Abstract
Counts of Escherichia coli cells in water indicate the potential presence of pathogenic microbes of intestinal origin but give no indication of the sources of the microbial pollution. The objective of this research was to evaluate methods for differentiating E. coli isolates of livestock, wildlife, or human origin that might be used to predict the sources of fecal pollution of water. A collection of 319 E. coli isolates from the feces of cattle, poultry, swine, deer, goose, and moose, as well as from human sewage, and clinical samples was used to evaluate three methods. One method was the multiple-antibiotic-resistance (MAR) profile using 14 antibiotics. Discriminant analysis revealed that 46% of the livestock isolates, 95% of the wildlife isolates, and 55% of the human isolates were assigned to the correct source groups by the MAR method. Amplified fragment length polymorphism (AFLP) analysis, the second test, was applied to 105 of the E. coli isolates. The AFLP results showed that 94% of the livestock isolates, 97% of the wildlife isolates, and 97% of the human isolates were correctly classified. The third method was analysis of the sequences of the16S rRNA genes of the E. coli isolates. Discriminant analysis of 105 E. coli isolates indicated that 78% of the livestock isolates, 74% of the wildlife isolates, and 80% of the human isolates could be correctly classified into their host groups by this method. The results indicate that AFLP analysis was the most effective of the three methods that were evaluated.
Water quality in many lakes and rivers has been impaired by the presence of high levels of fecal coliform bacteria, which is indicative of contamination by feces (1, 18). Such contamination brings the threat of infection for people who use the water for drinking, bathing, or watering fruits and vegetables. Underlying this concern are numerous reports of waterborne outbreaks of disease involving fecal organisms such as Escherichia coli O157:H7, Campylobacter jejuni, Salmonella, Vibrio cholerae, and shigellae (4, 10, 18, 22). Identification of the source of the bacterial contamination is an essential first step in seeking to control fecal contamination of water. In particular, it is important to determine whether the source of fecal contamination is of human, livestock, or wildlife origin, as microorganisms of human origin are regarded as having greater potential to cause disease in humans (29).
The multiple-antibiotic-resistance (MAR) test is based on detection of bacterial resistance to a panel of antibacterial agents. The MAR patterns reflect the selective pressures imposed on the gastrointestinal floras of humans and animals during antibiotic use. The MAR test has been reported to be capable of identifying the sources of fecal streptococcal contamination in water (14) and distinguishing between E. coli strains from specific point sources, such as industrial and municipal effluents, and strains from nonpoint sources, such as land runoff, that are dispersed over wide areas (27).
Rapid advances in nucleic acid-based technologies may permit discrimination among E. coli strains from different hosts (30). These methods include amplified fragment length polymorphism (AFLP) analysis and 16S rRNA gene sequencing (5, 16).
AFLP analysis is a relatively new and sensitive fingerprinting technique, which generates highly reproducible results and has superior discriminatory power (32). This method is based on selective amplification of a subset of DNA fragments from a digest of total genomic DNA (38). AFLP analysis has been used in the genotyping of a variety of bacterial species, such as Campylobacter spp. (8, 23), Pseudomonas spp. (32, 33), Chlamydia spp., V. cholerae, Mycoplasma spp. (20), and E. coli (3), and has been shown to discriminate bacteria down to the strain level. In addition, fluorescent AFLP analysis can be easily automated, digitized, and standardized for long-term database buildup, cross-referencing, and exchange of data between laboratories. For these reasons, AFLP analysis has been widely used for bacterial taxonomic, diagnostic, epidemiological, and source-tracking applications (6, 17, 42). However, the feasibility of using AFLP fingerprinting to predict the host origins of E. coli strains has not been investigated previously.
Sequence analysis of bacterial 16S rRNA genes, amplified directly from bacterial cultures or complex communities, provides an efficient strategy for differentiating bacterial species. This technique is becoming increasingly automated and, therefore, is a reasonable alternative for genotyping bacterial isolates. It has been successfully applied in various ecosystems to investigate the bacterial composition of complex microbiota, such as the human colon (40), to compare strains from different reference collections (28), and to identify bacterial pathogens (24, 41).
The objective of this study was to evaluate the three test procedures identified above for effectiveness in determining the species of origin of E. coli. This bacterium was selected because it is a common environmental bacterium and a specific indicator of fecal pollution in water.
MATERIALS AND METHODS
Sample collection and identification.
A total of 319 E. coli isolates were collected from known sources in widespread locations in southern Ontario (Table 1). The samples from which E. coli strains were isolated included fresh feces from cattle (dairy and beef), poultry (chickens and turkeys), and wildlife; lagoons of swine operations; and influents of municipal sewage treatment plants. All samples were streaked on MacConkey agar plates, which were incubated at 37°C overnight. A single E. coli colony was selected from each fecal sample and from each sample of municipal sewage. The initial selection was for lactose-fermenting colonies, which were confirmed to be E. coli by the Cathra RepliAnalyzer system (Oxoid Inc., Nepean, Ontario, Canada). It was often necessary to test two or three colonies from municipal sewage samples in order to obtain a single E. coli colony. The E. coli isolates and their host sources are shown in Table 1.
TABLE 1.
E. coli isolates used in this study
| Host | Designation prefix(es) | No. of isolates |
|---|---|---|
| Human | H, HAa | 96 |
| Bovine beef | BB | 25 |
| Bovine dairy | BD | 32 |
| Pig | P | 19 |
| Geese | G | 20 |
| Chicken | C | 10 |
| Turkey | T | 34 |
| Moose | M | 2 |
| Deer | DP, DRb | 81 |
| Total | 319 |
H, isolates from Guelph sewage plant; HA, isolates from human fecal samples.
DP, deer isolates from Pinery Provincial Park, Ontario, Canada; DR, deer isolates from Rondeau Provincial Park, Ontario, Canada.
MAR test.
The following antibiotics and concentrations (in micrograms per milliliter) were used in this study: ampicillin, 16; cephalothin, 16; streptomycin, 16; neomycin, 32; kanamycin, 16; gentamicin, 8; tetracycline, 16; chloramphenicol, 16; sulfathiazole, 500; cotrimoxazole, 10; apramycin, 8; ceftiofur, 2; spectinomycin, 16; and tilmycosin, 64. The 14 antibiotics were chosen in order to permit comparisons with previous studies involving the use of the MAR index and also to reflect the antibiotics to which farm animals are exposed. The MAR tests were performed by using the Cathra RepliAnalyzer system (Oxoid) according to the manufacturer's instructions. Cultures to be tested were replica plated on each of 14 antibiotic plates plus a control plate which lacked antibiotic. E. coli strain ATCC 25922 was used as a drug-sensitive negative control. After inoculation, the plates were incubated at 35°C for 18 to 24 h, and the results were recorded. The MAR patterns were converted to binary codes on the basis of sensitivity or resistance for discriminant analysis (DA) (the DISCRIM procedure) using Statistical Analysis System (SAS) software (version 8.1 for Unix; SAS Institute Inc.). The MAR index for each group was determined by the method of Kaspar et al. (19). The MAR index for a host group was calculated by summing the numbers of drugs to which each isolate was resistant and dividing the resulting number by the product of the number of antibiotics and the number of isolates tested. This index was a measure of the extent of drug resistance for isolates in the group.
Fluorescent AFLP fingerprinting.
E. coli cells were collected from 0.5-ml overnight cultures grown in brain heart infusion broth (Becton Dickinson, Oakville, Ontario, Canada) at 37°C, and the genomic DNA were extracted from the cell pellets with a DNeasy tissue kit (QIAGEN, Missisauga, Ontario, Canada). A 50-ng portion of each genomic DNA was digested with EcoRI and MseI, and the resulting DNA fragments were ligated to EcoRI and MseI adapters of an AFLP microbial fingerprinting kit (Applied Biosystems, Foster City, Calif.) by following the specifications of the manufacturer. The EcoRI-MseI fragments tagged with specific adapters were then selectively amplified by using the GeneAmp 9600 PCR system (Applied Biosystems). Two pairs of primers, EcoRI-A (labeled with the fluorescent dye FAM) plus MseI-G and EcoRI-C (labeled with the fluorescent dye NED) plus MseI-CA, were used in this work (38). The following thermocycling conditions were used: one cycle of 2 min at 96°C, 30 s at 65°C, and 2 min at 72°C; eight cycles of 1 s at 94°C, 30 s at 64 to 57°C (1°C touching down from the previous cycle), and 2 min at 72°C; 28 cycles of 1 s at 94°C, 30 s at 56°C, and 2 min at 72°C; and a final incubation at 60°C for 30 min. The amplified DNA products were separated with a 5% (wt/vol) Long Ranger gel (J. T. Baker, Toronto, Ontario, Canada) by using 0.6× Tris-borate-EDTA buffer (Bio-Rad, Missisauga, Ontario, Canada) for 3.5 h and an ABI 377 DNA sequencer (Applied Biosystems). The GSROX 500 size standard (Applied Biosystems) was included as an internal marker.
The AFLP results were captured by using ABI PRISM GeneScan 3.1 software (Applied Biosystems), and the fragment data were tabulated by size and fluorescent intensity by using the ABI PRISM GenoTyper 2.5 software (Applied Biosystems). All electropherograms were visually inspected for polymorphous peaks before the table file was produced. Only fragments in the range from 50 to 500 bp were considered further because the resolving capability of the sequencing gel is generally good in this range. The table files generated with the GenoTyper software were scanned for discriminatory bands by using a Perl script developed for this research. Bands that appeared predominantly in one or two of the three host groups (livestock, wildlife, and human) but were absent from or less frequent in the other groups were considered to be discriminatory bands. The band pattern of each isolate was converted to binary codes by using another Perl script based on the presence or absence of the discriminatory bands (1 for presence and 0 for absence). Further analysis of the AFLP data was performed by using other Perl programs in combination with SAS software and TreeCon, version 1.3b (37). An unrooted phylogenetic tree was visualized with TreeView 1.6.5 software (26).
Sequencing of 16S rRNA genes.
Bacterial genomic DNA was extracted from the same 105 bacterial isolates that were used for the AFLP analysis. PCR amplification of the partial 16S rRNA gene was performed by using a forward primer (5′-AATTGAAGAGTTTGATCATG-3′) and a reverse primer (5′-CTCTACGCATTTCACCGCTAC-3′). Each PCR mixture (25 μl) contained 10 mM Tris-HCl (pH 8.3), 2 mM MgCl2, 100 ng of genomic DNA, each primer at a concentration of 0.8 μM, each deoxynucleoside triphosphate at a concentration of 2 mM, and 2 U of Taq DNA polymerase (Applied Biosystems). The PCRs were performed with a PE-9600 thermocycler (Applied Biosystems) under the following conditions: denaturation for 5 min at 95°C; 30 cycles of 40 s at 94°C, 30 s at 56°C, and 30 s at 72°C; and a final extension step of 7 min at 72°C. The resultant purified PCR products were sequenced by using the forward primer. The amplicon size varies slightly among different E. coli isolates. In the case of E. coli K-12, PCR amplification of the 16S rRNA gene (GenBank accession number AE000452) with the designated primers produces a 704-bp DNA fragment. The 543 bases of nucleotide sequence corresponding to positions 68 to 610 of the E. coli K-12 16S rRNA gene were sequenced. Sequences were aligned and a phylogenetic tree was constructed by using ClustalW software, version 1.7 (35). Sequence data were then converted into numeric data by using the Replace function of Microsoft Word. Conversion from nucleotide bases to numbers was based on the following relationships: A = 1, C = 2, G = 3, T = 4, N = 5, and gap = 6. Spaces were used between numbers. For instance, the sequence ATGC-N was converted to numeric pattern 1 4 3 2 6 5. The resulting data were further analyzed by using the DA procedure of SAS (the DA procedure requires numeric data for analysis).
RESULTS
MAR test.
Of the 319 E. coli isolates examined, 178 (55.8%) were resistant to one or more antibiotics (Table 2). Eighty-two distinctive antibiotic resistance patterns were observed altogether. Livestock isolates, especially poultry and swine isolates, displayed higher percentages of resistance to most of the antibiotics tested than did human and wildlife isolates. The average MAR indices for human, livestock, and wildlife isolates were 0.1339, 0.0966, and 0.027, respectively. Among the livestock isolates, the MAR indices were highest for the turkey isolates (0.3298) and pig isolates (0.3008), intermediate for the chicken isolates (0.1286), and lower for the cattle isolates (0.0400 for beef isolates and 0.0513 for dairy isolates), whose MAR indices were similar to those of wildlife isolates. These results are consistent with other reports that the MAR indices of fecal E. coli from wild animals were generally low, while human and livestock isolates had higher indices (19, 21).
TABLE 2.
Number of E. coli isolates resistant to each antimicrobial agent
| Antimicrobial agent | No. of isolates resistanta
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Human (n = 96) | Beef (n = 25) | Dairy (n = 32) | Chicken (n = 10) | Pig (n = 19) | Turkey (n = 34) | Deer (n = 81) | Goose (n = 20) | Moose (n = 2) | |
| Ampicillin | 42 | 1 | 2 | 0 | 6 | 18 | 0 | 1 | 0 |
| Cephalothin | 14 | 4 | 1 | 0 | 2 | 3 | 4 | 1 | 0 |
| Streptomycin | 13 | 1 | 3 | 4 | 13 | 28 | 1 | 0 | 0 |
| Neomycin | 0 | 0 | 1 | 0 | 5 | 14 | 0 | 0 | 0 |
| Kanamycin | 2 | 0 | 1 | 0 | 6 | 15 | 0 | 0 | 0 |
| Gentamicin | 3 | 0 | 0 | 1 | 0 | 4 | 0 | 0 | 0 |
| Tetracycline | 16 | 1 | 4 | 2 | 17 | 29 | 2 | 0 | 0 |
| Chloramphenicol | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Sulfathiazole | 15 | 3 | 4 | 8 | 11 | 24 | 2 | 0 | 0 |
| Cotrimoxazole | 7 | 0 | 2 | 0 | 2 | 2 | 0 | 0 | 0 |
| Apramycin | 16 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 0 |
| Ceftiofur | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
| Spectinomycin | 22 | 4 | 2 | 3 | 11 | 8 | 5 | 2 | 0 |
| Tilmycosin | 21 | 0 | 3 | 0 | 6 | 9 | 15 | 4 | 0 |
The MAR indices for the human, beef, dairy, chicken, pig, turkey, deer, goose, and moose isolates are 0.1339, 0.04, 0.0513, 0.1286, 0.3008, 0.3298, 0.0265, 0.0321, and 0, respectively. The MAR indices for human and chicken isolates are each significantly different from the MAR indices for beef, dairy, pig, turkey, deer and goose isolates; the MAR indices for beef, dairy, deer, and goose isolates are each significantly different from the MAR indices for human, chicken, pig, and turkey isolates; and the MAR indices for pig and turkey isolates are each significantly different from the MAR indices for human, beef, dairy, chicken, deer, and goose isolates. The value for the moose isolates was not included in the analysis of significant differences because of the low number of samples. When the isolates of animal origin are grouped as livestock and wildlife isolates, the MAR indices for these groups are 0.0966 and 0.027, respectively.
The DISCRIM procedure of SAS classifies observations into two or more possible groups on the basis of quantitative variables. DA differs from cluster analysis (CA) in that all varieties of DA require prior knowledge of the classes, usually in the form of a sample from each class. In CA, the data do not include information on class membership; the purpose is simply to construct a classification. By using DA with the 319 E. coli isolates, the average rate of correct classification (ARCC) for all isolates was 33.9% when all isolates were reclassified into nine host groups (81 of 319 isolates). The ARCC was calculated by dividing the number of isolates which were correctly assigned to a given group by the total number of isolates in that group tested and multiplying by 100%. The probability that an isolate fell into one of nine categories by chance alone is 11.1%. Moose and chicken isolates were well classified (100 and 80%, respectively), while goose, bovine dairy, and deer isolates were classified poorly (0, 0, and 14.81%, respectively). Most of the goose and bovine dairy isolates were misclassified into moose categories because these groups displayed similar MAR profiles (i.e., zero resistance to many of the antibiotics) (Table 3).
TABLE 3.
Classification of E. coli isolates by source based on MAR profiles and DA analysis
| Source | % of isolates classified asa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Human (n = 96) | Beef (n = 25) | Dairy (n = 32) | Chicken (n = 10) | Pig (n = 19) | Turkey (n = 34) | Deer (n = 81) | Goose (n = 20) | Moose (n = 2) | |
| Human | 48.96 | 5.21 | 1.04 | 2.08 | 9.38 | 3.13 | 2.08 | 1.04 | 27.08 |
| Beef cattle | 0 | 24.00 | 0 | 8.00 | 0 | 4.00 | 0 | 0 | 64.00 |
| Dairy cattle | 3.13 | 6.25 | 0 | 0 | 9.38 | 3.13 | 9.38 | 0 | 68.75 |
| Chicken | 0 | 10.00 | 0 | 80.00 | 10.00 | 0 | 0 | 0 | 0 |
| Pig | 0 | 5.26 | 5.26 | 0 | 63.16 | 26.32 | 0 | 0 | 0 |
| Turkey | 2.94 | 0 | 0 | 5.88 | 14.71 | 61.76 | 2.94 | 0 | 11.76 |
| Deer | 1.23 | 7.41 | 0 | 1.23 | 1.23 | 1.23 | 14.81 | 2.47 | 70.37 |
| Goose | 5.00 | 10.00 | 0 | 0 | 0 | 0 | 15.00 | 0 | 70.00 |
| Moose | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100.00 |
When human, livestock, and wildlife species groups were distinguished, 55.21% of the isolates in the human species group (n = 96) were classified as human isolates, 10.42% of the isolates were classified as livestock isolates, and 34.38% of the isolates were classified as wildlife isolates; in the livestock species group (n = 120), 6.67% of the isolates were classified as human isolates, 45.83% of the isolates were classified as livestock isolates, and 47.50% of the isolates were classified as wildlife isolates; and in the wildlife species group (n = 103), 1.94% of the isolates were classified as human isolates, 2.91% of the isolates were classified as livestock isolates, and 95.15% of the isolates were classified as wildlife isolates. When human and nonhuman species groups were distinguished, 56.25% of the isolates in the human species group (n = 96) were classified as human isolates and 43.75% of the isolates were classified as nonhuman isolates; in the nonhuman species group (n = 223), 7.62% of the isolates were classified as human isolates and 92.38% of the isolates were classified as nonhuman isolates.
When deer, goose, and moose isolates were pooled together as the wildlife group and swine, turkey, chicken, and bovine isolates were pooled together as the livestock group, the ARCC were 55.21% for the human isolates, 45.83% for the livestock isolates, and 95.15% for the wildlife isolates (Table 3). The most common misclassifications were between livestock and wildlife isolates and between human and wildlife isolates. When all nonhuman sources were pooled so that each isolate was classified as either human or nonhuman and DA was performed, the ARCC for human increased to 56.25% and the ARCC for nonhuman isolates increased to 92.38% (Table 3).
Fluorescent AFLP analysis.
Two selective primer combinations (EcoRI-A plus MseI-G and EcoRI-C plus MseI-CA) were used with a subset of 105 E. coli isolates, which were randomly selected from every host group. Primers EcoRI-A and MseI-G generated 35 to 40 fragments that ranged in size from 50 to 500 bp, while primers EcoRI-C and MseI-CA generated 10 to 20 fragments. The average band intensities (represented by peak heights) were 2,038 fluorescence units for the EcoRI-A-MseI-G primer set and 2,166 fluorescence units for the EcoRI-C-MseI-CA primer set under the test conditions. This indicated that most of the bands were readily distinguishable from background noise, which was less than 200 fluorescence units.
To evaluate whether the AFLP results could be used to correctly predict the host origins of E. coli isolates, the AFLP data for the 105 E. coli isolates were converted to binary codes by using the Perl scripts referred to in Materials and Methods. Altogether, 101 distinct band patterns were observed with data obtained with primers EcoRI-A and MseI-G based on 87 selected discriminatory bands, whereas 85 distinct band patterns were observed with data obtained with primers EcoRI-C and MseI-CA based on 74 selected discriminatory bands. These observations indicated that most of the E. coli isolates produced unique band patterns.
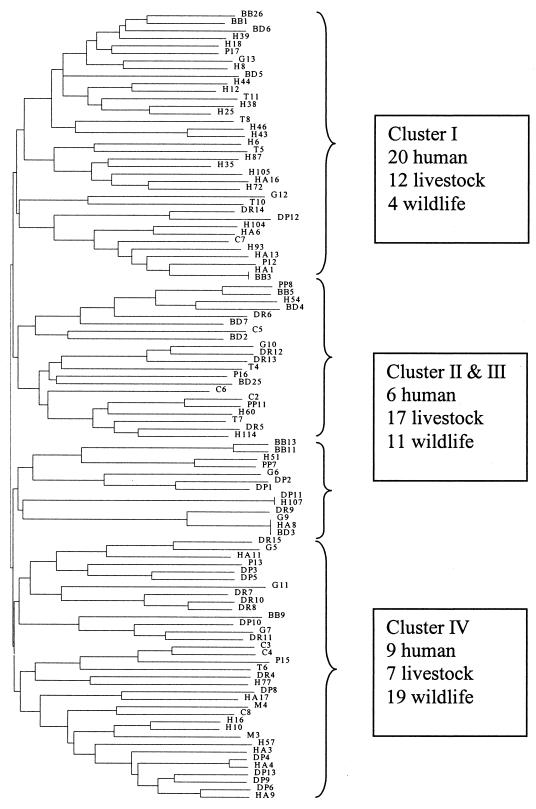
CA of the AFLP data obtained with primers EcoRI-A and MseI-G resulted in the dendrogram shown in Fig. 1. Four major phylogenetic groups (clusters I through IV) were evident. Twenty of the 35 human isolates were present in the largest cluster (cluster I), along with 12 livestock isolates and four wildlife isolates. The second largest branch was cluster IV, which contained nine human isolates, seven livestock isolates, and 19 wildlife isolates. Clusters II and III together contained six human isolates, 17 livestock isolates, and 11 wildlife isolates. Cluster I contained more than one-half of the human isolates, clusters II and III contained more livestock isolates, and cluster IV essentially favored wildlife isolates. The AFLP data obtained with primers EcoRI-C and MseI-CA were also examined by using CA, but no significant conclusions could be drawn (data not shown). These results indicated that CA of AFLP data might not be suitable for predicting fecal contamination sources.
FIG. 1.
CA of fluorescent AFLP results obtained by using primers EcoRI-A and MseI-G for 105 E. coli isolates from various host sources. Bacterial designation prefixes based on host origins are explained in Table 1. The tree was constructed with TREECON, version 1.3b.
Whereas CA of AFLP data is commonly reported, regrouping and predicting bacterial contamination sources based on AFLP results are generally not done. When the binary code files were analyzed by using the DISCRIM procedure of SAS, satisfactory grouping was achieved for the 105 E. coli isolates examined (Tables 4 and 5). For instance, 97.1% of the human isolates, 94.4% of the livestock isolates, and 97.1% of the wildlife isolates were correctly classified when the data generated with primers EcoRI-A and MseI-G were analyzed, whereas 71.4% of the human isolates, 77.8% of the livestock isolates, and 94.1% of the wildlife isolates were correctly classified when data obtained with primers EcoRI-C and MseI-CA were analyzed (Tables 4 and 5). These results suggest that the AFLP method could be used as an effective way to predict the sources of contaminating E. coli isolates.
TABLE 4.
DA of AFLP data obtained with primers EcoRI-A and MseI-G
| Source | % of isolates classified asa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Human (n = 35) | Beef (n = 7) | Dairy (n = 7) | Chicken (n = 7) | Pig (n = 8) | Turkey (n = 7) | Deer (n = 25) | Goose (n = 7) | Moose (n = 2) | |
| Human | 97.14 | 0 | 0 | 0 | 0 | 0 | 2.86 | 0 | 0 |
| Beef cattle | 14.29 | 85.71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Dairy cattle | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chicken | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 | 0 |
| Pig | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 | 0 |
| Turkey | 0 | 0 | 0 | 0 | 0 | 100 | 0 | 0 | 0 |
| Deer | 4.00 | 0 | 0 | 0 | 0 | 0 | 96.00 | 0 | 0 |
| Geese | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 | 0 |
| Moose | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 |
When human, livestock, and wildlife species groups were distinguished, 97.14% of the isolates in the human species group (n = 35) were classified as human isolates, and 2.86% of the isolates were classified as wildlife isolates; in the livestock species group (n = 36), 2.78% of the isolates were classified as human isolates, 94.44% of the isolates were classified as livestock isolates, and 2.78% of the isolates were classified as wildlife isolates; and in the wildlife species group (n = 34), 2.94% of the isolates were classified as human isolates, and 97.06% of the isolates were classified as wildlife isolates. When human and nonhuman species groups were distinguished, 97.14% of the isolates in the human species group (n = 35) were classified as human isolates, and 2.86% of the isolates were classified as nonhuman isolates; in the nonhuman species group (n = 70), 1.43% of the isolates were classified as human isolates, and 98.57% of the isolates were classified as nonhuman isolates.
TABLE 5.
DA of AFLP data obtained with primers EcoRI-C and MseI-CA
| Source | % of isolates classified asa:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Human (n = 35) | Beef (n = 7) | Dairy (n = 7) | Chicken (n = 7) | Pig (n = 8) | Turkey (n = 7) | Deer (n = 25) | Goose (n = 7) | Moose (n = 2) | |
| Human | 71.43 | 5.71 | 5.71 | 0 | 8.57 | 0 | 2.86 | 2.86 | 2.86 |
| Beef cattle | 0 | 71.43 | 0 | 0 | 0 | 0 | 14.29 | 14.29 | 0 |
| Dairy cattle | 0 | 14.29 | 71.43 | 0 | 0 | 0 | 0 | 14.29 | 0 |
| Pig | 0 | 0 | 0 | 87.50 | 0 | 0 | 0 | 12.50 | 0 |
| Turkey | 14.29 | 14.29 | 0 | 0 | 71.43 | 0 | 0 | 0 | 0 |
| Chicken | 0 | 14.29 | 0 | 0 | 0 | 85.71 | 0 | 0 | 0 |
| Deer | 0 | 4.00 | 8.00 | 0 | 0 | 0 | 84.00 | 4.00 | 0 |
| Geese | 0 | 0 | 0 | 0 | 0 | 0 | 14.29 | 85.71 | 0 |
| Moose | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100 |
When human, livestock, and wildlife species groups were distinguished, 71.43% of the isolates in the human species group (n = 35) were classified as human isolates, 8.57% of the isolates were classified as livestock isolates, and 20.0% of the isolates were classified as wildlife isolates; in the livestock species group (n = 36), 8.33% of the isolates were classified as human isolates, 77.78% of the isolates were classified as livestock isolates, and 13.89% of the isolates were classified as wildlife isolates; and in the wildlife species group (n = 34), 2.94% of the isolates were classified as human isolates, 2.94% of the isolates were classified as livestock isolates, and 94.12% of the isolates were classified as wildlife isolates. When human and nonhuman species groups were distinguished, 80% of the isolates in the human species group (n = 35) were classified as human isolates and 20% of the isolates were classified as nonhuman isolates; in the nonhuman species group (n = 70), 8.57% of the isolates were classified as human isolates and 91.43% of the isolates were classified as nonhuman isolates.
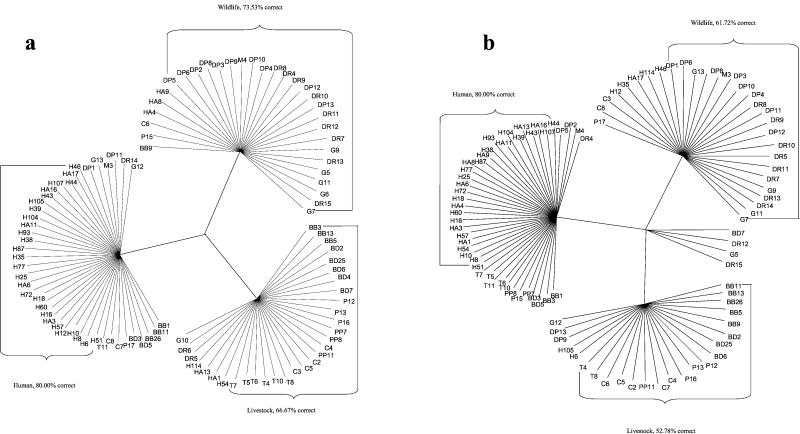
To further verify the predictive value of DA, another Perl program was developed to assign each individual sample to one of three possible groups (human, livestock, and wildlife) on the basis of AFLP band patterns. This program treats each band listed in the criterion file independently. The program first screened for discriminatory bands from all known samples and established a criterion file containing a list of selected bands. If a band was restricted to or predominantly present in one or two of the three possible groups, it was selected into the criterion file. The program then compared the band pattern of an unknown sample and the band list in the criterion file. If the unknown sample contained a band listed in the criterion file, the program calculated three likelihood values that this sample might fall into each of the three groups. After all the bands on the criterion band list were examined, the likelihood values corresponding to each of the three possible groups were combined. The unknown sample was classified into the group that had the greatest likelihood value. The best way to assess the reclassification accuracy of this Perl program is to examine the rate of correct classification of E. coli isolates from known sources. When the Perl program was used to reclassify the 105 E. coli isolates mentioned above, 80% of the human isolates, 67% of the livestock isolates, and 74% of the wildlife isolates could be correctly classified with data obtained with primers EcoRI-A and MseI-G (Table 6). When the results from the Perl program analysis were visualized by using the TreeView software, each individual isolate was shown in the category to which it most likely belonged (Fig. 2a). A slightly lower rate of correct classification was obtained when data obtained with primers EcoRI-C and MseI-CA were analyzed (Table 6 and Fig. 2b). The advantage of using the Perl program is that it does not require a large database to predict the results for unknown samples because it is a band-by-band-based method instead of a pattern-by-pattern-based method. It also displays a confidence rate that equals the number of isolates correctly classified into a given group divided by the total number of isolates classified into that group based on known samples (Table 6).
TABLE 6.
Reclassification analysis of AFLP data using a Perl program
| Group | Primers EcoRI-A and MseI-G
|
Primers EcoRI-C and MseI-CA
|
||
|---|---|---|---|---|
| ARCC (%) | Confidence (%)a | ARCC (%) | Confidence (%) | |
| Human | 80.00 | 65.12 | 80.00 | 63.64 |
| Livestock | 66.67 | 77.42 | 52.78 | 79.17 |
| Wildlife | 73.53 | 80.65 | 61.72 | 72.41 |
Confidence = (number of isolates which were correctly classified into one group/total number of isolates that were classified into that group) × 100%.
FIG. 2.
Unrooted tree generated by a Perl program showing that most of the E. coli isolates were correctly classified into the host groups. The tree was visualized by using TreeView 1.6.5 software (26). Bacterial designation prefixes based on host origins are explained in Table 1. (a) AFLP results generated with primers EcoRI-A and MseI-G; (b) AFLP results generated with primers EcoRI-C and MseI-CA.
16S rRNA gene sequencing.
The partial 16S rRNA genes of the 105 E. coli isolates used in the AFLP analysis were amplified. This part of the 16S rRNA gene was chosen for analysis because it is more variable and is routinely used in our lab for reliable identification of unknown bacterial species. All sequences showed a high degree of similarity to each other and to the 16S rRNA gene of E. coli K-12. CA using ClustalW or the neighbor-joining method of the Phylip package (9) indicated that the 16S rRNA gene sequencing analysis was not sufficiently discriminative. No obvious correlation was observed between 16S rRNA gene sequences and host sources of isolation, except that 13 of the human isolates were clustered together (data not shown). However, when the multiple-aligned-sequence file was converted into a numeric file and the numeric file was further analyzed by using the DA procedure of SAS, remarkably good rates of correct classification were achieved (Table 7). Pooling of related animal sources increased the rate of correct classification, although the rates were not as high as the AFLP analysis rates.
TABLE 7.
DA analysis of the partial 16S rRNA genes from 105 E. coli isolatesa
| Source | % of isolates classified asb:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Human (n = 35) | Beef (n = 7) | Diary (n = 7) | Chicken (n = 7) | Pig (n = 8) | Turkey (n = 7) | Deer (n = 25) | Goose (n = 7) | Moose (n = 2) | |
| Human | 77.14 | 0 | 0 | 0 | 0 | 11.43 | 8.57 | 2.86 | 0 |
| Beef cattle | 14.29 | 85.71 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Dairy cattle | 0 | 0 | 71.43 | 0 | 0 | 14.29 | 0 | 14.29 | 0 |
| Chicken | 0 | 0 | 0 | 85.71 | 0 | 0 | 14.29 | 0 | 0 |
| Pig | 37.50 | 0 | 0 | 0 | 37.50 | 12.50 | 12.50 | 0 | 0 |
| Turkey | 28.57 | 0 | 0 | 0 | 14.29 | 42.86 | 14.29 | 0 | 0 |
| Deer | 20.83 | 0 | 0 | 4.17 | 0 | 0 | 75.00 | 0 | 0 |
| Goose | 12.50 | 0 | 0 | 0 | 0 | 0 | 0 | 87.50 | 0 |
| Moose | 50.00 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50.00 |
Partial nucleotide sequences (543 bp) corresponding to positions 68 to 610 of the E. coli K-12 16S rRNA gene (GenBank accession number AE000452) were analyzed.
When human, livestock, and wildlife species groups were distinguished, 80.00% of the isolates in the human species group (n = 35) were classified as human isolates, 8.57% of the isolates were classified as livestock isolates, and 11.43% of the isolates were classified as wildlife isolates; in the livestock species group (n = 36), 11.11% of the isolates were classified as human isolates, 77.78% of the isolates were classified as livestock isolates, and 11.11% of the isolates were classified as wildlife isolates; and in the wildlife species group (n = 34), 14.71% of the isolates were classified as human isolates, 11.76% of the isolates were classified as livestock isolates, and 73.53% of the isolates were classified as wildlife isolates. When human and nonhuman species groups were distinguished, 80.00% of the isolates in the human species group (n = 35) were classified as human isolates and 20.00% of the isolates were classified as nonhuman isolates; in the nonhuman species group (n = 70), 11.43% of the isolates were classified as human isolates and 88.57% of the isolates were classified as nonhuman isolates.
DISCUSSION
The purpose of this study was to identify a procedure that could be used to differentiate E. coli isolates of various host origins, thereby allowing development of a system for determining the sources of fecal pollution. A total of 319 E. coli isolates were collected from nine known host sources in widespread locations in southern Ontario, and three typing techniques were evaluated. Our results indicate that fluorescent AFLP fingerprinting provided the highest level of discriminatory capacity for differentiating various E. coli isolates and for identifying the fecal contamination sources.
Of the three methods investigated in this work, the MAR test is the simplest and the least expensive approach. Several researchers have previously used the MAR profiles of streptococci to identify fecal pollution sources (13, 14, 39). Wiggins reported that the ARCC was 84% when streptococcal isolates were pooled into four possible categories (cattle, human, poultry, and wildlife) (39), whereas Hagedorn et al. (13) demonstrated that the average correct identification rate for fecal Streptococcus sources was 87% for 7,058 isolates. These results are better than the 64.58% accuracy obtained if all 319 E. coli isolates in the present study were pooled into three possible categories (human, livestock, and wildlife). Several factors may contribute to the difference in the results. First, fecal Streptococcus was investigated rather than E. coli, and second, different antibiotic panels were used. A third contributing factor was the difference in the diversities of the bacterial collections. In the present study, animal fecal samples were collected from farms or provincial parks located over a wide geographic area and only one E. coli isolate was selected from each animal fecal sample, so that repetitious selection of the same clone of E. coli was avoided. Consequently, a more diversified and representative collection of E. coli isolates was utilized. This sampling protocol could produce a more heterogeneous collection of bacterial isolates than other protocols, in which multiple bacterial isolates were derived from each fecal sample (39).
It is interesting that E. coli isolates from beef and dairy cattle exhibited very low MAR indices which were similar to those of wildlife. Most of the bovine isolates were not resistant to any of the antibiotics tested. This may reflect the fact that antibiotics are less widely used on dairy and beef farms than in poultry and swine operations. However, the overall rate of correct classification for the three sources was significantly weighted because most of the bovine isolates were misclassified and placed into the wildlife group. If bovines were considered members of the wildlife group, the rates of correct classification were dramatically increased for three sources (human, 54.17%; livestock, 73.02%; and wildlife, 94.38%). Interestingly, most (>80) of the wildlife samples were obtained from deer, which, like cattle, are ruminants. In another study of fecal streptococci, Hagedorn et al. also reported that the antibiotic resistance patterns of beef cow and wildlife isolates are very similar (13). It is unclear if the low antibiotic resistance of these E. coli isolates is associated with the diets of their hosts.
Bacteria gain resistance to antimicrobial agents primarily through three mechanisms: (i) acquisition of antibiotic resistance genes through mobile elements, such as plasmids and insertion sequences (31); (ii) mutations in genes responsible for antibiotic uptake or binding sites (34); and (iii) activation of the MAR locus (mar) in the bacterial chromosome (2, 12). Over time, the patterns of antibiotic resistance in bacterial communities may change dramatically depending on geographic location, farm management, and levels and kinds of antibiotics used in the local human population and livestock husbandry (11, 15, 25). For this reason, the MAR test might be more suitable for microbial source tracking or surveillance in designated geographic locations and self-contained systems, such as estuaries, provided that a comprehensive and representative criterion database is locally established beforehand.
AFLP fingerprinting has been shown to be a powerful tool for molecular characterization of various bacterial species (7, 36). However, the real power of this technique may rely on tailoring to account for genomic differences of various organisms (genome size, G+C content, and DNA modification). The choice of the restriction enzymes and selective primers used is crucial to the outcome of AFLP analysis. In this study, two restriction enzymes (EcoRI and MseI) and two sets of primers (EcoRI-A plus MseI-G and EcoRI-C plus MseI-CA) were selected. Since the genomes of three E. coli strains have been completely sequenced, the number of bands and fragments generated by using the designated adapters and selective primers could be predicted (i.e., electronic AFLP analysis). Based on the complete sequence information deposited in GenBank, the combination of restriction enzymes EcoRI and MseI should generate a total of 21,852 fragments for E. coli K-12 (accession number U00096), 26,131 fragments for E. coli O157:H7 (NC_002695), and 26,313 fragments for E. coli O157:H7 strain EDL933 (AE005174). Theoretically, 1,234, 1,502, and 2,939 of these fragments should contain one EcoRI end and one MseI end, respectively; the rest of the fragments should contain either two EcoRI ends or two MseI ends. Computer-assisted analysis suggested that 86, 82, and 78 fragments could be amplified if selective primers EcoRI-A and MseI-G were used, 49, 52, and 51 of which should be unique in size and should fall in the size range from 50 to 500 bp. In contrast, selective primers EcoRI-C and MseI-CA should amplify only 14, 23, and 22 fragments, and 9, 19, and 17 of them should be in the size range from 50 to 500 bp and be unique. Although the genomic sequences of E. coli strains vary, this electronic AFLP analysis suggested that digestion of E. coli genomic DNA with EcoRI and MseI was suitable for AFLP analysis.
The computer analysis also predicted that selective primers EcoRI-A and MseI-G should produce more bands that were evenly distributed in terms of length than primers EcoRI-C and MseI-CA should produce and hence should result in greater discrimination. Our experimental data were consistent with these expectations. The difference between the number of fragments detected and the number of fragments predicted could be explained by the genetic diversity of E. coli isolates, inefficient amplification of some of the fragments, and the inability of the software to detect some weak bands. The use of a Perl program to screen for discriminative bands could potentially lead to discovery of new DNA markers that are specifically associated with one host group.
In conclusion, the three different methods that were evaluated varied in the ability to differentiate the E. coli isolates from various sources. Fluorescent AFLP analysis provided the greatest discriminatory power, the highest rate of correct classification, and ease of standardization and automation, but this technique requires a major capital investment (an automated DNA sequencer and appropriate software). The two other methods (MAR test and 16S rRNA sequencing) also provided moderate to high degrees of correct classification. In particular, the MAR test is a simple and cost-effective approach, which is more suitable for surveillance of local self-contained water or environmental systems. Although extensive field testing is required to determine the efficacy of these assays and much larger referencing databases must be accumulated before these methods could be used for routine natural environmental monitoring, these assays appear to provide promising diagnostic tools for tracking nonpoint sources of fecal pollution.
Acknowledgments
We thank Michele Edwards for assistance.
This work was supported by the Agricultural Adaptation Council of the National Soil and Water Conservation Program and the Ontario Ministry of Agriculture, Food, and Rural Affairs.
REFERENCES
- 1.Ackman, D., S. Marks, P. Mack, M. Caldwell, T. Root, and G. Birkhead. 1997. Swimming-associated haemorrhagic colitis due to Escherichia coli O157:H7 infection: evidence of prolonged contamination of a fresh water lake. Epidemiol. Infect. 119:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alekshun, M. N., and S. B. Levy. 1999. The mar regulon: multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 7:410-413. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, C., L. Metherell, G. Willshaw, A. Maggs, and J. Stanley. 1999. Predictive fluorescent amplified-fragment length polymorphism analysis of Escherichia coli: high-resolution typing method with phylogenetic significance. J. Clin. Microbiol. 37:1274-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrouane, Y. F., L. A. McNutt, B. J. Buschelman, P. R. Rhomberg, M. D. Sanford, R. J. Hollis, M. A. Pfaller, and L. A. Herwaldt. 2000. Outbreak of severe Pseudomonas aeruginosa infections caused by a contaminated drain in a whirlpool bathtub. Clin. Infect. Dis. 31:1331-1337. [DOI] [PubMed] [Google Scholar]
- 5.de Bruijn, F. J. 1992. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 58:2180-2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai, M., A. Tanna, R. Wall, A. Efstratiou, R. George, and J. Stanley. 1998. Fluorescent amplified-fragment length polymorphism analysis of an outbreak of group A streptococcal invasive disease. J. Clin. Microbiol. 36:3133-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Desai, M., E. J. Threlfall, and J. Stanley. 2001. Fluorescent amplified-fragment length polymorphism subtyping of the Salmonella enterica serovar Enteritidis phage type 4 clone complex. J. Clin. Microbiol. 39:201-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duim, B., T. M. Wassenaar, A. Rigter, and J. Wagenaar. 1999. High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl. Environ. Microbiol. 65:2369-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Felsenstein, J. 1989. PHYLIP--phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 10.Gugnani, H. C. 1999. Some emerging food and water borne pathogens. J. Commun. Dis. 31:65-72. [PubMed] [Google Scholar]
- 11.Guillemot, D. 1999. Antibiotic use in humans and bacterial resistance. Curr. Opin. Microbiol. 2:494-498. [DOI] [PubMed] [Google Scholar]
- 12.Hachler, H., S. P. Cohen, and S. B. Levy. 1991. marA, a regulated locus which controls expression of chromosomal multiple antibiotic resistance in Escherichia coli. J. Bacteriol. 173:5532-5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagedorn, C., S. L. Robinson, J. R. Filtz, S. M. Grubbs, T. A. Angier, and R. B. Reneau, Jr. 1999. Determining sources of fecal pollution in a rural Virginia watershed with antibiotic resistance patterns in fecal streptococci. Appl. Environ. Microbiol. 65:5522-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harwood, V. J., J. Whitlock, and V. Withington. 2000. Classification of antibiotic resistance patterns of indicator bacteria by discriminant analysis: use in predicting the source of fecal contamination in subtropical waters. Appl. Environ. Microbiol. 66:3698-3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houndt, T., and H. Ochman. 2000. Long-term shifts in patterns of antibiotic resistance in enteric bacteria. Appl. Environ. Microbiol. 66:5406-5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iyoda, S., A. Wada, J. Weller, S. J. Flood, E. Schreiber, B. Tucker, and H. Watanabe. 1999. Evaluation of AFLP, a high-resolution DNA fingerprinting method, as a tool for molecular subtyping of enterohemorrhagic Escherichia coli O157:H7 isolates. Microbiol. Immunol. 43:803-806. [DOI] [PubMed] [Google Scholar]
- 17.Janssen, P., R. Coopman, G. Huys, J. Swings, M. Bleeker, P. Vos, M. Zabeau, and K. Kersters. 1996. Evaluation of the DNA fingerprinting method AFLP as an new tool in bacterial taxonomy. Microbiology 142:1881-1893. [DOI] [PubMed] [Google Scholar]
- 18.Jones, I. G., and M. Roworth. 1996. An outbreak of Escherichia coli O157 and campylobacteriosis associated with contamination of a drinking water supply. Public Health (London) 110:277-282. [DOI] [PubMed] [Google Scholar]
- 19.Kaspar, C. W., J. L. Burgess, I. T. Knight, and R. R. Colwell. 1990. Antibiotic resistance indexing of Escherichia coli to identify sources of fecal contamination in water. Can. J. Microbiol. 36:891-894. [DOI] [PubMed] [Google Scholar]
- 20.Kokotovic, B., N. F. Friis, J. S. Jensen, and P. Ahrens. 1999. Amplified-fragment length polymorphism fingerprinting of Mycoplasma species. J. Clin. Microbiol. 37:3300-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krumperman, P. H. 1983. Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 46:165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Licence, K., K. R. Oates, B. A. Synge, and T. M. Reid. 2001. An outbreak of E. coli O157 infection with evidence of spread from animals to man through contamination of a private water supply. Epidemiol. Infect. 126:135-138. [PMC free article] [PubMed] [Google Scholar]
- 23.Lindstedt, B. A., E. Heir, T. Vardund, K. K. Melby, and G. Kapperud. 2000. Comparative fingerprinting analysis of Campylobacter jejuni subsp. jejuni strains by amplified-fragment length polymorphism genotyping. J. Clin. Microbiol. 38:3379-3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linton, D., A. J. Lawson, R. J. Owen, and J. Stanley. 1997. PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35:2568-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mathew, A. G., A. M. Saxton, W. G. Upchurch, and S. E. Chattin. 1999. Multiple antibiotic resistance patterns of Escherichia coli isolates from swine farms. Appl. Environ. Microbiol. 65:2770-2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Applic. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 27.Parveen, S., R. L. Murphree, L. Edmiston, C. W. Kaspar, K. M. Portier, and M. L. Tamplin. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Postic, D., N. Riquelme-Sertour, F. Merien, P. Perolat, and G. Baranton. 2000. Interest of partial 16S rDNA gene sequences to resolve heterogeneities between Leptospira collections: application to L. meyeri. Res. Microbiol. 151:333-341. [DOI] [PubMed] [Google Scholar]
- 29.Puech, M. C., J. M. McAnulty, M. Lesjak, N. Shaw, L. Heron, and J. M. Watson. 2001. A statewide outbreak of cryptosporidiosis in New South Wales associated with swimming at public pools. Epidemiol. Infect. 126:389-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rademaker, J. L., B. Hoste, F. J. Louws, K. Kersters, J. Swings, L. Vauterin, P. Vauterin, and F. J. de Bruijn. 2000. Comparison of AFLP and rep-PCR genomic fingerprinting with DNA-DNA homology studies: Xanthomonas as a model system. Int. J. Syst. E vol. Microbiol. 50:665-677. [DOI] [PubMed] [Google Scholar]
- 31.Rubens, C. E., W. F. McNeill, and W. E. Farrar, Jr. 1979. Evolution of multiple-antibiotic-resistance plasmids mediated by transposable plasmid deoxyribonucleic acid sequences. J. Bacteriol. 140:713-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savelkoul, P. H., H. J. Aarts, J. de Haas, L. Dijkshoorn, B. Duim, M. Otsen, J. L. Rademaker, L. Schouls, and J. A. Lenstra. 1999. Amplified-fragment length polymorphism analysis: the state of an art. J. Clin. Microbiol. 37:3083-3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Speijer, H., P. H. Savelkoul, M. J. Bonten, E. E. Stobberingh, and J. H. Tjhie. 1999. Application of different genotyping methods for Pseudomonas aeruginosa in a setting of endemicity in an intensive care unit. J. Clin. Microbiol. 37:3654-3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spratt, B. G. 1994. Resistance to antibiotics mediated by target alterations. Science 264:388-393. [DOI] [PubMed] [Google Scholar]
- 35.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valsangiacomo, C., F. Baggi, V. Gaia, T. Balmelli, R. Peduzzi, and J. C. Piffaretti. 1995. Use of amplified fragment length polymorphism in molecular typing of Legionella pneumophila and application to epidemiological studies. J. Clin. Microbiol. 33:1716-1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van de Peer, Y., and R. De Wachter. 1994. TREECON for Windows: a software package for the construction and drawing of evolutionary trees for the Microsoft Windows environment. Comput. Applic. Biosci. 10:569-570. [DOI] [PubMed] [Google Scholar]
- 38.Vos, P., R. Hogers, M. Bleeker, M. Reijans, T. van de Lee, M. Hornes, A. Frijters, J. Pot, J. Peleman, M. Kuiper, et al. 1995. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 23:4407-4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wiggins, B. A. 1996. Discriminant analysis of antibiotic resistance patterns in fecal streptococci, a method to differentiate human and animal sources of fecal pollution in natural waters. Appl. Environ. Microbiol. 62:3997-4002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woo, P. C., P. K. Leung, K. W. Leung, and K. Y. Yuen. 2000. Identification by 16S ribosomal RNA gene sequencing of an Enterobacteriaceae species from a bone marrow transplant recipient. Mol. Pathol. 53:211-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao, S., S. E. Mitchell, J. Meng, S. Kresovich, M. P. Doyle, R. E. Dean, A. M. Casa, and J. W. Weller. 2000. Genomic typing of Escherichia coli O157:H7 by semi-automated fluorescent AFLP analysis. Microbes Infect. 2:107-113. [DOI] [PubMed] [Google Scholar]