Abstract
The gene (palI) encoding isomaltulose synthase (PalI) from a soil bacterial isolate, Klebsiella sp. strain LX3, was cloned and characterized. PalI converts sucrose into isomaltulose, trehalulose, and trace amounts of glucose and fructose. Sequence domain analysis showed that PalI contains an α-amylase domain and (β/α)8-barrel structures, suggesting that it belongs to the α-amylase family. Sequence alignment indicated that the five amino acid residues of catalytic importance in α-amylases and glucosyltransferases (Asp241, Glu295, Asp369, His145, and His368) are conserved in PalI. Purified recombinant PalI displayed high catalytic efficiency, with a Km of 54.6 ± 1.7 mM for sucrose, and maximum activity (approximately 328.0 ± 2.5 U/mg) at pH 6.0 and 35°C. PalI activity was strongly inhibited by Fe3+ and Hg2+ and was enhanced by Mn2+ and Mg2+. The half-life of PalI was 1.8 min at 50°C. Replacement of selected amino acid residues by proline significantly increased the thermostability of PalI. Simultaneous replacement of Glu498 and Arg310 with proline resulted in an 11-fold increase in the half-life of PalI at 50°C.
Isomaltulose (6-O-α-d-glucopyranosyl-d-fructose, commonly referred to as palatinose) is a sucrose isomer with physical and organoleptic properties similar to those of sucrose. It has been suggested as a noncariogenic alternative to sucrose (1) and has now been widely used as a sugar substitute in food. Unlike ingestion of sucrose, ingestion of isomaltulose has only a minor effect on the concentration of glucose in blood, indicating its potential as a parenteral nutrient acceptable to diabetics and nondiabetics (6). Several microorganisms have been found to form isomaltulose and trehalulose from sucrose, for example Protaminobacter rubrum (39), Serratia plymuthica (5), Erwinia rhapontici (4), Klebsiella planticola CCRC 19112 (7), Pseudomonas mesoacidophila MX-45 (18), and Agrobacterium radiobacter MX-232 (19).
Isomaltulose synthase, also known as sucrose isomerase, has been purified from S. plymuthica (14, 33), E. rhapontici (4), a Klebsiella sp. (24), and Pseudomonas mesoacidophila MX-45 (18). In addition to isomerizing sucrose to produce isomaltulose and trehalulose, the enzyme reaction also releases small amounts of glucose and fructose as by-products (7, 14, 31, 33). The isomaltulose synthase from S. plymuthica even converted sucrose to isomaltose and isomelezitose (33). The product composition varies depending on the bacterial strain used. Protaminobacter rubrum (15), S. plymuthica NCIB 8285 (5), E. rhapontici NCPPB 1579 (4), and K. planticola CCRC 19112 (7) produce mainly isomaltulose (75 to 85%), whereas Pseudomonas mesoacidophila MX-45 (17) and A. radiobacter MX-232 (19) produce more trehalulose (90%) than isomaltulose. The reaction of these enzymes is strongly influenced by temperature. The optimum temperature for isomaltulose production by the enzyme from S. plymuthica ATCC 15928 is around 30°C, and no product is produced at 55°C (33), indicating the thermolability of isomaltulose synthase. Isomaltulose synthase sequence information is required to clarify the molecular mechanism of isomerization and assess the feasibility of engineering thermostability.
We recently identified a new isomaltulose-producing bacterial isolate, Klebsiella sp. strain LX3 (L.-H. Zhang, unpublished data). Biochemical analyses showed that this isolate converts sucrose mainly into isomaltulose, with small amounts of trehalulose and trace amounts of glucose and fructose. In the present study, we report the cloning and characterization of the gene (palI) encoding isomaltulose synthase (PalI) from Klebsiella sp. strain LX3. The recombinant enzyme was overexpressed in Escherichia coli, purified, and characterized. In addition, site-directed mutagenesis was performed based on protein domain and secondary structure analysis to improve the thermostability of PalI by proline substitution.
MATERIALS AND METHODS
Bacterial strains and chemicals.
The bacterial strains and plasmids used in this study are listed in Table 1. Klebsiella sp. isolate LX3 was isolated from a soil sample collected in Singapore. Cosmid vector pLAFR3 and cloning vector pBluescript II SK(+) (Stratagene) were used for construction of a genomic library and for subcloning, respectively. E. coli DH5α was used as the host for cloning and overexpression of palI. E. coli was grown at 37°C in Luria-Bertani (LB) medium containing 100 μg of ampicillin/ml. Klebsiella sp. strain LX3 was grown at 30°C in SPY medium (4% sucrose, 0.5% peptone, and 0.4% yeast extract, pH 6.5).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or phenotype | Source |
|---|---|---|
| E. coli DH5α | recA1 endA1 hsdR17 supE4 gyrA96 relA1 Δ(lacZYAargF)U169 (φ80dlacZΔM15) | Laboratory collection |
| Klebsiella sp. strain LX3 | Isomaltulose producer | This study |
| pPLAFR3 | Cosmid vector; Tcr | Stratagene |
| pBluescript II SK(+) | Cloning vector; Ampr | Stratagene |
| pPLAK164 | pLAFR3 containing ∼6.1-kb BamHI fragment | This study |
| pPLAK169 | pLAFR3 plasmid with ∼6.1-kb BamHI fragment | This study |
| pBKBB | ∼6.1-kb BamHI fragment in pBluescript II SK(+) | This study |
| pBKBP | ∼4.8-kb BamHI/PstI fragment in pBluescript II SK(+) | This study |
| pBKCB | ∼3.6-kb ClaI/BamHI fragment in pBluescript II SK(+) | This study |
| pBKBX | ∼3.5-kb BamHI/XhoI fragment in pBluescript II SK(+) | This study |
| pBKEE | ∼2.5-kb EcoRV/EcoRV fragment in pBluescript II SK(+) | This study |
| pGEK | palI ORF without signal peptide-coding sequence fused with GST in pGEX-2T | This study |
| pGEK:E498P | Glu residue at position 498 in PalI was replaced by Pro in pGEK | This study |
| pGEK:E498P/R310 | Arg residue at position 310 in PalI:E498P was replaced by Pro in pGEK:E498P | This study |
DNA manipulation.
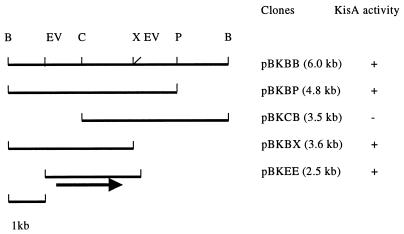
DNA manipulation, agarose gel electrophoresis, and transformation of E. coli were performed according to standard procedures. DNA fragments were isolated from agarose gels with the QIAEX II gel extraction kit (Qiagen), and plasmids were purified with the Qiagen plasmid minikit. A DNA library was constructed by partially digesting DNA from Klebsiella sp. strain LX3 with BamHI, cloning the fragments into the BamHI site of cosmid vector pLAFR3, and transfecting into E. coli DH5α after in vitro packaging with Gigapack II (Stratagene). Following transformation, individual bacterial colonies were incubated in liquid LB medium supplemented with sucrose (0.4%) for 15 h, and the reducing sugar released in the supernatant was tested using the dinitrosalicyclic acid method (16). The presence of isomaltulose was further verified by thin-layer chromatography (TLC), using authentic isomaltulose as control. The positive cosmid clone was subcloned into pBluescript SK(+) to reduce the size of the insert (Fig. 1). The resultant clone, pBKEE, was sequenced from both strands by the chain termination method with the ABI PRISM dRhodamine Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems).
FIG. 1.
Cloning of the palI gene from Klebsiella sp. strain LX3. B, BamHI; EV, EcoRV; C, ClaI; X, XhoI; P, PstI; +, PalI activity detected; −, no activity. The arrow indicates the relative position of the palI coding region. The size of the insert in each clone is listed in parentheses.
Computer analysis.
DNA and peptide sequence homologies were analyzed using the BLAST program to search major databases (GenBank, EMBL, and SWISSPROT databases). Sequence alignment was performed using the Genetics Computer Group program (http://www.bic.nus.edu.sg:8888/gcg/java/). Domain analysis was carried out with the Pfam program (http://pfam.wustl.edu). Signal peptide prediction was performed with the SignalP program (23). The secondary structure of PalI was predicted using the PredictProtein program (25) (http://dodo.cpmc.columbia.edu).
Construction of GST-PalI.
The palI gene without the coding sequence for the first 28-amino-acid signal peptide was amplified by PCR using the forward primer 5′-TTGGATCCGCACCATCCTTGAATCAGGATATTCAC-3′ and the reverse primer 5′-TTGAATTCTTACCGCAGCTTATACACACCTGCCTG-3′. After digestion with BamHI and EcoRI, the fragment was fused to the glutathione S-transferase (GST) gene in the same open reading frame (ORF) in fusion vector pGEX-2T to generate an expression construct, pGEK, which was verified by DNA sequencing.
Site-directed mutagenesis.
A point mutation was introduced into the palI gene by using the QuickChange site-directed mutagenesis kit (Stratagene). Mutant pGEK:E498P was produced using pGEK as a template. Double mutant pGEK:E498P/R310P was created using pGEK:E498P as a template. Mutations were verified by DNA sequencing.
Enzyme purification.
E. coli strains containing different constructs were incubated in LB medium at 28°C. The GST-PalI fusion protein and its derivatives were overexpressed by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM after the optical density at 600 nm of the bacterial culture reached 0.6. Purification of fusion proteins was carried out by a method described previously (40). Further purification was performed by fast protein liquid chromatography with a Protein Pak 300SW semipreparative column at a flow rate of 0.5 ml/min with sodium phosphate buffer (50 mM, pH 7.4). Purified enzymes were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% polyacrylamide).
Enzyme assay.
A sample (100 μl, 2 μg of protein) of enzyme was mixed with 400 μl of 0.1 M citrate-phosphate buffer (pH 6.0) containing sucrose (40 g/liter) and incubated at 35°C for 15 min with gentle agitation. The reaction was stopped by boiling for 5 min, and the total reducing sugar was assayed using the dinitrosalicylic acid method (16). One unit of enzyme is defined as the amount of protein that forms 1 μmol of reducing sugar (with isomaltulose as a standard) per min under the assay conditions specified. The protein concentration was determined using the Bradford method (3) with bovine serum albumin as a standard. For the kinetic analysis, 2 μg of the purified enzyme was mixed with different concentrations of sucrose (10 to 150 mM) dissolved in 0.1 M citrate-phosphate buffer (pH 6.0) at a final reaction volume of 500 μl. The initial reaction velocity was determined after incubation at 35°C for an appropriate reaction time. The Km and kcat values were calculated based on the Michaelis-Menten equation.
Influence of pH on activity and stability.
Reaction solutions were prepared by adding sucrose to a final concentration of 4% and 2.0 μg of PalI in 0.1 M citrate-sodium phosphate buffer at a range of pH values, except that 0.05 M Tris buffer was used at pH 8.5. To investigate the effect of pH on stability, the same amount of enzyme was added to each reaction buffer and maintained at room temperature for 1 h before addition of sucrose. Enzyme activity was assayed as described above.
Effect of temperature on enzyme activity.
The effect of temperature on enzyme activity was determined by incubating the reaction solution (pH 6.0) at the temperatures indicated for 15 min. To determine the effect of temperature on enzyme activity, the enzyme solution (pH 6.0) was subjected to different temperatures for 10 min and the residual activity was assayed. Enzyme thermoinactivation was performed by incubating the enzyme at 50°C for various periods, and the remaining activity was determined at different intervals as stated. The first-order constant, k, of irreversible thermoinactivation was obtained by linear regression in semilog coordinates. The enzyme half-life was calculated using the equation t1/2 = ln 2/k.
Effects of metal ions and other reagents on PalI activity.
Purified PalI was extensively dialyzed against a 50 mM citrate-phosphate buffer (pH 6.0). Samples (2.0 μg) were preincubated with metal ions and other reagents in various concentrations at room temperature for 10 min (in a 200-μl final volume). Aliquots (20 μl) were withdrawn and tested for activity as described above.
Effect of temperature and pH on product formation.
To investigate the influence of temperature on product formation, the conversion reaction was performed for 4 h in a test tube containing 2 ml of 4% sucrose solution (pH 6.0) and 10 μg of PalI at the temperatures indicated. The reaction mixture was boiled for 5 min and subjected to high-pressure liquid chromatography (HPLC) analysis. The pH experiments were conducted at a fixed temperature of 35°C.
Analytical methods.
Qualitative analysis of isomaltulose, glucose-fructose, and trehalulose was carried out using TLC in an ethyl acetate-acetic acid-water (4:3:1 by volume) solvent system. Sugar spots were visualized by spraying TLC plates with diphenylamine-aniline-85% phosphate reagent and incubating for 5 min at 80°C (27). Quantitative analysis of sugars was performed by HPLC (Waters 2690 separations module) on a Symmetry C18 column (4.6 by 250 mm) coupled to a Waters 2410 refractive index detector. Samples were eluted isocratically with water at a flow rate of 1 ml/min. Under these conditions, the retention times of trehalulose, glucose, fructose, isomaltulose, and sucrose were 2.20, 2.67, 2.87, 3.25, and 3.87 min, respectively. Each sugar was quantified based on peak area.
Nucleotide sequence accession number.
The nucleotide and peptide sequences of the Klebsiella sp. strain LX3 isomaltulose synthase gene (palI) were deposited in the GenBank database under accession number AY040843.
RESULTS
Cloning and sequence analysis of the palI gene, encoding isomaltulose synthase.
The cosmid library of a Klebsiella sp. was screened for isomaltulose biosynthesis activity. Two positive clones, designated pPLAK164 and pPLAK169, were selected from about 1,500 colonies for their ability to produce isomaltulose. Restriction enzyme analysis of the two clones revealed that they contained the same BamHI fragment of ∼6.0 kb. Deletion analysis narrowed down the region encoding isomaltulose biosynthesis to a 2.5-kb EcoRV fragment in the subclone pBKEE (Fig. 1).
DNA sequence analysis of pBKEE identified an ORF of 1,797 nucleotides. Deletion analysis confirmed that it was the coding region of isomaltulose synthase (data not shown). The ORF encodes a protein of 598 amino acids with a calculated molecular mass of 69.94 kDa and an isoelectric point at 6.62. The first 28 amino acids (Met1 to Ala28) at the N-terminal constitute a signal peptide, as predicted by the SignalP computer program (23). Deletion of the signal peptide resulted in accumulation of isomaltulose synthase in the cytoplasm of bacterial cells (data not shown).
Homology comparison of the isomaltulose synthase from Klebsiella sp. strain LX3.
The best homolog of the isomaltulose synthase from Klebsiella sp. strain LX3 is the isomaltulose synthase from Enterobacter sp. strain SZ62, reported in a patent publication (15). The two enzymes share 99.3% homology at the nucleotide sequence level and 99.7% homology at the peptide sequence level. There are differences in the C-terminal regions of the enzymes. The Klebsiella sp. strain LX3 isomaltulose synthase contains an extra amino acid, Ala577, and differs in two residues (Ala578 and Val594) from isomaltulose synthase of strain SZ62 (Gly577 and Ala593). The Klebsiella sp. strain LX3 enzyme also shares 70 and 67% homology with isomaltulose synthase encoded by the palI gene from E. rhapontici (2). The isomaltulose synthase gene from Klebsiella sp. strain LX3 is therefore designated palI.
PalI of Klebsiella sp. strain LX3 exhibits 47% identity to the oligo-1,6-glucosidase (OGL) from Bacillus thermoglucosidasius (35) and the α-glucosidase from a Bacillus sp. (22) and 45% identity to the OGL from Bacillus cereus (36). Despite high sequence similarity (62 to 65%) between α-glucosidase and the two OGLs, the enzymes exhibit significant differences in substrate specificity (22). OGL and α-glucosidase are hydrolases with α-1,6 and α-1,4 bond specificities, respectively, whereas isomaltulose synthase catalyzes a two-step reaction with sucrose as the substrate, i.e., hydrolysis of the α-1,2 bond and then formation of an α-1,6 bond between the glucose and fructose moieties.
The PalI protein is a member of the α-amylase family.
A domain search with the Pfam program revealed that PalI contains an α-amylase domain spanning residues 52 to 518, with an E value of 4.2e − 125. Secondary-structure prediction indicated that PalI contains a (β/α)8-barrel structure and a subdomain B located between Nβ3 and Nα3 (Fig. 2). A motif search showed a short stretch (209QPDLN) between the Sβ3 and Nα3 regions of PalI which is a proposed selection marker of the α-amylase family (9). The sequence alignment with the two OGLs showed that PalI also contains a catalytic triad consisting of three carboxylic amino acids and the two histidine residues essential for enzyme activity. The four regions (I, II, III, and IV) containing the potential essential residues (D241, E295, D369, H145, and H368) in PalI are shown in Fig. 2. Site-directed mutagenesis of these residues in PalI verified that they are essential for enzyme activity (D. Zhang, unpublished data).
FIG. 2.
Sequence comparison and assignment of secondary structure. The peptide sequence of PalI of Klebsiella sp. strain LX3 (K. si) is compared with the sequences of the thermolabile OGL of B. cereus ATCC 7064 (B. ce) (accession no. P21332) and the thermostable OGL of B. thermoglucosidasius KP1006 (B. th) (accession no. P29094). The secondary structural features, i.e., α (helix) and β (sheet), of B. cereus OGL, based on crystal structural analysis (11), are indicated above the sequence. The predicted secondary structural regions of PalI are underlined and labeled at the bottom (H, α-helix; E, β-sheet). The selection marker for the α-amylase family, QPDLN, is indicated by double lines. The potential amino acid residues for proline substitution are shown in boldface, and the two mutated residues, R310 and E498, are indicated by ♠. The crucial invariant residues (D241, E295, D369, H145, and H368 in PalI) conserved in α-amylase and glucosyltransferase are indicated by +. Boxes I to IV show the regions containing the five essential residues.
Overexpression and purification of isomaltulose synthase.
The palI without the coding sequence for the signal peptide was subcloned into expression vector PGEX-2T to generate the GST-PalI fusion gene under the control of an IPTG-inducible promoter, Ptac. To maximize the yield of the fusion gene, we tested different induction conditions and found that induction with 0.5 mM IPTG at 28°C for 15 h produced the maximum level of soluble active fusion protein (data not shown). A 500-ml culture of E. coli DH5α/pGEK produced a total of 840 U of PalI. Approximately 60% (540 U) of the total activity was found in the cell-free fraction. The GST-PalI fusion protein bound by glutathione-Sepharose affinity resins was digested by thrombin, and PalI was released from the affinity column. The product was found to be impure, containing one major band of ∼67 kDa (PalI) and several minor bands. These minor bands were eliminated after further purification using semipreparative fast protein liquid chromatography. The homogeneity of the final product, purified up to 350-fold with a 29% final yield, was confirmed by SDS-PAGE (data not shown).
Analysis of PalI reaction product.
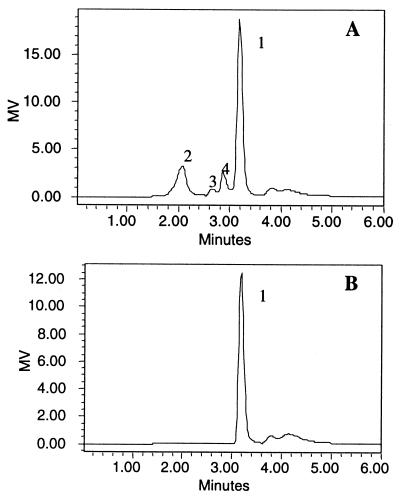
The PalI reaction product was separated by TLC and detected by diphenylamine-aniline-phosphate reagent, which reacts with glucosaccharides of varied linkages to yield spots with different colors (27). Isomaltulose, which appeared as a green spot, was the major product of PalI. The second product showed the red color typical of trehalulose. Glucose and fructose were hardly detected using this method. HPLC analysis of the reaction solution showed four peaks (Fig. 3A). By comparison with the HPLC profiles of the standards and the typical color reaction of each peak on TLC plate, peak 1, with a retention time of 3.25 min was identified as isomaltulose, and peaks 2, 3, and 4 were identified as trehalulose, glucose, and fructose, respectively.
FIG. 3.
HPLC analysis of the reaction products of PalI. Sucrose was incubated with PalI at 35°C for 4 h. (A) The reaction mixture was diluted, filtered through a 0.2-μm-pore-size membrane, and subjected to HPLC analysis. The sample was eluted isocratically with water at a flow rate of 1 ml/min and detected with a Waters 2410 refractive index detector. The peaks (1, isomaltulose; 2, trehalulose; 3, glucose; 4, fructose) were assigned based the retention times of the standard samples (isomaltulose, 3.18 min; trehalulose, 2.2 min; glucose, 2.67 min; fructose, 2.87 min). MV, recorder output in millivolts (MV = 2,000 × SF × S × Δn, where SF is the scale factor, S is the sensitivity setting, and Δn is the refractive index). (B) HPLC profile of isomaltulose.
General properties of PalI.
The optimal temperature and pH for PalI activity were examined. Maximal activity was observed at pH 6.0, and the enzyme was stable within a narrow pH range of 5.0 to 6.5. PalI rapidly lost activity at pH values higher than 6.5 (data not shown). PalI was most active at 35°C. At above 40°C PalI activity quickly dropped, indicating the poor thermostability of this enzyme under the assay conditions used. The values of Km and kcat/Km for sucrose were 54.6 ± 1.7 mM and 0.27 ± 0.02 mM−1 min−1, respectively, and the specific activity was 328.0 ±2.5 U/mg at pH 6.0 and 35°C.
Fe3+, Hg2+, and the detergent SDS completely abolished PalI activity, whereas Mg2+ and Mn2+ enhanced the activity of the enzyme (Table 2). Divalent cations such as Fe2+, Li2+, Ca2+, and Cu2+ partially inhibited the enzyme activity, and Zn2+ had no effect. EDTA (a chelating reagent for divalent cations) also inhibited PalI, confirming that the enzyme requires certain cations for activity.
TABLE 2.
Influence of different reagents on PalI activitya
| Reagent | Concn (mM) | PalI activity (%) |
|---|---|---|
| EDTA | 3 | 64.0 |
| 15 | 30.4 | |
| SDS | 3 | 0 |
| FeCl3 | 5 | 0 |
| FeCl2 | 1 | 75.3 |
| 5 | 48.6 | |
| LiCl2 | 5 | 93.7 |
| CaCl2 | 5 | 90.4 |
| HgCl2 | 1 | 0 |
| MgSO4 | 5 | 124.6 |
| ZnSO4 | 5 | 102.5 |
| CuSO4 | 1 | 85.6 |
| 5 | 76.9 | |
| MnCl2 | 1 | 126.7 |
| 5 | 145.7 |
Purified PalI protein was extensively dialyzed against 50 mM citrate-phosphate buffer (pH 6.0). The enzyme (2 μg in a 200-μl final volume) was preincubated with the indicated reagents and ions at room temperature for 10 min, and 20 μl of sample was taken for enzyme assay as described in Materials and Methods.
Effect of temperature and pH on product composition.
Changing the temperature or pH affects the ratio of monosaccharides (glucose and fructose) and trehalulose in the PalI reaction products. Analysis of the products formed at different temperatures showed that high temperatures promote monosaccharide release but suppress trehalulose formation (Table 3). An increase of the temperature from 25 to 50°C caused the amount of monosaccharides in the reaction products to double, while trehalulose formation decreased approximately threefold. Neutral pH favors trehalulose formation, with its content increasing from 11.0 ± 2.3% at pH 5.0 to 13.0 ± 3.1% at pH 7.0. In contrast, slightly more monosaccharides were produced at an acidic pH than at neutral pH (6.9 ± 1.4% at pH 5.0 and 4.5 ± 0.9% at pH 7.0). Under these assay conditions, the proportion of isomaltulose in the PalI reaction products was not significantly influenced by either temperature (Table 3) or pH (data not shown).
TABLE 3.
Influence of temperature on product formationa
| Sugar | % (mean ± SD) in product mixture at the following temp (°C):
|
||||||
|---|---|---|---|---|---|---|---|
| 15 | 20 | 25 | 30 | 35 | 45 | 50 | |
| Glucose + fructose | ND | ND | 5.5 ± 0.7 | 5.9 ± 0.8 | 5.9 ± 1.0 | 9.1 ± 0.4 | 12.7 ± 2.3 |
| Trehalulose | 20.9 ± 3.2 | 21.5 ± 2.7 | 17.0 ± 2.3 | 14.4 ± 0.8 | 12.2 ± 0.6 | 8.3 ± 0.6 | 5.2 ± 0.8 |
| Isomaltulose | 79.1 ± 2.8 | 78.9 ± 4.3 | 77.4 ± 2.7 | 78.7 ± 3.1 | 81.9 ± 1.4 | 82.6 ± 1.7 | 82.1 ± 3.3 |
Samples (2 ml) of sucrose (4%) in citrate-phosphate buffer (pH 6.0) containing 10 μg of PalI were incubated for 4 h at the indicated temperatures. The reaction mixture was boiled for 5 min and analyzed by HPLC on a Symmetry C18 column (4.6 by 250 mm).
ND, not determined.
Identification of potential sites for proline substitution.
The proline rule, i.e., the fact that the presence of proline at the second position of a β-turn makes a protein more stable by decreasing the entropy of unfolding of the backbone (29, 30), has been widely applied to enhance thermostability in α-amylases (8, 21, 28, 37). Given that PalI belongs to the α-amylase family and its predicted secondary structure, i.e., (β/α)8-barrel, is similar to that of B. cereus OGL (Fig. 2), we assumed that the proline rule could also be applied to PalI. We compared the primary sequence and the predicted secondary structure of the protein with those of thermolabile B. cereus ATCC 7064 OGL and thermostable B. thermoglucosidasius KP1006 OGL, using the proline rule. The comparison identified five residues (L301, R310, S330, R419, and E498) in PalI which are potential candidates for proline substitution (Fig. 2). Two residues, R310 in the loop and E498 in the β-turn, were selected to investigate the effect of proline substitution on the thermostability and catalytic activity of PalI.
Proline substitution increases thermostability.
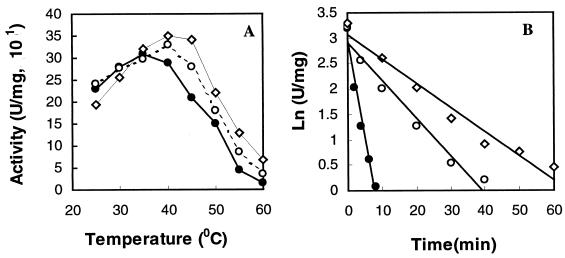
Site-directed mutagenesis was used to replace E498 of PalI with proline to produce pGEK:E498P, which was further mutated to change R310 to proline to form the double mutant pGEK:E498P/R310P. The two mutant enzymes were overexpressed and purified by the same procedure as for native PalI. Mutation did not affect the expression levels of these mutant versions, as shown by SDS-PAGE (data not shown). Proline substitution changed not only the optimum temperature but also the maximum specific activity of PalI (Fig. 4A). The optimum temperature was 35°C for PalI but increased to 40 and 45°C for PalI:E498P and PalI:E498P/R310P, respectively. In comparison to PalI, the maximum specific activity increased by 7% for PalI:E498P and 16% for PalI:E498P/R310P. The temperatures resulting in 50% loss of activity were estimated to be approximately 45.8°C for PalI, 48.3°C for PalI:E498P, and 51°C for PalI:E498P/R310P (data not shown). The half-lives of PalI, PalI:E498P, and PalI:E498P/R310P were 1.81, 9.45, and 13.61 min at 50°C (Fig. 4B), respectively. The half-life of PalI:E498P/R310P at 50°C is therefore about 11-fold longer than that of PalI.
FIG. 4.
Effect of temperature on PalI, PalI: E498P, and PalI: E498P/R310P. Enzymes were analyzed for activity (A) and in a thermoinactivation assay at 50°C (B). To test for thermoinactivation, enzyme samples (500 μl, 20 μg/ml) in 0.1 M citrate-phosphate buffer (pH 6.0) were incubated at 50°C, and the aliquots (50 μl) were removed at different times and assayed at 35°C for residual activity. Residual activities are shown as the logarithms of the specific activities. Solid circles, PalI; open circles, PalI:E498P; open rhombuses, PalI:E498P/R310P.
To determine the difference in catalytic activity, we measured Km (millimolar) and kcat (minutes−1) at pH 6.0 and 35°C. The kcat/Km values for PalI, PalI:E498P, and PalI:E498P/R310P were 0.27 ± 0.02, 0.29 ±0.04, and 0.31 ± 0.03 min−1 mM−1, respectively. This result indicates that replacement of E498 and/or R310 by proline slightly increases the apparent catalytic efficiency (kcat/Km) of PalI on sucrose.
Proline substitution facilitates isomaltulose formation.
The reaction products of PalI and its mutant versions were analyzed to test whether proline replacement affects the percent contents of PalI products. The percent content of monosaccharides decreased from 5.9 ± 1.0% for PalI to 3.4 ± 0.8% for PalI:E498P and 3.3 ± 0.8% for PalI:E498P/R310P, whereas the percentage of isomaltulose increased from 81.9 ± 1.4% for PalI to 84.8 ± 1.0% for PalI:E498P and 84.1 ± 2.0% for PalI:E498P/R310P. The percent content of trehalulose in the reaction products was not significantly influenced by proline substitution. It is likely that proline substitution enhances the stability of the glucosyl-enzyme complex and consequently diminishes the release of monosaccharides.
DISCUSSION
The gene (palI) encoding the isomaltulose synthase (PalI) has been cloned and characterized. palI encodes a peptide of 598 amino acids with a molecular mass of 69.94 kDa. Domain structure analysis indicates that PalI contains an α-amylase domain. The main homologs of PalI, besides several uncharacterized isomaltulose synthases from an Enterobacter sp., Protaminobacter rubrum (15), and E. rhapontici (2), are OGLs from B. thermoglucosidasius and B. cereus (35, 36) and the α-glucosidase from a Bacillus sp. (21). The α-amylase domain of PalI shows about 47.7, 47.5, and 48.4% identity to the same domain of α-glucosidase from the Bacillus sp., OGL from B. thermoglucosidasius, and OGL from B. cereus, respectively. The predicted (β/α)8-barrel secondary structure of PalI is similar to that of B. cereus OGL. These characteristics, together with the presence of the marker sequence 209QPDLN, indicate that PalI is a member of the α-amylase family.
Sequence comparison revealed that PalI contains a potential catalytic triad (Asp241, Glu295, and Asp369) and two histidine residues (His145 and His368) that are highly conserved in α-amylases and glucosyltransferases. The importance of the catalytic triad and the two invariable residues has been verified for a range of α-amylases. Watanabe et al. (38) reported that mutation of the catalytic triad (Asp199, Glu255, and Asp329) in the OGL of B. cereus to asparagine or glutamine resulted in a significant loss of enzyme activity. Mutation of the equivalent residues, Asp294 and Asp401, in Neisseria polysaccharea amylosucrase inactivated the enzyme activity (26). A crystallographic study of cyclomaltodextran glucanotransferase from Bacillus circulans revealed that D256 functions as a nucleophilic site, E284 functions as a proton donor, and D354 functions as a substrate binding site (12, 32). The two conserved histidine residues may participate in stabilization of the substrate-binding transition state at subsite −1, as in the case of the Taka-amylase of Aspergillus oryzae (10) and cyclomaltodextran glucanotransferase of alkalophilic Bacillus sp. strain 1011 (20, 32). We found that the potential catalytic triad and the two histidine residues are essential for PalI activity (D. Zhang, unpublished data). The similar protein domain structure and function of the conserved catalytic triad suggests that the catalytic mechanism of PalI may resemble that of α-amylases and glucosyltransferases, especially for the first part of the reaction, i.e., cleavage of the glycosidic linkage and formation of a glucosyl-enzyme covalent intermediate.
PalI efficiently converts sucrose to isomaltulose and trehalulose with trace amounts of glucose and fructose. Formation of multiple products seems to be a characteristic common to isomaltulose synthases from various sources (7, 14, 15, 31, 33). The sucrose isomerase of S. plymuthica converts sucrose into isomaltulose, trehalulose, glucose, fructose, isomaltose, and isomelezitose (33). We found no evidence of isomaltose or isomelezitose in the reaction products of PalI by either TLC or HPLC analysis. The optimum pH (6.0) of PalI is similar to that of sucrose isomerase from S. plymuthica (14, 33), a Klebsiella sp. (24), and E. rhapontici (2). The optimum temperature (35°C) for PalI is identical to that for the sucrose isomerase from the Klebsiella sp. (24) and higher than that (30°C) for the sucrose isomerases from S. plymuthica (14, 33) and E. rhapontici (2).
Our data show that formation of trehalulose and monosaccharides, but not isomaltulose, is significantly influenced by temperature. High temperature tends to stimulate release of monosaccharides but suppress formation of trehalulose. It is likely that low temperature favors tautomerization of fructofuranose into fructopyranose (13), a critical step for trehalulose formation. Our results are consistent with the finding that isomaltulose synthase from Pseudomonas mesoacidophila produces 90% trehalulose at 20°C but only 50% trehalulose at 50°C (18).
The thermal stability of a protein is determined by many factors, for example, packing efficiency, hydrophobic interaction, loop stabilization, reduction of entropy of unfolding, and electrostatic interaction (34). Proline replacement in the OGL of B. cereus ATCC 7064 demonstrated that proline residues, especially at the second site of a β-turn, at the first turn of α-helices, or in coils within the loop binding to adjacent secondary structure, contributed to protein thermostability (30, 35, 37). Structural similarities between PalI and OGLs from B. cereus ATCC 7064 and B. thermoglucosidasius KP1006 and differences in thermostability facilitate the identification of potential residues for proline substitution to engineer protein thermostability properties. Our results showed that the proline substitution of two identified residues, Glu498 and Arg310, significantly enhanced both the optimum temperature for enzyme activity and the thermostability. In particular, the half-life of the double mutant PalI:E498P/R310P is 11-fold higher at 50°C than the half-life of PalI. The similarity of the apparent catalytic efficiencies (kcat/Km) of PalI and its mutant versions implies that no extensive conformational change, especially in the active-site cleft, is induced by proline substitution. Our results indicate that comparison of primary and secondary structures of enzymes with similar domain structures is an effective approach for molecular design in protein engineering.
REFERENCES
- 1.Baer, A. 1989. Significance and promotion of sugar substitution for the prevention of dental caries. Lebensm. Wiss. Technol. 22:46-53. [Google Scholar]
- 2.Bornke, F., M. Hajirezaei, and U. Sonnewald. 2001. Cloning and characterization of the gene cluster for palatinose metabolism from the phytopathogenic bacterium Erwinia rhapontici. J. Bacteriol. 183:2425-2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 4.Cheetham, P. S. J. 1984. The extraction and mechanism of a novel isomaltulose-synthesizing enzyme from Erwinia rhapontici. Biochem. J. 220:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujii, S., S. Kishihara, M. Komoto, and J. Shimizu. 1983. Isolation and characterisation of oligosaccharides produced from sucrose by transglucosylation action of Serratia plymuthica. Nippon Shokuhin Kogyo Gakkaishi 30:339-344. [Google Scholar]
- 6.Goda, T., and N. Hosoya. 1983. Hydrolysis of palatinose by rat intestinal sucrase-isomaltase complex. J. Jpn. Soc. Food Sci. 36:169-173. [Google Scholar]
- 7.Huang, J. H., L. H. Hsu, and Y. C. Su. 1998. Conversion of sucrose to isomaltulose by Klebsiella planticola CCRC 19112. J. Ind. Microbiol. Biotechnol. 21:22-27. [Google Scholar]
- 8.Igarashi, K., T. Ozawa, K. Ikawa-Kitayama, Y. Hayashi, H. Araki, K. Endo, H. Hagihara, K. Ozaki, S. Kawai, and S. Ito. 1999. Thermostabilization by proline substitution in an alkaline, liquefying alfa-amylase from Bacillus sp. strain KSM-1378. Biosci. Biotechnol. Biochem. 63:1535-1540. [DOI] [PubMed] [Google Scholar]
- 9.Janecek, S. 1995. Close evolutionary relatedness among functionally distantly related members of the (α/β)8-barrel glycosyl hydrolases suggested by the similarity of their fifth conserved sequence region. FEBS Lett. 377:6-8. [DOI] [PubMed] [Google Scholar]
- 10.Jespersen, H. M., E. A. MacGregor, B. Henrissat, M. R. Sierks, and B. Svensson. 1993. Starch and glycogen-debranching and branching enzymes: prediction of structural features of the catalytic (β/α)8-barrel domain and evolutionary relationship to other amylolytic enzymes. J. Protein Chem. 12:791-805. [DOI] [PubMed] [Google Scholar]
- 11.Kizaki, H., Y. Hata, K. Watanabe, Y. Katsube, and Y. Suzuki. 1993. Polypeptide folding of Bacillus cereus ATCC7064 oligo-1,6-glucosidase revealed by 3.0 A resolution X-ray analysis. J. Biochem. 113:646-649. [DOI] [PubMed] [Google Scholar]
- 12.Knegtel, R. M., B. Strokopytov, D. Penninga, O. G. Fuber, H. J. Rozeboom, K. H. Kalk, L. Dijkhuizen, and B. W. Dijkstra. 1995. Crystallographic studies of the interaction of cyclodextrin glycosyltransferase from Bacillus circulans strain 251 with natural substrate and products. J. Biol. Chem. 270:29256-29264. [DOI] [PubMed] [Google Scholar]
- 13.Lichtenthaler, F. W., and S. Ronninger. 1990. α-d-Glucopyranosyl-d-fructoses: distribution of furanoid and pyranoid tautomers in water, dimethyl sulfoxide, and pyridine. Studies on kestoses. Part 4. J. Chem. Soc. Perkin Trans. 2:1489-1497. [Google Scholar]
- 14.MacAllister, M., C. T. Kelly, E. Doyle, and W. M. Fogarty. 1990. The isomaltulose-synthesizing enzyme of Serratia plymuthica. Biotechnol. Lett. 12:667-672. [Google Scholar]
- 15.Mattes, R., K. Klein, H. Schiwech, M. Kunz, and M. Munir. July1998. DNAs encoding sucrose isomerase and palatinase. U.S. patent 5,786,140.
- 16.Miller, G. L. 1959. Use of dinitrosalicylic acid reagent for the determination of reducing sugar. Anal. Chem. 31:426-428. [Google Scholar]
- 17.Miyata, Y., T. Sugitani, K. Tsuyuki, T. Ebashi, and Y. Nakajima. 1992. Isolation and characterisation of Pseudomonas mesoacidophila producing trehalulose. Biosci. Biotechnol. Biochem. 54:1680-1681. [Google Scholar]
- 18.Nagai, Y., T. Sugitani, and K. Tsuyuki. 1994. Characterization of α-glucosyltransferase from Pseudomonas mesoacidophila MX-45. Biosci. Biotechnol. Biochem. 58:1789-1793. [DOI] [PubMed] [Google Scholar]
- 19.Nagai-Miyata, J., K. Tsuyuki, T. Sugitani, T. Ebashi, and Y. Nakajima. 1993. Isolation and characterization of a trehalulose-producing strain of Agrobacterium. Biosci. Biotechnol. Biochem. 57:2049-2053. [Google Scholar]
- 20.Nakamura, A., K. Haga, and K. Yamane. 1993. Three histidine residues in the active center of cyclodextrin glucanotransferase from alkaliphilic Bacillus sp. 1011: effect of replacement on pH dependence and transition state stabilization. Biochemistry 32:6624-6631. [DOI] [PubMed] [Google Scholar]
- 21.Nakamura, S., T. Tanaka, T. Y. Yada, and S. Nakai. 1997. Improving the thermostability of Bacillus stearothermophilus neutral protease by introducing proline into the active site helix. Protein Eng. 10:1263-1269. [DOI] [PubMed] [Google Scholar]
- 22.Nakao, M., T. Nakayama, A. Kakudo, M. Inohara, M. Harada, F. Omura, and Y. Shibano. 1994. Structure and expression of a gene coding for thermostable α-glucosidase with a broad substrate specificity from Bacillus sp. SAM1606. Eur. J. Biochem. 220:293-300. [DOI] [PubMed] [Google Scholar]
- 23.Neilson, H., J. Engelbrecht, S. Brunak, and G. Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage site. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 24.Park, Y. K., R. T. Vegina, and A. M. Pupin. 1992. Conversion of sucrose to somaltulose by microbial glucosyltransferase. Biotechnol. Lett. 14:547-551. [Google Scholar]
- 25.Rost, B. 1996. PHD: predicting 1D protein structure by profile based neural networks. Methods Enzymol. 266:525-539. [DOI] [PubMed] [Google Scholar]
- 26.Sarcabal, P., M. Remaud-Simeon, R. M. Willemot, G. P. de Montalk, B. Svensson, and P. Monsan. 2000. Identification of key amino acid residues in Neisseria polysaccharea amylosucrase. FEBS Lett. 474:33-37. [DOI] [PubMed] [Google Scholar]
- 27.Schwimmer, S., and A. Bevenue. 1956. Reagent for differentiation of 1,4- and 1,6-linked glucosaccharides. Science 123:543-544. [DOI] [PubMed] [Google Scholar]
- 28.Sriprapundh, D., C. Vieille, and J. G. Zeikus. 2000. Molecular determinants of xylose isomerase thermal stability and activity: analysis of thermozymes by site-directed mutagenesis. Protein Eng. 13:259-265. [DOI] [PubMed] [Google Scholar]
- 29.Suzuki, Y. 1989. A general principle of increasing protein thermostability. Proc. Jpn. Acad. Ser. B 65:146-148. [Google Scholar]
- 30.Suzuki, Y., K. Oishi, H. Nakano, and T. Nagayama. 1987. A strong correlation between the increase in number of proline residues and the rise in thermostability of five Bacillus oligo-1,6-glucosidase. Appl. Microbiol. Biotechnol. 26:546-551. [Google Scholar]
- 31.Tsuyuki, K., T. Sugitani, Y. Miyata, T. Ebashi, and Y. Nakajima. 1992. Isolation and characterization of isomaltulose- and trehalulose-producing bacteria from Thailand soil. J. Gen. Appl. Microbiol. 38:483-490. [Google Scholar]
- 32.Uitdehaag, J. C. M., R. Mosi, K. H. Kalk, B. A. Van der Veen, L. Dijkhuizen, S. G. Withers, and B. W. Dijkstra. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat. Struct. Biol. 6:432-436. [DOI] [PubMed] [Google Scholar]
- 33.Veronese, T., and P. Perlot. 1999. Mechanism of sucrose conversion by the sucrose isomerase of Serratia plymuthica ATCC 15928. Enzyme Microb. Technol. 24:263-269. [Google Scholar]
- 34.Vieille, C., and J. G. Zeikus. 1996. Thermozymes: identifying molecular determinants of protein structural and functional stability. Trends Biotechnol. 14:183-190. [Google Scholar]
- 35.Watanabe, K., K. Chishiro, K. Kitamura, and Y. Suzuki. 1991. Proline residues responsible for thermostability occur with high frequency in the loop regions of an extremely thermostable oligo-1,6-glucosidase from Bacillus thermoglucosidasius KP 1006. J. Biol. Chem. 266:24287-24294. [PubMed] [Google Scholar]
- 36.Watanabe, K., K. Kitamura, H. Iha, and Y. Suzuki. 1990. Primary structure of the oligo-1,6-glucosidase of Bacillus cereus ATCC7064 deduced from the nucleotide sequence of the gene. Eur. J. Biochem. 192:609-620. [DOI] [PubMed] [Google Scholar]
- 37.Watanabe, K., T. Masuda, H. Ohashi, H. Mihara, and Y. Suzuki. 1994. Multiple proline substitutions cumulatively thermostabilize Bacillus cereus ATCC 7064 oligo-1.6-glucosidase. Eur. J. Biochem. 226:277-283. [DOI] [PubMed] [Google Scholar]
- 38.Watanabe, K., Y. Hata, H. Kizaki, Y. Katsube, and Y. Suzuki. 1997. The refined crystal structure of Bacillus cereus oligo-1,6-glucosidase at 2.0A resolution: structural characterization of proline-substitution sites for protein thermostabilization. J. Mol. Biol. 269:142-153. [DOI] [PubMed] [Google Scholar]
- 39.Weidenhagen, R., and S. Lorenz. 1957. Palatinose [6-(α-Glucopyranoside)-fructofuranose], ein neues bakterielles umwandlungsprodukt der Saccharose. Z. Zuckerindust. 7:533-534. [Google Scholar]
- 40.Zhang, L. H., J. L. Xu, and R. G. Birch. 1998. High affinity binding of albicidin phytotoxins by the AlbA protein from Klebsiella oxytoca. Microbiology 144:555-559. [DOI] [PubMed] [Google Scholar]