene sequence, and significance of a bacterioferritin from Mycobacterium leprae. J. Exp. Med. 180:319-327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Redenbach, M., H. M. Kieser, D. Denapaite, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 45.Rezzonico, E., N. Flury, F. Meins, and R. Beffa. 1998. Transcriptional down-regulation by abscisic acid of pathogenesis-related β-1,3-glucanase genes in tobacco cell cultures. Plant Physiol. 117:585-592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saito, N., and M. Nei. 1987. A neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 14:266-269. [DOI] [PubMed] [Google Scholar]
- 47.Saito, Y. 1973. Studies on green spot disease of Porphyra, p. 21-27. In Japanese Society of Fisheries Science (ed.), The disease of cultivated Porphyra. Koseisha Koseikaku, Tokyo, Japan.
- 48.Sasaki, M., and S. Sato. 1969. Composition of medium and cultural temperature of Pythium sp., a pathogenic fungus, of the “Akagusare” disease of cultivated Porphyra. Bull. Tohoku Reg. Natl. Fish. Res. Inst. 29:125-132. [Google Scholar]
- 49.Sasaki, M., and S. Sakurai. 1972. Comparative observation on the growth among the five strains in Pythium porphyrae under the same cultural condition. Bull. Tohoku Reg. Natl. Fish. Res. Inst. 32:83-87. [Google Scholar]
- 50.Sato, S., and M. Sasaki. 1973. Studies on red rot disease of Porphyra, p. 59-69. In Japanese Society of Fisheries Science (ed.), The disease of cultivated Porphyra. Koseisha Koseikaku, Tokyo, Japan.
- 51.Seal-Buurlage, M. B., A. S. Ponstein, S. A. Bres Vloemans, L. S. Melchers, P. J. M. Van den Elzen, and B. J. C. Cornelissen. 1993. Only specific tobacco (Nicotiana tabacum) chitinase and beta-1,3-glucanase exhibit antifungal activity. Plant Physiol. 101:857-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Takahashi, M. 1970. Identification of genus Pythium. Plant Prot. 24:339-346. [Google Scholar]
- 53.Takeuchi, T., H. Sawada, F. Tanaka, and I. Matsuda. 1996. Phylogenetic analysis of Streptomyces spp. causing potato scab based on 16S rDNA sequences Int. J. Syst. Bacteriol. 46:476-479. [DOI] [PubMed] [Google Scholar]
- 54.Tercero, J. A., R. A. Lacalle, and A. Jimenez. 1993. The pur8 gene from the pur cluster of Streptomyces alboniger encodes a highly hydrophobic polypeptide which confers resistance to puromycin. Eur. J. Biochem. 218:963-971. [DOI] [PubMed] [Google Scholar]
- 55.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uppalapati, S. R., and Y. Fujita. 1999. Carbohydrate regulation of attachment, encystment, and appressorium formation by Pythium porphyrae (Oomycota) zoospores on Porphyra yezoensis (Rhodophyta). J. Phycol. 36:359-366. [Google Scholar]
- 57.White, J. G., N. F. Lyons, A. J. Wakeham, A. Mead, and J. R. Green. 1994. Serological profiling of the fungal genus Pythium. Physiol. Mol. Plant Pathol. 44:349-361. [Google Scholar]
- 58.Williams, S. T., M. Goodfellow, and G. Alderson. 1989. Genus Streptomyces Waksman and Henrichi 1943, 339AL, p. 2451-2492. In S. T. Williams, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 4. The Williams and Wilkins Co., Baltimore, Md. [Google Scholar]
- 59.Young, N. D. 2000. The genetic architecture of resistance. Curr. Opin. Plant Biol. 3:285-290. [DOI] [PubMed] [Google Scholar]
- 60.Zhu, Q., E. A. Maher, S. Masoud, R. A. Dixon, and C. J. Lamb. 1994. Enhanced protection against fungal attack by constitutive co-expression of chitinase and glucanase genes in transgenic tobacco. Bio/Technology 12:807-812. [Google Scholar]